Abstract
Over the past 500 years, North America has been the site of ongoing mixing of Native Americans, European settlers, and Africans (brought largely by the trans-Atlantic slave trade), shaping the early history of what became the United States. We studied the genetic ancestry of 5,269 self-described African Americans, 8,663 Latinos, and 148,789 European Americans who are 23andMe customers and show that the legacy of these historical interactions is visible in the genetic ancestry of present-day Americans. We document pervasive mixed ancestry and asymmetrical male and female ancestry contributions in all groups studied. We show that regional ancestry differences reflect historical events, such as early Spanish colonization, waves of immigration from many regions of Europe, and forced relocation of Native Americans within the US. This study sheds light on the fine-scale differences in ancestry within and across the United States and informs our understanding of the relationship between racial and ethnic identities and genetic ancestry.
Introduction
Over the last several hundred years, the United States has been the site of ongoing mixing of peoples of continental populations that were previously separated by geography. Native Americans, European immigrants to the Americas, and Africans brought to the New World largely via the trans-Atlantic slave trade came together in the New World. Mating between individuals with different continental origins, which we refer to here as “population admixture,” results in individuals who carry DNA inherited from multiple populations. Although US government census surveys and other studies of households in the US have established fine-scale self-described ethnicity at the state and county level (see the US 2010 Census online), the relationship between genetic ancestry and self-reported ancestry for each region has not been deeply characterized. Understanding genetic ancestry of individuals from a self-reported population, and differences in ancestry patterns among regions, can inform medical studies and personalized medical treatment.1 The genetic ancestry of individuals can also shed light on the history of admixture and migrations within different regions of the US, which is of interest to historians and sociologists.
Previous studies have shown that African Americans in the US typically carry segments of DNA shaped by contributions from peoples of Europe, Africa, and the Americas, with variation in African and European admixture proportions across individuals and differences in groups across parts of the country.2–4 More recent studies that utilized high-density genotype data provide reliable individual ancestry estimates, illustrate the large variability in African and European ancestry proportions at an individual level, and are able to detect low proportions of Native American ancestry.3–11 Latinos across the Americas have differing proportions of Native American, African, and European genetic ancestry, shaped by local historical interactions with migrants brought by the slave trade, European settlement, and indigenous Native American populations.12–18 Individuals from countries across South America, the Caribbean, and Mexico have different profiles of genetic ancestry molded by each population’s unique history and interactions with local Native American populations.1,19–25 European Americans are often used as proxies for Europeans in genetic studies.26 European Americans, however, have a history of admixture of many genetically distinct European populations.27,28 Studies have shown that European Americans also have non-European ancestry, including African, Native American, and Asian, though it has been poorly quantified with some discordance among estimates even within studies.29–32
That genetic ancestry of self-described groups varies across geographic locations in the US has been documented in anecdotal examples but has not previously been explored systematically. Most early studies of African Americans had limited resolution of ancestry because of small sample sizes and few genetic markers, and recent studies typically have limited geographic scope. Though much work has been done to characterize the genetic diversity among Latino populations from across the Americas, it is unclear the extent to which Latinos within the US share or mirror these patterns on a national or local scale. Most analyses have relied on mitochondrial DNA, Y chromosomes, or small sets of ancestry-informative markers, and few high-density genome-wide SNP studies have explored fine-scale patterns of African and Native American ancestry in individuals living across the US.
Here, we describe a large-scale, nationwide study of African Americans, Latinos, and European Americans by using high-density genotype data to examine subtle ancestry patterns in these three groups across the US. To improve the understanding of the relationship between genetic ancestry and self-reported ethnic and racial identity, and to characterize heterogeneity in the fine-scale genetic ancestry of groups from different parts of the US, we inferred the genetic ancestry of 5,269 self-reported African Americans, 8,663 Latinos, and 148,789 European Americans who are 23andMe customers living across the US, by using high-density SNPs genotype data from 650K to 1M arrays. 23andMe customers take an active role in participating in research by submitting saliva samples, consenting for data to be used for research, and completing surveys. We generated cohorts of self-reported European American, African American, and Latino individuals from self-reported ethnicity and identity. We obtained ancestry estimates from genotype data by using a Support Vector Machine-based algorithm that infers population ancestry with Native American, African, and European reference panels, leveraging geographic information collected through surveys (see Durand et al.33). For details on genotyping and ancestry deconvolution methods, see Subjects and Methods.
Subjects and Methods
Human Subjects
All participants were drawn from the customer base of 23andMe, Inc., a consumer personal genetics company. This data set has been described in detail previously.34,35 Participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRFP-accredited IRB, Ethical & Independent Review Services (E&I Review).
Genotyping
Participants were genotyped as described previously.36 In short, DNA extraction and genotyping were performed on saliva samples by National Genetics Institute (NGI), a CLIA-licensed clinical laboratory and a subsidiary of Laboratory Corporation of America. Samples have been genotyped on one of four genotyping platforms. The V1 and V2 platforms were variants of the Illumina HumanHap550+ BeadChip, including about 25,000 custom SNPs selected by 23andMe, with a total of about 560,000 SNPs. The V3 platform was based on the Illumina OmniExpress+ BeadChip, with custom content to improve the overlap with our V2 array, with a total of about 950,000 SNPs. The V4 platform in current use is a fully custom array, including a lower redundancy subset of V2 and V3 SNPs with additional coverage of lower-frequency coding variation and about 570,000 SNPs. Samples that failed to reach 98.5% call rate were reanalyzed. Individuals whose analyses failed repeatedly were recontacted by 23andMe customer service to provide additional samples, as is done for all 23andMe customers. Customer genetic data have been previously utilized in association studies and studies of genetic relationships.34–43
Research Cohorts
23andMe customers were invited to fill out web-based questionnaires, including questions on ancestry and ethnicity, on state of birth, and current zip code of residence. They were also invited to allow their genetic data and survey responses to be used for research. Only data of customers who signed IRB-approved consent documents were included in our study. Survey introductions are explicit about their applications in research. For example, the ethnicity survey introduction text states that the survey responses will be used in ancestry-related research (Table S1 available online).
Self-Reported Ancestry
It is important to note that ancestry, ethnicity, identity, and race are complex labels that result both from visible traits, such as skin color, and from cultural, economic, geographical, and social factors.23,44 As a result, the precise terminology and labels used for describing self-identity can affect survey results, and care in choice of labels should be utilized. However, we chose to maximize our available self-reported ethnicity sample size by combining information from questions asking for customer self-reported ancestry. We used two survey questions, with different nomenclature, to gauge responses about identity, which here we view as “the subjective articulation of group membership and affinity.”45
The first question is modeled after the US census nomenclature and is a multiquestion survey that allows for choice of “Hispanic” or “Not Hispanic,” and participants were asked “Which of these US Census categories describe your racial identity? Please check all that apply” from the following list of ethnicities: “White,” “Black,” “American Indian,” “Asian,” “Native Hawaiian,” “Other,” “Not sure,” and “Other racial identity.” For inclusion into our European American cohort, individuals had to select “Not Hispanic” and “White,” but not any other identity. For inclusion into our Latino cohort, individuals had to select “Hispanic,” with no other restrictions. For inclusion into our African American cohort, individuals had to select “Not Hispanic” and “Black” and no other identity.
The second question on identity is a single-choice question, where respondents were asked to choose “What best describes your ancestry/ethnicity?” from “African,” “African American,” “Central Asian,” “Declined,” “East Asian,” “European,” “Latino,” “Mideast,” “Multiple ancestries,” “Native American,” “Not sure,” “Other,” “Pacific Islander,” “South Asian,” and “Southeast Asian.” Because individuals could select only one response, we included individuals who selected “European” in our European American cohort, those who selected “African American” in our African American cohort, and those who selected “Latino” in our Latino cohort.
Some African American participants included in this study were recruited through 23andMe’s Roots into the Future project (accessed October 2013), which aimed to increase understanding of how DNA plays a role in health and wellness, especially for diseases more common in the African American community. Individuals who self-identified as African American, black, or African were recruited through 23andMe’s current membership, at events, and via other recruitment channels.
In the present work, we do not include individuals who self-report as having multiple identities, because this represents only a small fraction of individuals in our data set. Low rates of reporting as multiracial or multiethnic is in line with previous studies; an analysis of the 2000 US Census shows that 95 percent of blacks and 97 percent of whites acknowledge only a single identity.45 Future studies including multiracial individuals might further illuminate patterns of genetic ancestry and the complex relationship with self-identity.
Differences among states, where different proportions of people self-report as mixed race, might explain some regional differences in genetic ancestry. However, we note that, first, proportionally fewer people identify as mixed race than as a single identity, and second, it remains important to establish regional differences in genetic ancestry of self-reported groups even if these differences are driven, to some degree, by regional changes in self-reported identity. More work is needed to determine to what extent regional differences are a result of how people today report their ancestry. Lastly, when available, we excluded individuals who answered “No” to a question whether they are living in the US. In total, our final sets included 5,269 African Americans, 8,663 Latinos, and 148,789 European Americans.
Notes on Terminology and Selection of Populations
Throughout the manuscript, the term “Native American ancestry” refers to estimates of genetic ancestry from indigenous Americans found across North, Central, and South America, and we distinguish this term from present-day Native Americans living in the US. We use the term “Native American” to refer to indigenous peoples of the Americas, acknowledging that some people may prefer other terms such as “American Indian.” Our estimates of African ancestry specifically aim to infer ancestry of sub-Saharan Africa and does not include ancestry from North Africa. We note that the term “Latino” has many meanings in different contexts, and in our case, we use it to refer to individuals living in the US who self-report as either “Latino” or “Hispanic.”
