Abstract
Growth-associated protein 43 (GAP43), is a strictly conserved protein among vertebrates implicated in neuronal development and neurite branching. Since GAP43 structure contains a calmodulin-binding domain, this protein is able to bind calmodulin and gather it nearby membrane network, thus regulating cytosolic calcium and consequently calcium-dependent intracellular events. Even if for many years GAP43 has been considered a neuronal-specific protein, evidence from different laboratories described its presence in myoblasts, myotubes and adult skeletal muscle fibers. Data from our laboratory showed that GAP43 is localized between calcium release units (CRUs) and mitochondria in mammalian skeletal muscle suggesting that, also in skeletal muscle, this protein can be a key player in calcium/calmodulin homeostasis. However, the previous studies could not clearly distinguish between a mitochondrion- or a triad-related positioning of GAP43. To solve this question, the expression and localization of GAP43 was studied in skeletal muscle of Xenopus and Zebrafish known to have triads located at the level of the Z-lines and mitochondria not closely associated with them. Western blotting and immunostaining experiments revealed the expression of GAP43 also in skeletal muscle of lower vertebrates (like amphibians and fishes), and that the protein is localized closely to the triad junction. Once more, these results and GAP43 structural features, support an involvement of the protein in the dynamic intracellular Ca2+ homeostasis, a common conserved role among the different species.
Key words: GAP43, RyR, skeletal muscle, Xenopus, Zebrafish
Introduction
The growth-associated protein 43 (GAP43) was originally found in neurons and its expression is particularly high during axonal growth and during development and regeneration in both central and peripheral nervous systems.1
This protein is strictly conserved among vertebrates thus indicating that it plays an essential role. Its mRNA is translated into a protein of 194-238 amino acid residues in length, that is unusually rich in acidic amino acids. The protein primary structure contains two conserved domains with a high degree of homology in vertebrates. The first domain located at the N-terminal tail is the most highly conserved and contains sites for post-translational attachment to fatty acids and binding to membranes.2 A second domain is a calmodulin binding region.3 The two domains are functionally interdependent. In the nervous system, GAP43 is considered as a calmodulin sponge, capable of binding and releasing calmodulin in response to intracellular calcium variations or to protein kinase C (PKC) activating signals. Interestingly, within the conserved first 57 amino acid-domain, there is a critical serine-41 residue that, when phosphorylated by PKC, allows the release of calmodulin from its GAP43 binding site. These evidences imply a regulatory role for the protein in all those events triggered by calcium variations or by the activation of PKC. It is noteworthy that the region containing the calmodulin binding site is strictly conserved in fish and in all known mammalian GAP43 sequences, thus strengthening its importance.4 Even though GAP43 was classified as neuron-specific protein, increasing evidences indicate that GAP43 is not confined to the nervous system. Early data were collected in embryonic chicken limb and in human skeletal muscle biopsies.5-9 The skeletal muscle is characterized by an extremely high degree of intracellular organization. In muscle fiber, the sarcolemmal T-tubules and sarcoplasmic reticulum are functionally coupled by regularly localized structures, named triads,10 each consisting in two enlargements of sarcoplasmic reticulum (terminal cisternae) opposed to a T-tubule, hosting protein complexes and constituting the calcium release units (CRUs), key structures of the excitation-contraction coupling process. Within CRUs the dihydropyridine receptor (DHPR) drives the calcium release from the ryanodine receptor (RyR) triggering fiber contraction.11 Data from our laboratory showed that GAP43 is expressed in mouse myoblasts, myotubes and in adult skeletal muscle fibers. Interestingly, both in myotubes and in extensor digitorum longus (EDL) fibers, GAP43 is localized at well defined periodic spots forming a doublet of lines at each sarcomere in the position corresponding to the location of triads and mitochondria.12 This evidence, and the functional role of GAP43 in the nervous system, as described above, supported the hypothesis that the protein may also have a key role in calcium/calmodulin homeostasis also in skeletal muscle. A limit of our previous studies was to not distinguish clearly between a mitochondrion- or a triad-related positioning of GAP43, due to the close proximity, and almost obligatory pairing, of mitochondria and triads at the edges of the A-band in murine muscle. Aim of the experimental plan presented here was to solve this question investigating the expression and localization of GAP43 in skeletal muscle of an amphibian and a fish as the skeletal fibers of these lower vertebrates are known to have triads located at the level of the Z-lines and mitochondria not closely associated with them.13,14
Materials and Methods
Chemicals and materials
Unless otherwise indicated, cell culture media, sera, antibiotics were obtained from Life Technologies (Monza, Italy), cell culture dishware from Becton Dickinson Falcon™ (Sacco Srl, Cadorago, Italy) and reagents and standards from Sigma-Aldrich (Milan, Italy).