Our work represents a snapshot in time of genetic ancestry and identity, and future work is needed to inform the dynamic changes and forces that shape social interactions.
We note that our cohorts are likely to have ancestry from many African populations, but because of current reference sample availability, our resolution of West African ancestries is outside the scope of our study. Likewise, our estimates of Native American ancestry arise from a summary over many distinct subpopulations, but we are limited in scope because of insufficient sample sizes from subpopulations, so we currently use individuals from Central and South American together as a reference set (see Durand et al.33 for a list of populations and sample sizes).
Validation of Self-Reported Identity Survey Results
To verify that our self-reported ethnicities were reliable, we examined the consistency of ethnicity survey responses when individuals completed both ancestry and ethnicity surveys. Because the structure of the two surveys is different and multiple selections were allowed in one survey but not the other, we examined the replication rate of the primary ethnicity from the single-choice ethnicity survey in the multiple-selection survey.
In addition to structural differences, the survey content used very different nomenclature, and therefore we believe our estimated error rates to be overestimates of the true error rate, because it is likely that some individuals choose to identify with one label but not the other (i.e., “African American” but not “black”). Discrepancies in the question nomenclatures are likely to increase the error rate. Furthermore, because the two surveys could be completed at different times, either before or after obtaining personal ancestry results, it is possible that viewing genetic ancestry results might have led to a change in self-reported ancestry. Such a change would be tallied as an error in our estimates, but instead reflects a true change in perceived self-identity over time. Overall, we expect that our survey data represent highly reliable ancestry information, with errors affecting fewer than 1% of survey responses.
Geographic Location Collection
Self-reported state-of-birth survey data was available for 47,473 customers of 23andMe. However, because overlap of these customers with our cohorts was poor, we also chose to include data from a question on current zip code of residence. This provided an additional 34,351 zip codes of current residence. In cases where both the zip code of residence and state of birth were available, we used state-of-birth information. To obtain state information from zip codes, we translated zip codes to their state locations via an online zip code database (accessed October 2013).
In total, we had 50,697 individuals with available location information. About one third of each of our cohorts had location information: 1,970 African Americans, 2,944 Latinos, and 45,783 European Americans were used in our geographic analyses.
Ancestry Analyses
Ancestry Composition
We apply Ancestry Composition, a three-step pipeline that efficiently and accurately identifies the ancestral origin of chromosomal segments in admixed individuals, which is described in Durand et al.33 We apply the method to genotype data that have been phased via a reimplementation of Beagle.46 Ancestry Composition applies a string kernel support vector machines classifier to assign ancestry labels to short local phased genomic regions, which are processed via an autoregressive pair hidden Markov model to simultaneously correct phasing errors and produce reconciled local ancestry estimates and confidence scores based on the initial assignment. Lastly, these confidence estimates are recalibrated by isotonic regression models. This results in both precision and recall estimates that are greater than 0.90 across many populations, and on a continental level, have rates of 0.982–0.994 for precision and recall rates of 0.935–0.993, depending on populations (see Table 1 from Durand et al.33). We note that here, and throughout the manuscript, African ancestry corresponds to sub-Saharan African ancestry (including West African, East African, Central, and South African populations, but excluding North African populations from the reference set). For more details on our ancestry estimation method, see Durand et al.33
Aggregating Local Ancestry Information
23andMe’s Ancestry Composition method provides estimates of ancestry proportions for several worldwide populations at each window of the genome. To estimate genome-wide ancestry proportions of European, African, and Native American ancestry, we aggregate over populations to estimate the total likelihood of each population, and with a majority threshold of 0.51, if any window has a majority of a continental ancestry, we include it in the calculation of genome-wide ancestry, which is estimated as the number of windows passing the threshold for each ancestry over the total number of windows. Some windows might not pass our threshold for any population, so they remain unassigned, making it possible for estimates for all ancestries to not sum to 100%, resulting in population averages that likewise might not sum to 100%. We allow for this unspecified ancestry to reduce the error rates of our assignments, so, in some sense, our estimates might be viewed as lower bounds on ancestry, and it is possible that individuals carry more ancestry than estimated. In practice, we typically assign nearly all windows, with an average of about 1%–2% unassigned ancestry, so we do not expect it to affect our results, with the exception of Native American ancestry, which we discuss below.
Generating the Distribution of Ancestry Tracts
We generate ancestry segments as defined as continuous blocks of ancestry, estimating the best guess of ancestry at each window to define segments of each ancestry. Assigning the most likely ancestry at each window results in fewer spurious ancestry breaks and allows for a smaller upward bias in admixture dates, because breaks in ancestry segments push estimates of dates further back in time. We measure segment lengths by using genetic distances, by mapping segment start and end physical positions to the HapMap genetic map.
Admixture Dating
To estimate the time frame of admixture events, we test a simple two-event, three-population admixture model via TRACTS.47 We use a grid-search optimization to find four optimal parameters for the times of two admixture events and the proportions of admixture. We are limited to simple admixture models resulting from the computationally intensive grid search, because we were unable to obtain likelihood convergence with any of the built-in optimizers. The model tested is as follows: two populations admix t1 generations ago, with proportion frac1 and 1 − frac1, respectively. A third population later mixes in t2 generations ago, with proportion frac2.
Both our ancestry segments and prior results supported a model with an earlier date of Native American admixture.25,47 We estimated likelihoods over plausible grid of admixture times and fractions for African Americans, Latinos, and European Americans to estimate dates of initial Native American and European admixture and subsequent African admixture. These dates are estimated as the best fit for a pulse admixture event: because they represent an average over more continuous or multiple migrations, initial admixture is likely to have begun earlier.
Lower Estimates of African Ancestry in 23andMe African Americans
Unlike previous estimates of the mean proportion of African ancestry, which typically have ranged from 77% to 93% African ancestry,2–4,48–62 our estimates, depending on exclusions, are 73% or 75%. There are several possible explanations for our low mean African ancestry. If our Ancestry Composition estimates are downward biased, then the African Americans might have levels of African ancestry consistent with other studies, and our results are simply underestimates. However, our Ancestry Composition estimates are extremely well calibrated for African Americans from the 1000 Genomes Project and their consensus estimates, and we see no evidence of a downward bias (see Figure 5 from Durand et al.33).
The mean ancestry proportion of 23andMe self-reported African Americans is about 73%. A small fraction, about 2%, of African Americans carry less than 2% African ancestry, which is far less than typically seen in most African Americans (Figure S18A available online). Further investigation reveals that the majority of these individuals (88%) have predominantly European ancestry, and others carry East Asian, South Asian, and Southeast Asian ancestry, roughly in proportion to the frequencies found in the 23andMe database overall. Given the large number of non-African American individuals in the 23andMe database, even an exceeding low survey error rate of 0.02% could be sufficient to account for the number of outlier individuals we detect. Hence, we posit that these individuals represent survey errors rather than true self-reported African Americans. Exclusion of these 108 self-reported African Americans with less than 2% African ancestry from mean ancestry calculations results in a moderate rise, to 74.8%, of the mean proportion of African ancestry in African Americans.
To quantify differences in African ancestry driving mean state differences, we examined the distributions of estimates of African ancestry in African Americans from the District of Columbia (D.C.) and Georgia, which had at least 50 individuals with the lowest and highest mean African ancestry proportions (Figure S1E). We find a qualitative shift in the two distributions of African ancestry, with D.C. showing a reduced mode, higher variance, and a heavier lower tail of African ancestry, corresponding to more African Americans with below-average ancestry than Georgia. Qualitative differences in the distributions of African ancestry proportions in African Americans from states with higher and lower mean ancestry appear to be driven by both a shift in the mode of the distribution as well as a heavier left tail reflecting more individuals with a minority of African ancestry (Figure S1). We posit that differences among states could be due to differences in admixture, differences in self-identity, or differences in patterns of assortative mating, whereby individuals with similar ancestry might preferentially mate. For example, greater levels of admixture with Europeans would both shift the mode and result in more African American individuals who have a minority of African ancestry. Alternatively, a shift toward African American self-identity for individuals with a majority of European ancestry (possibly because of changes in cultural or social forces) would likewise result in lower estimates of mean African ancestry. Lastly, assortative mating would work to maintain or increase the variance in ancestry proportions, though assortative mating alone could not shift the mean proportion of African ancestry in a population.
Sex Bias in Ancestry Contributions
Sex bias in ancestry contributions, often assessed through ancestry of mtDNA and Y chromosome haplogroups, is also manifested in unequal estimates of ancestry proportions on the X chromosome, which has an inheritance pattern that differs between males and females. The X chromosome more closely follows female ancestry contributions because males contribute half as many X chromosomes. Comparing ancestry on the X chromosome to the autosomal ancestry allows us to infer whether that ancestry historically entered via males (lower X ancestry) or by females (higher X ancestry). Under equal ancestral contributions from both males and females, the X chromosome should show the same levels of admixture as the genome-wide estimates. To look for evidence of unequal male and female ancestry contributions in our cohorts, we examined ancestry on the X chromosome (NRY region), which follows a different pattern of inheritance from the autosomes. In particular, estimates of ancestry on the X chromosome have been shown to have higher African ancestry in African Americans.9 We calculate ancestry on the X chromosome as the estimate of ancestry on just windows on the X, and we compare to genome-wide estimates (which do themselves include the X chromosome). It should be noted that these calculations differ among males and females, because the X chromosome is diploid in females and thus has twice as many windows in calculation of genome-wide mean proportions. However, our results still allow a peek into sex bias because the overall contribution of the X chromosome to the genome-wide estimates is small. We note that because our ancestry estimation method conservatively assigns Native American ancestry, we expect that much of the remaining unassigned ancestry might be due to Native American ancestry assigned as broadly East Asian/Native American, which is not included in these values (see Figure 5 in Durand et al.33).
To infer estimates of male and female contributions from each ancestral population, we estimated the male and female fractions of ancestry that total the genome-wide estimates and minimize the mean square error of the X chromosome ancestry estimates. We assume that overall male and female contributions are each 50% ( and ). We assume that the total contribution from males and females of a population gives rise to the autosomal ancestry fraction (fpop,male + fpop,female = autopop). We then compute, via a grid search, the predicted X chromosome estimates from fpop,male, fpop,female for each , which are calculated, as in Lind et al.,6 as
We choose the parameters of male and female contributions that minimize the mean squared error of the X ancestry estimates and the predicted . These are the estimates of male and female ancestry fractions under a single simplistic population mixture event that best fit our X chromosome ancestry estimates observed.
Population Size Correlations
From the 2010 Census Brief “The Black Population” available online, we calculated the correlation between the number of reported African Americans living in a state and our sample of African Americans from that state. The correlation is strong, with p value of 9.5 × 10−14, suggesting that our low sample sizes from states in the US Mountain West is expected from estimates of population sizes.
African ancestry in European Americans most frequently occurs in individuals from states with high proportions of African Americans and is rare in states with few African Americans. This observation led us to look at the correlation between population size (as a percent of state population using self-reported ethnicity from the 2010 US Census) and state mean levels of ancestry.
To examine the interaction between proportions of minorities and ancestry, we used the 2010 US Census demographic survey by state. We compare the state population proportion to the mean estimated admixture proportion of individuals from that state, fitting linear regressions, and generating figures with geom_smooth(method = “lm,” formula = y ∼ x) from the ggplot2 package in R.
We find that African ancestry in European Americans is strongly correlated with the population proportion of African Americans in each state. We find that the higher the state proportion of African Americans, the more African ancestry is found in European Americans from that state, reflecting the complex interaction of genetic ancestry, historical admixture, culture, and self-identified ancestry.
Logistic Regression Modeling of Self-Identity
We examine the probabilistic relationship between self-identity and genetically inferred ancestry. To explore the interaction between genetic ancestry and self-reported identity, we estimated the proportion of individuals that identify as African American and European American, partitioned by levels of African ancestry. Jointly considering the cohorts of European Americans and African Americans, we examined the relationship between an individual’s genome-wide African ancestry proportion and whether they self-report as European American or African American. We note a strong dependence on the amount of African ancestry, with individuals carrying less than 20% African ancestry identifying largely as European American, and those with greater than 50% reporting as African American. To test the significance of this relationship, we fit a logistic regression model, using Python’s statsmodels package, predicting self-reported ancestry by using proportion African ancestry, sex, age, intercept, and interaction variables.
Validation of Non-European Ancestry in African Americans and European Americans
Although our Ancestry Composition estimates are well calibrated and have been shown to accurately estimate African, European, and Native American ancestry in tests of precision and recall,33 we were concerned that low levels of non-European ancestry in European Americans that we detected might represent an artifact of Ancestry Composition. Hence, we pursued several lines of investigation to provide evidence that estimates of African and Native American ancestry in European Americans are robust and not artifacts.
Comparison with 1000 Genomes Project Consensus Estimates
Comparisons of our estimates with those published by the 1000 Genomes Consortium show the high consistency across populations and individuals. We compare estimates across Americans of African Ancestry in SW USA (ASW), Colombians from Medellin, Colombia (CLM), Mexican Ancestry from Los Angeles USA (MXL), and Puerto Ricans from Puerto Rico (PUR). We note that our estimates of Native American ancestry are conservative. Indeed, when our Ancestry Composition assignment probabilities do not pass over the confidence threshold, including signals of Native American ancestry together with general East Asian/Native American ancestry (but not East Asian) recapitulates estimates from the 1000 Genomes Project consensus estimates. Five individuals from the ASW population from the 1000 Genomes Project have poor consistency in their estimates. These individuals have a large amount of Native American ancestry that was not modeled by the 1000 Genomes Project estimates. That these particular individuals were sampled in Oklahoma, and carry significant Native American ancestry, is supported by our own high estimates of Native American ancestry in 23andMe self-reported African Americans from Oklahoma.
Estimates of African and Native American Ancestry in Europeans
We looked at whether all individuals who are expected to carry solely European ancestry also have similar rates of detection of non-European ancestry. To this end, we generated a cohort of 15,289 customers of 23andMe who reported that all four of their grandparents were born in the same European country. The use of four-grandparent birth-country has been utilized as a proxy for assessing ancestry.27,63 We then examined Ancestry Composition results for these individuals and calculated at what rate we detected at least 1% African and at least 1% Native American ancestry.
Independent Validation of African Ancestry in European Americans via f4 Statistics
We used f4 statistics from the ADMIXTOOLS software package to confirm the presence of African ancestry.64 We used the f4 ratio test, designed to estimate the proportion of admixture from a related ancestral population, to compare admixture in European Americans versus reference European individuals. We tested whether European Americans with estimated African ancestry showed any admixture from Africans by using our cohorts of individuals with estimated African ancestry and reference populations from the 1000 Genomes Project data set. Admixture would be expected to result in estimates of α significantly different from 1.
Detection of Native American mtDNA in European Americans and African Americans
The mitochondrial DNA (mtDNA) haplogroups A2, B2, B4b, C1b, C1c, C1d, and D1 are most prevalently found in the Americas and are likely to be Native-American-specific haplogroups because they are rarely found outside of the Americas. We assessed the fraction of individuals that carry these haplogroups to validate the likelihood of Native American ancestry in European Americans and African Americans and show that these haplogroups are virtually absent in European controls. Because mtDNA haplogroups are assigned by classification with SNPs that segregate on these lineages, these orthogonal results provide an independent line of support for our estimated Native American ancestry in European Americans and African Americans.
Distribution of Ancestry Segment Start Positions
Regions of the genome that have structural variation or show strong linkage disequilibrium (LD) have been shown both to confound admixture mapping and to influence the detection of population substructure in studies using Principal Components Analysis (PCA).27,63,65 If such regions were to drive artifacts of spurious ancestry, we would expect that segments of local ancestry would probably occur around these regions, rather than in a uniform distribution across the genome. To this end, we examined the starting positions of all African and Native American ancestry segments in European Americans and Native American ancestry in African Americans.
Comparison with ADMIXTURE Genome-wide Estimates
We applied ADMIXTURE,66 a model-based estimation of ancestry proportions, to estimate proportions of European, Native American, East Asian, sub-Saharan African, Middle Eastern, and Oceanian ancestry proportions. We use the supervised algorithm for K = 6, with 9,694 reference individuals representing the six aforementioned populations. We ran ADMIXTURE on 269,229 autosomal markers after pruning SNPs to have r2 < 0.5, via PLINK.67 To reduce computation time, we examined consistency of methods on the African Americans whom we estimated to have at least 1% Native American ancestry, European Americans estimated to have at least 1% Native American ancestry, and European Americans estimated to have at least 1% African ancestry.
Results
Self-reported survey data was used to generate cohorts of African Americans, Latinos, and European Americans. Out of 35,524 self-reported “European” individuals, 35,279 selected “white” on the ethnicity survey, yielding a per-survey error estimate of 0.2%. Out of 1,560 self-reported “Latino” individuals, 1,540 selected “Hispanic,” giving a per-survey error estimate of 0.7%. Lastly, out of 1,327 self-reported “African American” individuals, 1,287 selected “black,” resulting in a per-survey error rate estimate of 1.1%. For more details on our cross-survey validation, see Subjects and Methods.
The Genetic Landscape of the US
Patterns of Genetic Ancestry of Self-Reported African Americans
Genome-wide ancestry estimates of African Americans show average proportions of 73.2% African, 24.0% European, and 0.8% Native American ancestry (Table 1). We find systematic differences across states in the US in mean ancestry proportions of self-reported African Americans (Figure 1 and Table S2). On average, the highest levels of African ancestry are found in African Americans living in or born in the South, especially South Carolina and Georgia (Figure 1Aand Table S3). We find lower proportions of African ancestry in the Northeast, the Midwest, the Pacific Northwest, and California. The amount of Native American ancestry estimated for African Americans also varies across states in the US. More than 5% of African Americans are estimated to carry at least 2% Native American ancestry genome-wide (Figures S1 and 1D). African Americans in the West and Southwest on average carry higher levels of Native American ancestry, a trend that is largely driven by individuals with less than 2% Native American ancestry (Figure 1B). With a lower threshold of 1% Native American ancestry, we estimate that about 22% of African Americans carry some Native American ancestry (Figure S2).
Table 1.
Comparison of Genome-wide Ancestry Estimates and X Chromosome Estimates in African Americans, Latinos, and European Americans
| Estimate |
Ancestry |
||
|---|---|---|---|
| African | Native American | European | |
| African Americans | |||
| Genome-wide | 73.2% | 0.8% | 24.0% |
| X chromosome | 76.9% | 0.9% | 19.8% |
| Relative increase or decrease on X | +5.1% | +13.6% | −17.7% |
| p value | 4.4 × 10−17∗∗∗ | 0.078 | 7.8 × 10−24∗∗∗ |
| Latinos | |||
| Genome-wide | 6.2% | 18.0% | 65.1% |
| X chromosome | 6.8% | 19.4% | 56.7% |
| Relative increase or decrease on X | +9.0% | +7.4% | −13.0% |
| p value | 0.008∗∗ | 2.4 × 10−10∗∗∗ | 4.2 × 10−94∗∗∗ |
| European Americans | |||
| Genome-wide | 0.19% | 0.18% | 98.6% |
| X chromosome | 0.19% | 0.22% | 98.4% |
| Relative increase or decrease on X | −0.04% | +23.73% | −0.1% |
| p value | 0.99 | 6.6 × 10−10∗∗∗ | 8.0 × 10−5∗∗∗ |
Mean estimates of African, Native American, and European ancestry are shown. p values provided are calculated by two-sided Student’s t test on individual ancestry estimates for each cohort per ancestry, with no multiple testing correction. Significance is assigned as ∗p < 0.05, ∗∗p value < 0.01, and ∗∗∗p value < 0.001. Relative increase on the X chromosome is calculated as the absolute difference, X chromosome estimate minus genome-wide estimate, divided by the genome-wide estimate.