Ethics statement
The care and use of mice C57BL/6, Xenopus laevis and Danio rerio strictly followed The Guiding Principles for the Care and Use of Animals, in accordance with the principles of the Declaration of Helsinki and with the European Community Council (86/609/CEE) and the Italian Government law on the protection of animals for experimental procedures in research laboratory (92/116). Animals were housed in a facility c/o D’Annunzio Foundation Ce.S.I. (Center for Research on Ageing, Chieti, Italy) and sacrificed as approved by the local University Committee on Animal Resources (Comitato di Etica Interateneo per la Sperimentazione Animale - CEISA Università G. d’Annunzio Chieti-Pescara - Universita’ di Teramo. Prot. n. 15/2011/CEISA/COM).
Western blot analysis
Protein extracts for Western blot were isolated from muscle fibers of Zebrafish, Xenopus and mouse, and whole murine brain. Isolated muscle fibers were washed in PBS and homogenized in ice-cold lysis buffer (50 mM Tris–HCl, 100 mM NaCl, 50 mM NaF, 40 mM β-glycerophosphate, 5 mM EDTA, 1% Triton X-100, 200 μM sodium orthovanadate, 100 μg/mL phenylmethylsulfonyl fluoride, 10 μg/mL leupeptin, 5 μg/mL pepstatin A, 10 μg/mL benzamidine, pH 7.4), using a Polytron tissue disrupter (Janke and Kunkel, Germany), at high speed (three 10 s pulses) on ice. C57BL/6 whole brain proteins were obtained by tissue homogenization in the ice-cold lysis buffer using a Dounce tight-fitting homogenizer. All homogenates were centrifuged at 400× g at 4°C, for 5 min to remove cell debris. The supernatants were collected and protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories Srl, Segrate, MI, Italy). Samples, containing 40 or 2.5 µg of proteins for muscle or brain respectively, were suspended in Laemmli buffer (8% (w/v) SDS, 10% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, 25 mM Tris-HCl, pH 6.5, and 0.003% (w/v) bromophenol blue), boiled for 5 min and separated by SDS-PAGE on a 10% (w/v) homogeneous slab gel. Proteins were electroblotted onto a nitrocellulose membrane (Amersham™ Hybond™-ECL; GE Healthcare, Milan, Italy). Membranes were blocked in TBS-T (Trisbuffered saline with 0.1% (v/v) Tween 20) containing 5% (w/v) fat-free milk and then incubated with the primary antibody anti-GAP43 (HPA-GAP43, diluition 1:1000, Sigma-Aldrich). The membranes were then incubated with horseradish peroxidase-conjugated anti-IgG and detected by chemiluminescence (Pierce®; EuroClone SpA Pero, MI, Italy).
Danio rerio (Zebrafish) myocyte isolation
Zebrafish larvae (72 hpf) were anesthetized using 0.03% tricainemethanesulfonate (MS-222 from Spectrum chemical, USA) in E3 medium. After decapitation the distal, less-developed, about 1/3 portion of the tail was cut away and 20-50 proximal tails were washed in ice cold 0.5x Hank’s Balanced Salt Solution (HBSS). The proximal tails were digested in 200 U/ml collagenase (from Clostridium histolyticum; Sigma-Aldrich) in 1x HBSS for 1.5 h with gently shaking at 28°C and triturated every 15 min with a fine tip plastic transfer pipette. Digestion was stopped by adding 10 volumes of culture medium (60% L-15, 3% fetal bovine serum, 3% horse serum, 4 mM glutamine, 100 U/mL-100 µg/mL penicillin-streptomycin). The cells were pelleted by centrifugation (200x g, 5 min) and re-suspended in a minimum of 200 µL of culture medium. Cells were then plated on a matrigel-coated glass coverslip into a 35 mm culture dish.