Figure 1.
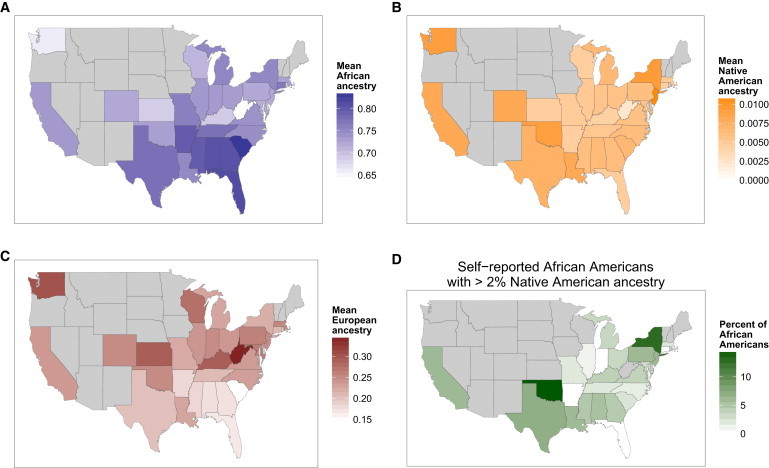
The Distribution of Ancestry of Self-Reported African Americans across the US
(A) Differences in levels of African ancestry in African Americans (blue).
(B) Differences in levels of Native American ancestry in African Americans (orange).
(C) Differences in levels of European ancestry of African Americans (red), from each state. States with fewer than ten individuals are excluded in gray.
(D) The geographic distribution of self-reported African Americans with Native American ancestry. The proportion of African Americans in each state who have 2% or more Native American ancestry is shown by shade of green. States with fewer than 20 individuals are excluded in gray.
We used the lengths of segments of European, African, and Native American ancestry to estimate a best-fit model of admixture history among these populations for African Americans (Figure S3). We estimate that initial admixture between Europeans and Native Americans occurred 12 generations ago, followed by subsequent African admixture 6 generations ago, consistent with other admixture inference methods dating African American admixture. A sex bias in African American ancestry, with greater male European and female African contributions, has been suggested through mtDNA, Y chromosome, and autosomal studies.6 On average, across African Americans, we estimate that the X chromosome has a 5% increase in African ancestry and 18% reduction in European ancestry relative to genome-wide estimates (see Table 1). Through comparison of estimates of X chromosome and genome-wide African and European ancestry proportions, we estimate that approximately 5% of ancestors of African Americans were European females and 19% were European males (Table S4).
Patterns of Genetic Ancestry of Self-Reported Latinos
Latinos encompass nearly all possible combinations of African, Native American, and European ancestries, with the exception of individuals who have a mix of African and Native American ancestry without European ancestry (see Figures S4A and S1). On average, we estimate that Latinos in the US carry 18.0% Native American ancestry, 65.1% European ancestry, and 6.2% African ancestry. We find the highest levels of estimated Native American ancestry in self-reported Latinos from states in the Southwest, especially those bordering Mexico (Figure 2C). We find the highest mean levels of African ancestry in Latinos living in or born in states in the South, especially Louisiana, the Midwest, and Atlantic (Figure 2A). Further stratification of individuals by their self-reported population affiliation (e.g., “Mexican,” “Puerto Rican,” or “Dominican”) reveals a diversity in genetic ancestry, consistent with previous work studying these populations (see Figure S5 and Table S5).10,20,24,25,68,69 We find that Latinos who, besides reporting as “Hispanic,” also self-report as Mexican or Central American, carry more Native American ancestry than Latinos overall; those also who self-report as black, Puerto Rican, or Dominican have higher levels of African ancestry; and those who additionally self-report as white, Cuban, or South American have on average higher levels of European ancestry.
Figure 2.
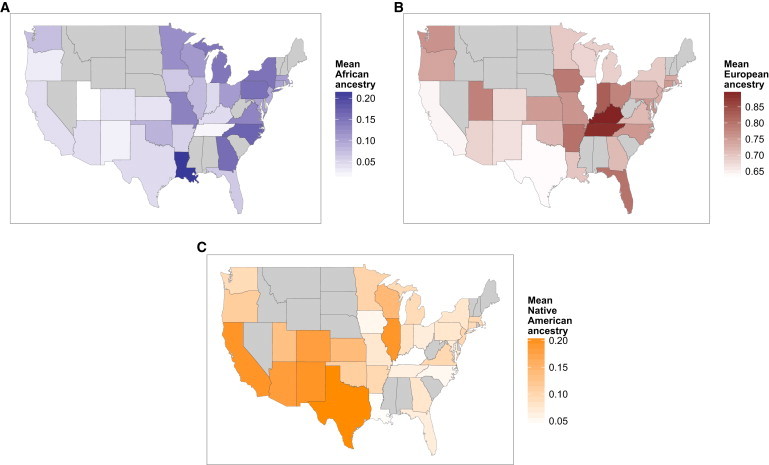
The Distribution of Ancestry of Self-Reported Latinos across the US
Differences in mean levels of African (A), European (B), and Native American (C) ancestry in Latinos from each state is shown by shade of blue, red, and orange, respectively. States with fewer than ten individuals are excluded in gray.
Admixture date estimates for Latino admixture suggest that Native American and European mixture occurred first, about 11 generations ago, followed by African admixture 7 generations ago. Consistent with previous studies that show a sex bias in admixture in Latino populations,12–18 we estimate 13% less European ancestry on the X chromosome than genome-wide (Table 1), showing proportionally greater European ancestry contributions from males. We inferred elevated African and Native American ancestry on the X chromosome, corresponding to higher female ancestry contributions from both Africans and Native Americans. Lastly, Latinos show higher proportions of inferred Iberian ancestry than both European Americans and African Americans (Figure S6).
Patterns of Genetic Ancestry of Self-Reported European Americans
We find that many self-reported European Americans, predominantly those living west of the Mississippi River, carry Native American ancestry (Figure 3B). We estimate that European Americans who carry at least 2% Native American ancestry are found most frequently in Louisiana, North Dakota, and other states in the West. Using a less stringent threshold of 1%, our estimates suggest that as many as 8% of individuals from Louisiana and upward of 3% of individuals from some states in the West and Southwest carry Native American ancestry (Figure S7).
Figure 3.
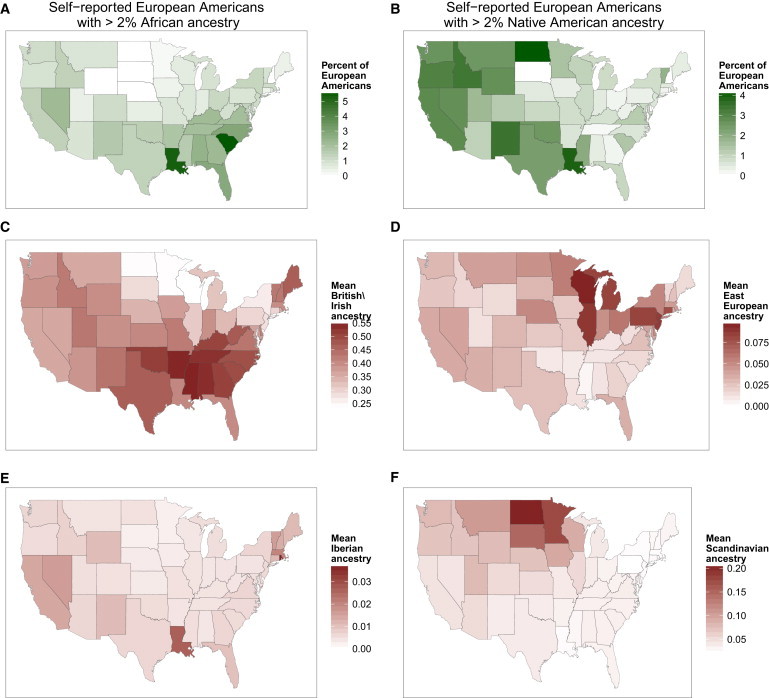
Differences in African, Native American, and European Subpopulation Ancestry among Self-Reported European Americans from Different States
(A) The geographic distribution of self-reported European Americans with African ancestry. The proportion of individuals with at least 2% African ancestry, out of the total number of European Americans per state, is shown by shade of green.
(B) The geographic distribution of self-reported European Americans with Native American ancestry. The proportion of European Americans who have 2% or more Native American ancestry is shown for each state.
(C–F) The mean British/Irish (C), Eastern European (D), Iberian (E), and Scandinavian (F) ancestry proportions among self-reported European Americans from each state are shown by shade of red.
Consistent with previous anecdotal results,32 the frequency of European American individuals who carry African ancestry varies strongly by state and region of the US (Figure 3A). We estimate that a substantial fraction, at least 1.4%, of self-reported European Americans in the US carry at least 2% African ancestry. Using a less conservative threshold, approximately 3.5% of European Americans have 1% or more African ancestry (Figure S8). Individuals with African ancestry are found at much higher frequencies in states in the South than in other parts of the US: about 5% of self-reported European Americans living in South Carolina and Louisiana have at least 2% African ancestry. Lowering the threshold to at least 1% African ancestry (potentially arising from one African genealogical ancestor within the last 11 generations), European Americans with African ancestry comprise as much as 12% of European Americans from Louisiana and South Carolina and about 1 in 10 individuals in other parts of the South (Figure S8).
Most individuals who have less than 28% African ancestry identify as European American, rather than as African American (Figures 4 and 5A). Logistic regression of self-identified European Americans and African Americans reveals that the proportion of African ancestry predicts self-reported ancestry significantly, with a coefficient of 20.1 (95% CI: 18.0–22.2) (Table S6 and Figure S9). For a full characterization of terms and logistic models, see Table S6 and Figure S9.
Figure 4.
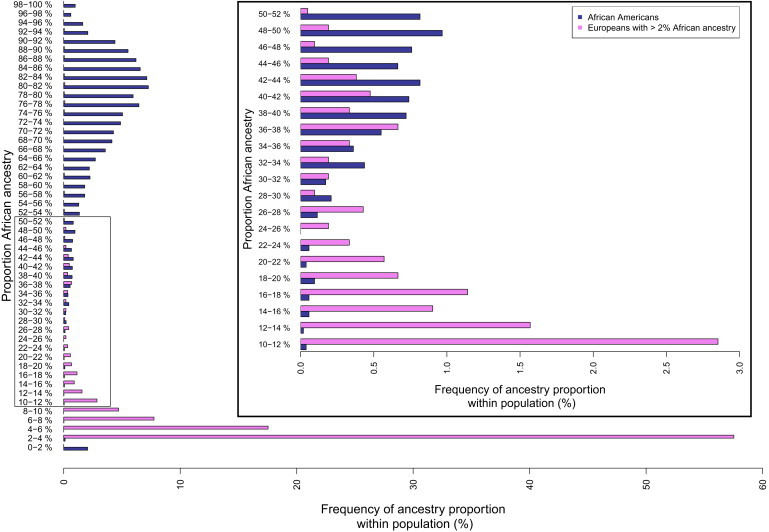
Distribution of African Ancestry in African Americans and European Americans
Histogram of African Americans (blue bars) and European Americans with ≥2% African ancestry (violet bars). Inset: Fine-scale histogram showing the region of greatest overlap between African Americans and European Americans, where African ancestry ranges from 10% and 52%. Both histograms have been normalized for each cohort to total 100%.
Figure 5.
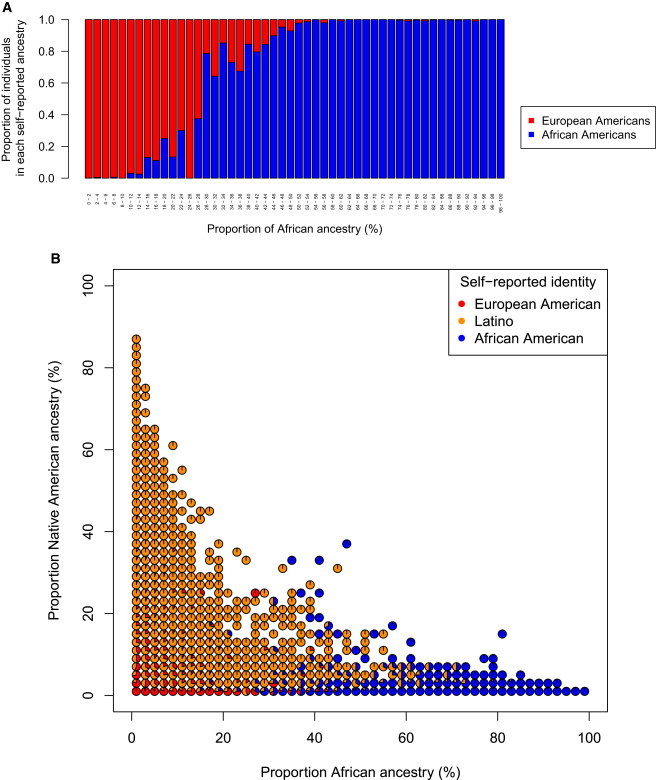
Proportions of Individual Self-Identities by Genome-wide Ancestry Proportions
(A) The proportion of individuals that self-report as African American versus European American for each 2% bin of African ancestry. Each vertical bar corresponds to the individuals that carry that bin of ancestry, and is colored by the proportion of African American and European American identities. Proportions are estimated from absolute numbers of individuals, not scaled by total cohort size.
(B) The proportion of individuals that self-report as European American, Latino, and African American for each 2% bin of African ancestry and Native American ancestry. The proportion for each 2% bin is shown as a pie chart, with slices colored in proportion to the absolute numbers of individuals from each self-reported identity that carry those levels of genome-wide ancestry. Pie charts are omitted for bins where there were no individuals with those corresponding levels of Native American and African ancestry.
Fitting a model of European and Native American admixture followed later by African admixture, we find the best fit with initial Native American and European admixture about 12 generations ago and subsequent African gene flow about 4 generations ago.
Non-European ancestry in European Americans follows a sex bias in admixture contributions from males and females, as seen in African Americans and Latinos. The ratio between X chromosome and genome-wide Native American ancestry estimates in European Americans shows greater Native American female and higher European male ancestry contributions (Tables 1 and S4). Though we do not observe evidence of a sex bias in African ancestry contributions in European Americans overall, analysis of only those individuals with at least 1% African ancestry reveals 15% higher African ancestry on the X chromosome relative to genome-wide estimates (p value 0.013). This increase suggests female-African and male-European sex bias in European Americans that follows the same direction as in African Americans and Latinos, with greater male European and female African and Native American contributions.
Finally, we estimate, for self-reported European Americans, proportions of British/Irish, Eastern European, Iberian, and Scandinavian ancestry (Figure 3) and other European subpopulation ancestries (Figure S10).
Correlations with Population Proportions
We find that levels of Native American and African ancestry in 23andMe customers in each state are significantly correlated with the proportion of African Americans and Latinos in each state (Figures S11–S13). For example, levels of African ancestry in European Americans and Latinos in a state are highly correlated with proportion of African Americans in each state (both p values < 10−4). Levels of Native American ancestry in European Americans and Latinos in a state are highly correlated with proportion of Latinos in each state (p values < 10−6 and < 10−2, respectively).
Validation of Ancestry Estimates
Robust and Consistent Ancestry Estimates
Estimates from Ancestry Composition are extremely well calibrated, with correlations of African, European, and Native American ancestry estimates showing r2 > 0.98 with 1000 Genomes Project African American and Latino consensus estimates (Figure 5 from Durand et al.33). Admixture tests via an independent admixture software package, ADMIXTOOLS,64 confirm significant signals of African admixture in European Americans (Table S7). Ancestry Composition estimates are highly concordant with ADMIXTURE66 estimates, with r2 values of 0.94, 0.98, and 0.91, for the three groups, respectively (Figure S14).
Evidence that the Great Majority of Ancestry Segments that We Detect Are Real
We show that positions of segments of non-European ancestry start uniformly across the genome (see Figure S15). Although some regions, including the HLA region containing the MHC complex on chromosome 6, show higher ancestry switches reflecting difficulties in assignment because of genetic diversity (as likewise seen in African Americans and Latinos; Figures S16 and S17), the majority of segments are uniformly distributed. Only 4% of all segment starts of African ancestry lie within the HLA region, and only about 1.4% of Native American segment starts lie in the HLA region.
We find very low levels of African and Native American ancestry in Europeans with four grandparents born in Europe. We estimate that only 0.98% of Europeans carry African ancestry and 0.26% of Europeans carry Native American ancestry. These levels are substantially lower than the 3.5% and 2.7% of European Americans who carry African and Native American ancestry, respectively. Furthermore, for most European countries we observed no individuals with substantial non-European ancestry, and the presence of individuals with African and Native American ancestry is limited to countries that had major ports in the Atlantic trade and were known to have been highly connected to the trans-Atlantic slave trade. Indeed, African ancestry in individuals from Europe is not unexpected; approximately 9,000 Africans were brought to Europe between 1501 and 1867 (as documented by Eltis and Richardson’s maps of the slave trade, accessible at Emory University’s database). Excluding countries that had major and minor ports in the Atlantic with strong connections to the slave trade (namely Portugal, Spain, France, and United Kingdom) and Malta, which has been the site of migrations from Africa and the Middle East, we obtain a data set of 9,701 Europeans, where we find African and Native American ancestry is virtually absent, with only 0.04% of individuals carrying 1% or more African ancestry and 0.01% carrying 1% or more Native American ancestry, within the margins of survey error estimates.
Native American mtDNA in European Americans and African Americans and Not in Europeans
The frequency of Native American mtDNA haplogroups in European Americans and African Americans correlate with our estimates of genome-wide ancestry in European Americans and African Americans and are found in appreciable fractions of individuals who are estimated to carry Native American ancestry. The frequencies of haplogroups are shown in Table S8. These haplogroups are virtually absent in individuals with four grandparents from a European country (21 individuals out of 15,651). Furthermore, the majority of these Native American haplogroups in Europeans are found in individuals from Spain. Though it is possible these represent non-Native American haplogroups, prior literature and studies of genetic, archaeological, and paleontological evidence suggest that these haplogroups have Native American origins and is evidence of gene flow from the Americas to Spain. Excluding Spain, Native-American-specific haplogroups are detected in fewer than 0.05% of individuals with four grandparents from Europe and can be explained by survey errors in reporting all four grandparents’ birth places.