After adhesion, cells were fixed with 4% buffered paraformaldehyde for immunofluorescence experiments.
Xenopus laevis (Xenopus) myocyte isolation
Xenopus embryos were used from stage 19 to 22 of their development. To obtain muscle culture, the neuronal tube and adjacent somites were excised and treated with calcium/magnesium-free medium (CMF), containing in mM: 115 NaCl, 2.6 KCl, 0.4 EDTA, 10 HEPES, pH 7.6. After 10 min, the ectodermal tissue was peeled off and discarded. The remaining tissue (somites and notocord) were then incubated for 10 min in CMF. To obtain neuron-free cultures, the tissue fragments were treated with collagenase (Type I, Sigma-Aldrich, 1 mg/mL in medium containing [v/v]: 50% Ringer solution [containing in mM: 115NaCl, 2.6 KCl, 2 CaCl2, 10 HEPES, pH 7.6] and 50% Leibovitz medium) for 15 min. At the end, the neuronal tube was gently removed and the somitic tissue transferred into CMF for cell dissociation. Dissociated cells were seeded on glass coverslip and maintained at room temperature (20-22 °C) in humidified atmosphere with medium containing (v/v) 49% Ringer solution, 50% Leibovitz medium and 1% fetal calf serum.
Isolation of skeletal muscle samples from adult Zebrafish and Xenopus
Adult Zebrafish were anesthetized with 0.03% tricaine, killed by decapitation, and then fixed in 4% paraformaldehyde in PBS (pH 7.4) for 15 min. After fixation, skin was carefully removed and strips of tail muscles dissected. Xenopus were killed by decapitation and the tibialis posterior muscle dissected and fixed in 4% paraformaldehyde in PBS for 15 min. After fixation bundles of fibers were obtained by longitudinal (tendon-tendon) muscle dissociation. Fibers obtained from Zebrafish or Xenopus muscles were processed for confocal microscopy observation.
Confocal microscopy
All samples were fixed with 4% paraformaldehyde for 10 min at room temperature, then washed three times in PBS, permeabilized with a 0.2% Triton X-100 solution for 10 min, incubated in blocking buffer (PBS containing 10% goat serum) for 1 h at room temperature, and incubated overnight at 4°C with primary antibodies anti-GAP43 (rabbit polyclonal anti-GAP43, HPA-GAP43, dilution 1:500 Sigma-Aldrich) and anti-RyR1/RyR2 (34C antibody, dilution 1:20, Developmental Studies Hybridoma Bank - University of Iowa). Primary antibodies were revealed by a 2 h incubation with secondary goat anti-rabbit Alexa 488 alone or together with goat anti-mouse Alexa 543 (dilution 1:200, Invitrogen, Monza, Images were acquired using a Zeiss LSM510 META system (Jena, Germany) equipped with a Zeiss Axiovert 200 inverted microscope and a Plan Neofluar oil-immersion objective (40X/1.3 NA and 100X/1.3 NA). Negative controls for each immunostaining assay were performed by sample immunolabeling with only secondary antibodies. Quantitative analyses on acquired images were obtained using Zeiss LSM software ver. 3.0, Manders’ coefficient was calculated using ImageJ software (W.S., ImageJ, National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2004).
Results
The expression of GAP43 was assayed in protein extracts from adult Zebrafish and Xenopus muscle samples, using mouse muscle and brain protein extracts as GAP43-positive control (Figure 1). The Western blotting revealed specific immuno-reactive bands in all muscle and brain extracts at about 43 kDa when the membrane was probed with anti-GAP43 HPA-antibody directed against the GAP43 conserved IQ domain, containing Ser-41 residue. GAP43 expression levels appeared lower in muscle samples compared to the brain extract, considering that 40 μg of total proteins for each muscle sample and only 2.5 μg of total proteins for brain extract were subjected to electrophoresis (see Materials and Methods). Only Zebrafish muscle sample shows two close bands with approximately the same intensity at ≈43 kDa, probably due to a species-specific genomic duplication (see Discussion). Using confocal microscopy, we analyzed the cellular localization of GAP43 in isolated primary muscle cultures derived from Zebrafish and Xenopus. GAP43 localized in myocytes of both species, depicting a regular punctuate pattern. The puncta were aligned along transversely orientated lines in respect to the longitudinal axe of the myocytes (Figure 2 A,B). The averaged distances between striations were 1.78±0.016 µm (n=31) and 1.79±0.021 µm (n=47) in samples from Zebrafish and Xenopus, respectively (Figure 2C) and represent a sarcomeric spacing. This disposition is clearly different from the one previously observed in mammalian myotubes and fibers in which GAP43 showed a regular doublets of aligned puncta within each sarcomere.12 A further specificity of the GAP43-positive foci location was obtained by comparing them with those of RyRs, the sarcoplasmic reticulum calcium-release channels, that are a major component of the CRUs. In fibers isolated from adult Zebrafish and Xenopus, double-immunolabeling for RyR/GAP43, showed single transverse rows of foci for both proteins with a detectable overlap (Figure 3).
Figure 1.
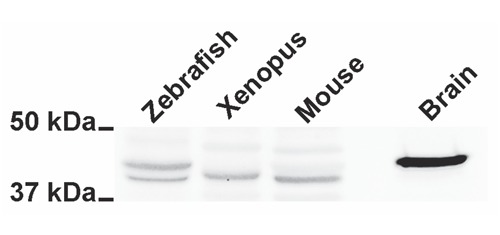
GAP43 is expressed in skeletal muscle of lower vertebrates. Western blot analysis of skeletal muscle samples deriving from Danio rerio (Zebrafish), Xenopus leavis (Xenopus), Mus musculus (Mouse) and a sample from mouse brain (Brain).
Figure 2.
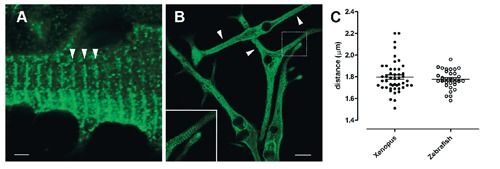
Localization of GAP43 in Zebrafish and Xenopus myotubes has a periodic sarcomeric spacing. Representative immunofluorescence images of muscle cell cultures deriving from Zebrafish (A) and Xenopus (B) samples. Confocal microscopy analysis shows a single striation cross-orientated as indicated by arrowheads (A and B). C) Measurements of distances between striations. Scale bars: A) 20 µm; B) 2 µm.
Figure 3.
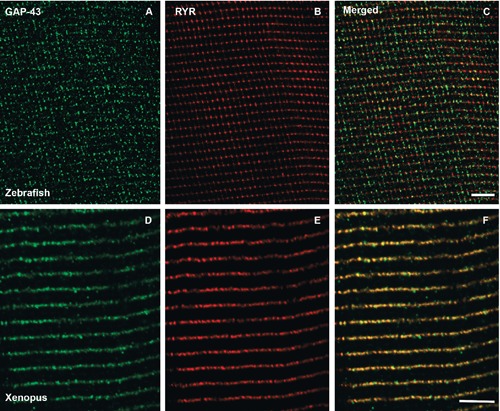
GAP43 is localized close to CRUs, in adult fibers. Representative immunofluorescence images of double-immunolabeled muscle fibers from Zebrafish (A-C) and Xenopus (D-F) samples. In adult Zebrafish and Xenopus muscle fibers, the staining of CRUs (marked with an anti-RyR antibody, B and E) forms dotted single rows along the Z-lines. Also GAP43 staining produces a single dotted cross-striation (A and D), which appears to co-localize with that of RyR (C and F, merged images). Scale bars: 5 µm.
Quantitative analyses of the degree of RyRGAP43 co-localization were performed calculating the Manders’ coefficient that indicates the overlap of the fluorescence signals.15 These analyses revealed overlapping coefficients of r =0.65 and r =0.77 for Zebrafish and Xenopus, respectively, showing a signal overlap higher than 60%. These data were also supported by fluorescence image profile analyses (Figure 4).
Figure 4.
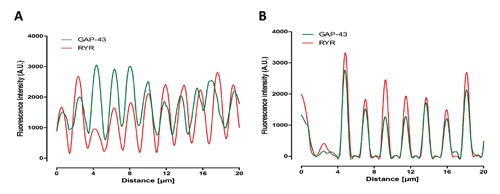
Fluorescence image profile. Graphs represent fluorescence intensity profiles calculated on images obtained from both Zebrafish (A) and Xenopus (B) muscle fiber samples co-immunostained for RyR and GAP43. The GAP43 fluorescence peak appears close to that of RyR.