Discussion
Selection of Populations
The ancestries of 23andMe customers, and therefore the demographics of the database used for this study, largely reflect the demographics of the US, as tallied in the 2010 US census. Our study considers three cohorts that comprise the three largest self-identified groups in the US, which are likewise well represented in the 23andMe database. In this study, we focus on the distribution of European, African, and Native American ancestries and European subpopulation ancestries. These populations were selected because we had available reference data sets, allowing for accurate estimation of ancestry proportions, they reflect the major waves of migration into the US just after the era of transcontinental travel began, and they are found at mean frequencies of more than 1% in our cohorts. At present, we are unable to delve deeper into the complexity of, and subancestries within, Native American and West African populations. Our resolution reflects the current availability of reference data sets from different regions.
However, we emphasize that these groups and ancestries are only a fraction of the diversity found within individuals living in the US, and as data set sizes grow, future work should extend to include analyses of other worldwide ancestries and populations and their distributions across the US.
Patterns of Genetic Ancestry of Self-Reported African Americans
Consistent with previous studies,2,70 the diversity of ancestry profiles of 23andMe African Americans reveal that individuals comprise the full range from 0% to 100% African ancestry, but, further, that there are differences in estimates of ancestry proportions among regions. Namely, we find differences between states that were slave-holding and those that were “free” at the time of the US Civil War. Reflected in these ancestry patterns are migration routes, such as the trans-Atlantic slave trade that brought Africans through important Southern seaports (as documented online in American FactFinder and American Community Survey Summary File). The small sample sizes from some areas of the US, including parts of the Midwest and Mountain regions, reflects the lower population density of African Americans residing in these regions (see the “The Black Populations” Census Brief).
Though mean estimates of Native American ancestry are low, many African Americans carry detectable levels of Native American ancestry. Consistent with historical narratives and family histories, our estimate suggests that one in every five African Americans carries Native American ancestry, a higher rate than we detected in self-reported European Americans. An individual that carries more than 1% Native American ancestry can arise from one genetically Native American ancestor within the last 11 or so generations, or multiple genealogical Native American ancestors (for discussion, see “How Many Genetic Ancestors Do I Have?” online). Oklahoma shows the highest proportion of African Americans with substantial Native American ancestry, where more than 14% of African Americans from Oklahoma carry at least 2% Native American ancestry (Figures 1B and S2). Oklahoma was the site of contact between Native Americans and African Americans after the Trail of Tears migration in the 1830s,71,72 where black slaves comprised a significant part of the population in the 1860s (according to the US 1860 Census), and the location of the slave-holding “Five Civilized Tribes.” In contrast, we do not observe higher rates of Native American ancestry in African Americans in Florida, which is potentially notable in light of the known history of Seminole intermarriage with blacks according to the 1860 US Census (information available online).
Even excluding individuals with no African ancestry, which are probably the result of survey errors, we still estimate a higher European, and corresponding lower African, mean genetic ancestry proportion in 23andMe African Americans compared to previous studies of African Americans. A significant difference between the 23andMe cohort of African Americans and many groups previously studied is geographic sampling. Our cohort reflects heavier sampling of individuals living in or born in California and New York, probably driven by population density as well as awareness of genetic testing or 23andMe. Both are regions where African Americans have lower mean African ancestry than other studies of African Americans, which are often drawn from locations in the South. However, participation in 23andMe is not free and requires online access, so therefore it is important to note that other social, cultural, or economic factors might interact to affect ancestry proportions of those individuals who choose to participate in 23andMe.
Our admixture dates for African Americans provide evidence that African and European mixture occurred prior to 1860, suggesting that gene flow between these groups might predate the Great Migration of African Americans from the South into the North beginning around 1910, though more complex models (that capture more continuous gene flow) are needed to resolve African and European mixture timing.73
Patterns of Genetic Ancestry of Self-Reported Latinos
We estimated that Iberian ancestry composes as much as a third of the European ancestry in Latinos in Florida, New Mexico, and other parts of the Southwest, probably reflecting either early Spanish influence and rule in these regions or recent immigration from Latin America, which might also be associated with higher levels of Iberian ancestry in New York and New Jersey. Regions with higher Iberian ancestry also correspond to regions with greater Native American ancestry; disentangling whether higher levels of Native American ancestry in the Southwest reflects the legacy of indigenous Native American ancestors or is the result of recent Latino immigrants into the Southwest might be possible through future studies of admixture dating or more Native American subpopulation reference data.
Patterns of Genetic Ancestry of Self-Reported European Americans
Our estimated rates of non-European ancestry in European Americans suggest that more than six million Americans, who self-identify as European, might carry African ancestry. Likewise, as many as five million Americans who self-identify as European might have at least 1% Native American ancestry. Louisiana’s high levels of African ancestry in European Americans are consistent with historical accounts of intermarriage in the New Orleans area.74,75
Regional differences in European subpopulation ancestry across states reflect known major historical migrations from Europe. Inferred British/Irish ancestry is found in European Americans from all states at mean proportions of more than 20% and represents a majority of ancestry (more than 50% mean proportion) in states such as Mississippi, Arkansas, and Tennessee. These states are similarly highlighted in the map of the self-reported “American” ethnicity in the US 2010 Census survey, which might reflect regions with lower subsequent migration from other parts of Europe. Inferred Eastern European ancestry is found at its highest levels in Illinois, Michigan, and Pennsylvania, potentially stemming from immigration during the late 19th and early 20th centuries, settling in metropolitan areas in the Northeast and Midwest. Inferred Iberian ancestry, found overall at lower mean proportions, still represents a measurable ancestry component in Florida, Louisiana, California, and Nevada, and might point to the early Spanish rule and colonization of the Americas. Scandinavian ancestry in European Americans is highly localized; most states show only trace mean proportions of Scandinavian ancestry, but it comprises a significant proportion, upward of 10%, of ancestry in European Americans from Minnesota and the Dakotas. The distributions of the European subpopulation ancestries in European Americans illustrate that the distribution of within-European ancestry is not homogenous among individuals from different states, and instead, reflects differences in population migrations and settlement patterns across the US.
Sex Bias in Ancestry Contributions
We find evidence that sex-biased admixture processes are widespread in US history in European Americans as well as in African American and Latino populations. Estimates of proportions of males and females from each ancestral population (Table S4) suggest that under a simple demographic model of admixture, European Americans might have ten times as many female Native American ancestors as male, and African Americans might have four times as many female Native American ancestors as male. Sex bias in ancestry contributions might have been driven by unbalanced sex ratios in immigration frontier settings,76 exploitation,77 or other social factors.
Robust Estimates of African and Native American Ancestry in African Americans and European Americans
Several lines of evidence suggest that Native American and African segments represent true signals of Native American and African introgression that occurred after the transcontinental migrations beginning in the 1500s. Validation of our self-reported survey data across two independent surveys shows that self-reported ancestry consistency is remarkably high. African ancestry in European Americans is not likely to be driven by survey errors because the number of European Americans with African ancestry is ten times larger than our estimates of survey error rates. Furthermore, the ancestry profiles of self-reported European Americans with African ancestry are distinct from all other cohorts: their African ancestry is much lower than for a random sample of African Americans, and the majority of these individuals do not carry any appreciable amount of Native American ancestry, distinguishing their ancestry profiles from Latinos (see Figure S1C).
A potential source of bias in our estimates is from errors in the ancestry inference algorithm. To show that our estimates are not the result of Ancestry Composition errors or biases, we validated the estimates of low levels of African ancestry in European Americans comparing to f4 statistics,64 1000 Genomes Project consensus estimates,78 and ADMIXTURE estimates.66 Another line of evidence supporting our estimates of non-European ancestry in European Americans in the US is that we observe a substantially lower occurrence of Native American and African ancestry in individuals who self-report four grandparents born in the same European country. The inferred segments of African and Native American are uniformly distributed across the genome. Although we expect that some of the inferred ancestry might arise from difficulties in assigning ancestry in complex regions of the genome, only a small fraction of the estimated African and Native American ancestry in European Americans can be explained through such biases and is not expected to give rise to any substantial (more than 1%) ancestry from any population.
Lastly, our recent dates for admixture suggest that introgression probably occurred in the Americas within the last 500 years. Hence, our estimates do not support that the African ancestry in European Americans stems from ancient population events that predate the migrations to the Americas. (For example, gene flow from Africa coinciding with the Moor invasion of the Mediterranean might have introduced African ancestry into the ancestral population of some European Americans.) Though such ancient events would probably not lead to inferred African ancestry because our supervised learning algorithm would apply a European label to such segments, it is possible that European population substructure could lead to inferred segments of African ancestry in some European Americans that derive from older historical admixture events, which are not seen in modern Europeans. However, these events would lead to admixture or introgression of segments several hundred or thousand years old, and our admixture dates for both Native American ancestry and African ancestry point to gene flow within the last 20 generations and is not consistent with any known historical migrations within Europe during this time period.
Correlations with Population Proportions
Correlations between state population proportions and mean ancestry proportions suggest that the numbers of African and Native American individuals in a state might have shaped the ancestries of present-day individuals. For African Americans, the states with the highest mean levels of African ancestry, such as South Carolina, Georgia, and Florida are not those with the highest proportions of African Americans. Given the highly significant statistics in European Americans, surprisingly, in African Americans, the correlation of African ancestry with proportions of African Americans is only marginally significant (p value 0.025). The correlation of Native American ancestry in African Americans with Latino state population proportion also has a marginal p value of 0.026. Not all correlations are strongly significant, suggesting that other social or cultural factors influenced levels of ancestry, especially in African Americans.
Relationship of Self-Identity and Genetic Ancestry
Contrary to expectations under a social one-drop rule, or “Rule of Hypodescent,” which would mandate that individuals who knowingly carry African ancestry identify as African American, the probability of self-reporting as African American given a proportion of African ancestry follows a logistic probability curve (Figure S9A, Table S6), suggesting that individuals identify roughly with the majority of their genetic ancestry (Figures 4 and 5A). Individuals with more than 5% Native American ancestry are most likely to self-identify as Latino (Figures S9C and 5B), suggesting differences in sociological or historical factors associated with identifying with these groups. The transitions between Latino, African American, and European American self-reported identity by proportions of African and Native American ancestry illustrate both the complexity of how one self identifies as well as the overlapping ancestry profiles among groups (Figure 5B).
Conclusion
This work demonstrates that the legacy of population migrations and interactions over the last several hundred years is visible in the genetic ancestry of modern individuals living in the US. Our results suggest that genetic ancestry can be leveraged to augment historical records and inform cultural processes shaping modern populations. The relationship between self-reported identity and genetic African ancestry, as well as the low numbers of self-reported African Americans with minor levels of African ancestry, provide insight into the complexity of genetic and social consequences of racial categorization, assortative mating, and the impact of notions of “race” on patterns of mating and self-identity in the US. Our results provide empirical support that, over recent centuries, many individuals with partial African and Native American ancestry have “passed” into the white community,79,80 with multiple lines of evidence establishing African and Native American ancestry in self-reported European Americans (see Subjects and Methods). Though the majority of European Americans in our study did not carry Native American or African ancestry, even a small proportion of this large population that carry non-European ancestry translates into millions of European Americans who carry African and Native American ancestry. Our results suggest that the early US history, beginning in the 17th century (around 12 generations ago), might have been a time of many population interactions resulting in admixture.
Large sample sizes, high-density genotype data, and accurate and robust local ancestry estimates allowed us to discern subtle differences in genetic ancestry. In spite of present-day high mobility of individuals, the genetic ancestry of present-day individuals recapitulates historical migration events, known settlement patterns, and admixture processes. Perhaps most importantly, however, our results reveal the impact of centuries of admixture in the US, thereby undermining the use of cultural labels that group individuals into discrete nonoverlapping bins in biomedical contexts “which cannot be adequately represented by arbitrary ‘race/color’ categories.”81
Our findings can inform medical genetic studies. Introgressed Native American and African haplotypes in European Americans might have implications for studies of complex diseases, especially for diseases that vary in prevalence among ancestral populations, can produce subtle population structure that should be carefully controlled for in GWASs, and might impact the distribution of rare variants in studies of whole-genome sequence. Our results also suggest new avenues for research, such as the potential for including European Americans in admixture mapping.
Acknowledgments
We thank the customers of 23andMe who answered surveys and participated in this research. We are grateful to Dr. Jeffrey C. Long at the University of New Mexico, Dr. Claudio Saunt at the University of Georgia, and Sarah Abel at Centre International des Recherches sur les Esclavages, CNRS, Paris for invaluable discussions and comments on a manuscript draft. We thank Nick Patterson and Priya Moorjani for helpful statistical discussions on f statistics. Of course, all mistakes and inaccuracies are our own. Thanks to all the employees of 23andMe, who together have made this research possible, especially Emma Pierson, Robin Smith, Youna Hu, and Scott Hadly. K.B., E.Y.D., and J.L.M. are current employees (and J.M.M. is a former employee) of 23andMe, Inc., and have private equity interest. This work was supported by NIH award 2R44HG006981-02. D.R. was supported by NSF HOMINID award BCS-1032255 and NIH grant GM100233, and D.R. is a Howard Hughes Medical Institute investigator.
Published: December 18, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
1860 Census, https://www.census.gov/history/www/through_the_decades/overview/1860.html
2010 Census, http://www.census.gov/2010census/
American Community Survey Summary File 2007–2010, http://www2.census.gov/acs2011_5yr/summaryfile/
American FactFinder, http://factfinder2.census.gov/
Free Zipcode Database, http://federalgovernmentzipcodes.us/
How Many Genetic Ancestors Do I Have?, http://gcbias.org/2013/11/11/how-does-your-number-of-genetic-ancestors-grow-back-over-time/
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
Python, https://www.python.org/
R statistical software, http://www.r-project.org/
Roots into the Future, https://www.23andme.com/roots/
The Black Population: 2010, http://www.census.gov/prod/cen2010/briefs/c2010br-06.pdf
Trans-Atlantic Slave Trade Database, Introductory Maps, http://www.slavevoyages.org/tast/assessment/intro-maps.faces
References
- 1.Moreno-Estrada A., Gignoux C.R., Fernández-López J.C., Zakharia F., Sikora M., Contreras A.V., Acuña-Alonzo V., Sandoval K., Eng C., Romero-Hidalgo S. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parra E.J. Admixture in North America. In: Suarez-Kurtz G., editor. Pharmacogenomics in Admixed Populations. Landes Bioscience; Austin: 2007. pp. 28–46. [Google Scholar]
- 3.Parra E.J., Marcini A., Akey J., Martinson J., Batzer M.A., Cooper R., Forrester T., Allison D.B., Deka R., Ferrell R.E., Shriver M.D. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith M.W., Patterson N., Lautenberger J.A., Truelove A.L., McDonald G.J., Waliszewska A., Kessing B.D., Malasky M.J., Scafe C., Le E. A high-density admixture map for disease gene discovery in african americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra E.J., Kittles R.A., Argyropoulos G., Pfaff C.L., Hiester K., Bonilla C., Sylvester N., Parrish-Gause D., Garvey W.T., Jin L. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Lind J.M., Hutcheson-Dilks H.B., Williams S.M., Moore J.H., Essex M., Ruiz-Pesini E., Wallace D.C., Tishkoff S.A., O’Brien S.J., Smith M.W. Elevated male European and female African contributions to the genomes of African American individuals. Hum. Genet. 2007;120:713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 7.Salas A., Richards M., Lareu M.-V., Sobrino B., Silva S., Matamoros M., Macaulay V., Carracedo A. Shipwrecks and founder effects: divergent demographic histories reflected in Caribbean mtDNA. Am. J. Phys. Anthropol. 2005;128:855–860. doi: 10.1002/ajpa.20117. [DOI] [PubMed] [Google Scholar]
- 8.Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.-M., Doumbo O. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryc K., Auton A., Nelson M.R., Oksenberg J.R., Hauser S.L., Williams S., Froment A., Bodo J.-M., Wambebe C., Tishkoff S.A., Bustamante C.D. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd J.M., Gravel S., Byrnes J., Moreno-Estrada A., Musharoff S., Bryc K., Degenhardt J.D., Brisbin A., Sheth V., Chen R. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am. J. Hum. Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dipierri J.E., Alfaro E., Martínez-Marignac V.L., Bailliet G., Bravi C.M., Cejas S., Bianchi N.O. Paternal directional mating in two Amerindian subpopulations located at different altitudes in northwestern Argentina. Hum. Biol. 1998;70:1001–1010. [PubMed] [Google Scholar]
- 13.González-Andrade F., Sánchez D., González-Solórzano J., Gascón S., Martínez-Jarreta B. Sex-specific genetic admixture of Mestizos, Amerindian Kichwas, and Afro-Ecuadorans from Ecuador. Hum. Biol. 2007;79:51–77. doi: 10.1353/hub.2007.0024. [DOI] [PubMed] [Google Scholar]
- 14.Green L.D., Derr J.N., Knight A. mtDNA affinities of the peoples of North-Central Mexico. Am. J. Hum. Genet. 2000;66:989–998. doi: 10.1086/302801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendizabal I., Sandoval K., Berniell-Lee G., Calafell F., Salas A., Martínez-Fuentes A., Comas D. Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol. Biol. 2008;8:213. doi: 10.1186/1471-2148-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrero A.R., Bravi C., Stuart S., Long J.C., Pereira das Neves Leite F., Kommers T., Carvalho C.M., Pena S.D., Ruiz-Linares A., Salzano F.M., Cátira Bortolini M. Pre- and post-Columbian gene and cultural continuity: the case of the Gaucho from southern Brazil. Hum. Hered. 2007;64:160–171. doi: 10.1159/000102989. [DOI] [PubMed] [Google Scholar]
- 17.Sans M., Weimer T.A., Franco M.H.L.P., Salzano F.M., Bentancor N., Alvarez I., Bianchi N.O., Chakraborty R. Unequal contributions of male and female gene pools from parental populations in the African descendants of the city of Melo, Uruguay. Am. J. Phys. Anthropol. 2002;118:33–44. doi: 10.1002/ajpa.10071. [DOI] [PubMed] [Google Scholar]
- 18.Carvajal-Carmona L.G., Ophoff R., Service S., Hartiala J., Molina J., Leon P., Ospina J., Bedoya G., Freimer N., Ruiz-Linares A. Genetic demography of antioquia (colombia) and the central valley of costa rica. Hum. Genet. 2003;112:534–541. doi: 10.1007/s00439-002-0899-8. [DOI] [PubMed] [Google Scholar]
- 19.Sans M. Admixture studies in Latin America: from the 20th to the 21st century. Hum. Biol. 2000;72:155–177. [PubMed] [Google Scholar]
- 20.Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seldin M.F., Tian C., Shigeta R., Scherbarth H.R., Silva G., Belmont J.W., Kittles R., Gamron S., Allevi A., Palatnik S.A. Argentine population genetic structure: large variance in Amerindian contribution. Am. J. Phys. Anthropol. 2007;132:455–462. doi: 10.1002/ajpa.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva-Zolezzi I., Hidalgo-Miranda A., Estrada-Gil J., Fernandez-Lopez J.C., Uribe-Figueroa L., Contreras A., Balam-Ortiz E., del Bosque-Plata L., Velazquez-Fernandez D., Lara C. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. USA. 2009;106:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimentidis Y.C., Miller G.F., Shriver M.D. Genetic admixture, self-reported ethnicity, self-estimated admixture, and skin pigmentation among Hispanics and Native Americans. Am. J. Phys. Anthropol. 2009;138:375–383. doi: 10.1002/ajpa.20945. [DOI] [PubMed] [Google Scholar]
- 24.Bryc K., Velez C., Karafet T., Moreno-Estrada A., Reynolds A., Auton A., Hammer M., Bustamante C.D., Ostrer H. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. USA. 2010;107(2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno-Estrada A., Gravel S., Zakharia F., McCauley J.L., Byrnes J.K., Gignoux C.R., Ortiz-Tello P.A., Martínez R.J., Hedges D.J., Morris R.W. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shriner D. Overview of admixture mapping. Curr. Protoc. Hum. Genet. 2013;Chapter 1:23. doi: 10.1002/0471142905.hg0123s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 29.Halder I., Shriver M., Thomas M., Fernandez J.R., Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum. Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 30.Halder I., Yang B.-Z., Kranzler H.R., Stein M.B., Shriver M.D., Gelernter J. Measurement of admixture proportions and description of admixture structure in different U.S. populations. Hum. Mutat. 2009;30:1299–1309. doi: 10.1002/humu.21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lao O., Vallone P.M., Coble M.D., Diegoli T.M., van Oven M., van der Gaag K.J., Pijpe J., de Knijff P., Kayser M. Evaluating self-declared ancestry of U.S. Americans with autosomal, Y-chromosomal and mitochondrial DNA. Hum. Mutat. 2010;31:E1875–E1893. doi: 10.1002/humu.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes B. Liveright; New York: 2012. DNA USA: A Genetic Portrait of America. [Google Scholar]
- 33.Durand, E.Y., Do, C.B., Mountain, J.L., and Macpherson, J.M. (2014). Ancestry composition: A novel, efficient pipeline for ancestry deconvolution. bioRxiv pp. 010512.