The image profiles, derived from green and red channels, showed a high degree of correlation between the periodic location of GAP43-and RyR-fluorescence peaks, particularly in Xenopus.
Discussion
Our results provide novel information about the fine sub-cellular distribution of the GAP43 and its spatial relationship with the CRUs’ components in skeletal muscle specimens of lower vertebrates, confirming and extending previously reported data in mammalian skeletal muscles.5,12
Stoker and co-workers had showed the presence of GAP43 in embryonic chick limb, demonstrating that the protein co-localized in cells that are immunoreactive for meromyosin, a muscle-specific marker. For this reason the authors hypothesized that GAP43 could be involved in myoblasts fusion process to form myotubes.8,9 The presence of GAP43 in mammalian muscle was also reported by other Authors. Heuss and colleagues showed that in human myopathies, regenerating muscle cells were GAP43-positive, and proposed that GAP43 could be involved in the morphogenesis of regenerating myocytes.5-7 In a previous work, we reported that GAP43 was expressed in skeletal muscle satellite cells and fibers, and acquires its sarcomeric-like disposition in mammalian in vitro differentiated myotubes and in isolated mature fibers. Moreover, immunofluorescence experiments and co-localization studies in isolated mouse EDL muscle fibers showed that GAP43 is located close to CRUs and mitochondria showing a robust degree of overlapping with RyR.12
The results showed here revealed that GAP43 is also expressed in skeletal muscle of non mammalian vertebrates. Western blot analysis demonstrated the presence of GAP43 in skeletal muscle homogenates from Xenopus and Zebrafish. Interestingly the latter showed a peculiar two-band pattern. The reason for this behavior is not known, but may be explained by the genomic duplication occurred during teleost fish evolution.16 Some of the duplicated genes evolved separately and eventually led to the codification, and consequently translation, of two slightly different variants of the same original protein.17 In addition we can also suppose that the revealed pattern could be due also to a possible different post-translational processing of GAP43. Conversely, this could be essentially due to the different immunoreactivity of the anti-GAP43 antibody used, since different anti-GAP43 antibodies can show different immunoreactive patterns as also previously reported.12
Localization of GAP43 by immunofluorescence analysis were performed both on in vitro differentiated myotubes (Xenopus), in isolated differentiated myocytes (Zebrafish larvae) and isolated adult fibers (Zebrafish and Xenopus) resembling skeletal muscle models at different maturation degree and in different conditions.18-20 In all these models, GAP43 localized in single transverse fairly narrow bands at the Z-line. This localization reflects the different disposition of CRUs in amphibians and fishes, compared to the one observed in mammalians,21-23 and confirms the direct correlation between GAP43 and the calcium release units, as we previously suggested.12
The RyR-GAP43 double-labeled images show a slightly different fluorescence pattern in Xenopus compared to Zebrafish muscle fibers. In the former, the signal appears to be diffuse along the Z-line creating an almost continuous strip of fluorescence, while in the latter, clearly separated dots are visible. This difference is due to the different spatial organization of myofibrils in fish compared to the other vertebrates. In the fish, ribbon-shaped myofibrils extend radially from the center of the fiber to the periphery. If the longitudinal optical section happens to cut in the middle of the myofibril no multiple myofibril overlaps will occur in the z direction, even in a relatively thick section (Figure 5A). In this situation the sarcoplasmic reticulum will be cut perpendicularly to the section ensuring that CRUs from levels at different depths in the section overlap in a relatively narrow area. This narrow area is visible as a single spot at light microscopy resolution (Figure 5C). On the other hand, in Xenopus (and in the other vertebrates) the presence of more rounded myofibrils and the lack of radial arrangement cause the occurrence of multiple myofibrils overlap in the z-direction (Figure 5B). In this scenario, the sarcoplasmic reticulum will be visible in both the perpendicular and parallel orientations to the section plan, resulting in a fluorescent signal spreading almost continuously along the Z-line (Figure 5D). Therefore, the fish muscle provides us a more precise localization of GAP43 relatively to RyR. The double-labeled images of fish muscle show that GAP43 fluorescent signal is often off centered in respect to the expected position of the CRUs (marked by the RyR signal). This would indicate a peripheral location of GAP43 relative to RyR, suggesting that the protein is not within the junctional region of the triad, but adjacent to it. As already documented in the nervous system, one of the functions of GAP43 is to sequester calmodulin molecules altering Ca2+/CaM homeostasis and thus modulating the numerous pathways triggered by this complex.24,25
Figure 5.

Schematic representation of a muscle fiber cross section from fish (A) and frog (B) cut at the Z-line level. At this level, the sarcoplasmic reticulum (black lines inside the fiber perimeter) is engaged in the formation of the triads together with the T-tubules. The images at the bottom C and D represent the projection of a longitudinal optical section of the same fibers (indicated by the green squares). The colored sarcoplasmic reticulum is the portion that is included into the optical section and actually visualized by the observer, the pink color represents RyR (centered in the triad) and the green color represents GAP43 (off-centered from the triad). The overlap between RyR and GAP43 signals is colored in yellow. The particular shape and disposition of the myofibrils in the fish (A) ensure that the sarcoplasmic reticulum delimiting the myofibrils mostly overlaps in the same direction of the laser beam (z).
It is hard to actually point out the GAP43 real function/s in the skeletal muscle. Considering its sub-cellular localization and ability to interact with calmodulin, GAP43 may play local functions involving trans-membrane signaling by means of calmodulin and/or calmodulin-binding proteins and may also play a key role in the dynamic handling of intracellular Ca2+.12 The presence of a calmodulin-binding domain in GAP43 structure suggests the hypothesis that, in skeletal muscle, GAP43 could sequester a large fraction of calmodulin to the sub-membranous regions and release free calmodulin in response to PKC activation or calcium increase. Recently, Stroffekova demonstrated, using skeletal muscle myotubes, that an intracellular Ca2+ increase triggered the association of calmodulin to its binding site on the DHPR mediating the Ca2+-dependent inhibition of DHPR currents.26 These findings were also supported by the evidence that endogenous calmodulin was primarily found to reside near the Z-line and about 25% of the calmodulin-immunoreactivity appeared at the level of the triads.27 In this scenario it is reasonable to speculate that GAP43, localized adjacently to the triads, could act as a buffer of calmodulin and play a role in RyR and/or DHPR modulation. In conclusion, data presented here demonstrated that GAP43 is expressed and localizes at, or very close to, the CRUs in skeletal muscle of lower vertebrates, but further investigations are required to detail and explain its physiological role. The recurrent pattern of GAP43 localization at CRUs in Xenopus and Zebrafish in addition to mammals including humans,7,12 suggests an involvement of the protein in the dynamic intracellular Ca2+ homeostasis, a common conserved role among the different species.
Acknowledgments
We thank Dr. Clara Franzini-Armstrong for encouragement and helpful advice. This work was supported by G. d’Annunzio University research funds to SG and MAM.