- 34.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe’er I., Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tung J.Y., Do C.B., Hinds D.A., Kiefer A.K., Macpherson J.M., Chowdry A.B., Francke U., Naughton B.T., Mountain J.L., Wojcicki A., Eriksson N. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS ONE. 2011;6:e23473. doi: 10.1371/journal.pone.0023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eriksson N., Benton G.M., Do C.B., Kiefer A.K., Mountain J.L., Hinds D.A., Francke U., Tung J.Y. Genetic variants associated with breast size also influence breast cancer risk. BMC Med. Genet. 2012;13:53. doi: 10.1186/1471-2350-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Do C.B., Tung J.Y., Dorfman E., Kiefer A.K., Drabant E.M., Francke U., Mountain J.L., Goldman S.M., Tanner C.M., Langston J.W. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson N., Tung J.Y., Kiefer A.K., Hinds D.A., Francke U., Mountain J.L., Do C.B. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS ONE. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henn B.M., Hon L., Macpherson J.M., Eriksson N., Saxonov S., Pe’er I., Mountain J.L. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS ONE. 2012;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson N., Wu S., Do C.B., Kiefer A.K., Tung J.Y., Mountain J.L., Hinds D.A., Francke U. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour. 2012;1:22. [Google Scholar]
- 41.Kiefer A.K., Tung J.Y., Do C.B., Hinds D.A., Mountain J.L., Francke U., Eriksson N. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013;9:e1003299. doi: 10.1371/journal.pgen.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinds D.A., McMahon G., Kiefer A.K., Do C.B., Eriksson N., Evans D.M., St Pourcain B., Ring S.M., Mountain J.L., Francke U. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durand E.Y., Eriksson N., McLean C.Y. Reducing pervasive false-positive identical-by-descent segments detected by large-scale pedigree analysis. Mol. Biol. Evol. 2014;31:2212–2222. doi: 10.1093/molbev/msu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bamshad M., Guthery S.L. Race, genetics and medicine: does the color of a leopard’s spots matter? Curr. Opin. Pediatr. 2007;19:613–618. doi: 10.1097/MOP.0b013e3282f163ca. [DOI] [PubMed] [Google Scholar]
- 45.Perez A.D., Hirschman C. The changing racial and ethnic composition of the US population: emerging American identities. Popul. Dev. Rev. 2009;35:1–51. doi: 10.1111/j.1728-4457.2009.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glass B., Li C.C. The dynamics of racial intermixture; an analysis based on the American Negro. Am. J. Hum. Genet. 1953;5:1–20. [PMC free article] [PubMed] [Google Scholar]
- 49.Glass B. On the unlikelihood of significant admixture of genes from the North American Indians in the present composition of the Negroes of the United States. Am. J. Hum. Genet. 1955;7:368–385. [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts D.F. The dynamics of racial intermixture in the American Negro-some anthropological considerations. Am. J. Hum. Genet. 1955;7:361–367. [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts D.F., Hiorns R.W. The dynamics of racial intermixture. Am. J. Hum. Genet. 1962;14:261–277. [PMC free article] [PubMed] [Google Scholar]
- 52.Reed T.E. Caucasian genes in American Negroes. Science. 1969;165:762–768. doi: 10.1126/science.165.3895.762. [DOI] [PubMed] [Google Scholar]
- 53.Adams J., Ward R.H. Admixture studies and the detection of selection. Science. 1973;180:1137–1143. doi: 10.1126/science.180.4091.1137. [DOI] [PubMed] [Google Scholar]
- 54.Chakraborty R. Gene admixture in human populations: models and predictions. Am. J. Phys. Anthropol. 1986;29:1–43. [Google Scholar]
- 55.Chakraborty R., Kamboh M.I., Nwankwo M., Ferrell R.E. Caucasian genes in American blacks: new data. Am. J. Hum. Genet. 1992;50:145–155. [PMC free article] [PubMed] [Google Scholar]
- 56.Reiner A.P., Ziv E., Lind D.L., Nievergelt C.M., Schork N.J., Cummings S.R., Phong A., Burchard E.G., Harris T.B., Psaty B.M., Kwok P.Y. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am. J. Hum. Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Workman P.L., Blumberg B.S., Cooper A.J. Selection, gene migration and polymorphic stability in a U. S. White and Negro population. Am. J. Hum. Genet. 1963;15:429–437. [PMC free article] [PubMed] [Google Scholar]
- 58.Pollitzer W.S. The Negroes of Charleston (SC); a study of hemoglobin types, serology, and morphology. Am. J. Phys. Anthropol. 1958;16:241–263. [Google Scholar]
- 59.Pollitzer W.S. Analysis of a tri-racial isolate. Hum. Biol. 1964;36:362–373. [Google Scholar]
- 60.Pollitzer W.S., Boyle E., Jr., Cornoni J., Namboodiri K.K. Physical anthropology of the Negroes of Charleston, S. C. Hum. Biol. 1970;42:265–279. [PubMed] [Google Scholar]
- 61.Blumberg B.S., Hesser J.E. Loci differentially affected by selection in two American black populations. Proc. Natl. Acad. Sci. USA. 1971;68:2554–2558. doi: 10.1073/pnas.68.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollitzer W.S. The relationship of the Gullah-speaking people of coastal South Carolina and Georgia to their African ancestors. Hist. Methods. 1993;26:53–67. [Google Scholar]
- 63.Nelson M.R., Bryc K., King K.S., Indap A., Boyko A.R., Novembre J., Briley L.P., Maruyama Y., Waterworth D.M., Waeber G. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price A.L., Weale M.E., Patterson N., Myers S.R., Need A.C., Shianna K.V., Ge D., Rotter J.I., Torres E., Taylor K.D. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. author reply 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Via M., Gignoux C.R., Roth L.A., Fejerman L., Galanter J., Choudhry S., Toro-Labrador G., Viera-Vera J., Oleksyk T.K., Beckman K. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS ONE. 2011;6:e16513. doi: 10.1371/journal.pone.0016513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zakharia F., Basu A., Absher D., Assimes T.L., Go A.S., Hlatky M.A., Iribarren C., Knowles J.W., Li J., Narasimhan B. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10:R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wells S.J., Tubby R., editors. After Removal: The Choctaw in Mississippi. Univ. Press of Mississippi; Jackson: 2004. [Google Scholar]
- 72.Green, L. (1978). Choctaw removal was really a “trail of tears.” Bishinik, November, 1978. pp. 8–9.
- 73.Lemann N. Random House LLC; 2011. The Promised Land: The Great Black Migration and How It Changed America. [Google Scholar]
- 74.Williamson J. Free Press New York; 1980. New People: Miscegenation and Mulattoes in the United States. [Google Scholar]
- 75.Piersen W.D. Twayne Publishers; 1996. From Africa to America: African American History from the Colonial Era to the Early Republic, 1526-1790. [Google Scholar]
- 76.Davis F.J. Penn State Press; 2001. Who Is Black?: One Nation’s Definition. [Google Scholar]
- 77.Fredrickson G.M. Oxford University Press; 1982. White Supremacy: A Comparative Study of American and South African History. [Google Scholar]
- 78.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burma J.H. The measurement of Negro passing. Am. J. Sociol. 1946;52:18–22. doi: 10.1086/219915. [DOI] [PubMed] [Google Scholar]
- 80.Broyard B. Little, Brown and Co.; New York: 2007. One Drop: My Father’s Hidden Life–A Story of Race and Family Secrets. [Google Scholar]
- 81.Suarez-Kurtz G. Pharmacogenomics and the genetic diversity of the Brazilian population. Cad. Saude Publica. 2009;25:1650–1651. doi: 10.1590/s0102-311x2009000800001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