References
- 1.Skene JH. Axonal growth-associated proteins. Annu Rev Neurosci 1989;12:127-56. [DOI] [PubMed] [Google Scholar]
- 2.Skene JH, Virag I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J Cell Biol 1989;108:613-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander KA, Wakim BT, Doyle GS, Walsh KA, Storm DR. Identification and characterization of the calmodulin-binding domain of neuromodulin, a neurospecific calmodulin-binding protein. J Biol Chem 19885;263:7544-9. [PubMed] [Google Scholar]
- 4.LaBate ME, Skene JH. Selective conservation of GAP-43 structure in vertebrate evolution. Neuron 1989;3:299-310. [DOI] [PubMed] [Google Scholar]
- 5.Heuss D, Engelhardt A, Gobel H, Neundorfer B. Light-microscopic study of phosphoprotein B-50 in myopathies. Virchows Arch 1995;426:69-76. [DOI] [PubMed] [Google Scholar]
- 6.Heuss D, Engelhardt A, Lochmuller H, Gobel H, Neundorfer B. Expression of growth associated protein 43 and neural cell adhesion molecule in congenital fibre type disproportion with interstitial myositis. Virchows Arch 1994;425:101-5. [DOI] [PubMed] [Google Scholar]
- 7.Heuss D, Schlotzer-Schrehardt U. Subcellular localization of phosphoprotein B-50 in regenerating muscle. An immunoelectron microscopic study. Neurol Res 1998;20:360-4. [PubMed] [Google Scholar]
- 8.Stocker KM, Baizer L, Ciment G. Transient expression of GAP-43 in nonneuronal cells of the embryonic chicken limb. Dev Bio. 1992;149:406-14. [DOI] [PubMed] [Google Scholar]
- 9.Stocker KM, Ciment G, Baizer L. GAP-43 in non-neuronal cells of the embryonic chick limb: clues to function. Perspect Dev Neurobiol 1992;1:53-62. [PubMed] [Google Scholar]
- 10.Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J Biophys Biochem Cytol 1957;3:269-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flucher BE, Franzini-Armstrong C. Formation of junctions involved in excitation-contraction coupling in skeletal and cardiac muscle. Proc Natl Acad Sci USA 1996;93):8101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarnieri S, Morabito C, Paolini C, Boncompagni S, Pilla R, Fanò-Illic G, et al. Growth associated protein 43 is expressed in skeletal muscle fibers and is localized in proximity of mitochondria and calcium release units. PLoS One 2013;8:e53267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzini-Armstrong C. Veratti and beyond: structuralcontributions to the study of muscleactivation. Rend Fis Acc Lincei 2002;13: 289-323. [Google Scholar]
- 14.Franzini-Armstrong C, Boncompagni S. The evolution of the mitochondria-to-calcium release units relationship in vertebrate skeletal muscles. J Biomed Biotechnol 2011;2011: 830573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem 2007;40:101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate protokaryotype. Nature 2004;431:946-57. [DOI] [PubMed] [Google Scholar]
- 17.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 1999; 11:699-704. [DOI] [PubMed] [Google Scholar]
- 18.Biga PR, Goetz FW. Zebrafish and giant danio as models for muscle growth: determinate vs. indeterminate growth as determined by morphometric analysis. Am J Physiol Regul Integr Comp Physiol 2006;291:R1327-37. [DOI] [PubMed] [Google Scholar]
- 19.Pistocchi A, Gaudenzi G, Foglia E, Monteverde S, Moreno-Fortuny A, Pianca A, et al. Co nserved and divergent functions of Nfix in skeletal muscle development during vertebrate evolution. Development 2013;140:1528-36. [DOI] [PubMed] [Google Scholar]
- 20.Yamane H, Nishikawa A. Differential muscle regulatory factor gene expression between larval and adult myogenesis in the frog Xenopus laevis: adult myogenic cell-specific myf5 upregulation and its relation to the notochord suppression of adult muscle differentiation. In Vitro Cell Dev Biol Anim 2013;49: 524-36. [DOI] [PubMed] [Google Scholar]
- 21.Franzini-Armstrong C. Studies of the triad. IV. Structure of the junction in frog slow fibers. J Cell Biol 1973;56:120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzini-Armstrong C, Porter KR. Sarcolemmal Invaginations Constituting the T System in Fish Muscle Fibers. J Cell Biol 1964;22:675-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzini-Armstrong C, Protasi F, Ramesh V. Shape, size, and distribution of Ca(2+) release units and couplons in skeletal and cardiac muscles. Biophys J 1999;77:1528-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int Rev Cytol 2005;245:245-325. [DOI] [PubMed] [Google Scholar]
- 25.Slemmon JR, Feng B, Erhardt JA. Small proteins that modulate calmodulin-dependent signal transduction: effects of PEP-19, neuromodulin, and neurogranin on enzyme activation and cellular homeostasis. Mol Neurobiol 2000;22:99-113. [DOI] [PubMed] [Google Scholar]
- 26.Stroffekova K. Ca2+/CaM-dependent inactivation of the skeletal muscle L-type Ca2+ channel (Cav1.1). Pflugers Arch 2008;455: 873-84. [DOI] [PubMed] [Google Scholar]
- 27.Rodney GG. Calmodulin in adult mammalian skeletal muscle: localization and effect on sarcoplasmic reticulum Ca2+ release. Am J Physiol Cell Physiol 2008;294:C1288-97. [DOI] [PMC free article] [PubMed] [Google Scholar]


