Abstract
Phylogenetic systematics is heading for a renaissance where we shift from considering our phylogenetic estimates as a static image in a published paper and taxonomies as a hardcopy checklist to treating both the phylogenetic estimate and dynamic taxonomies as metadata for further analyses. The Open Tree of Life project (opentreeoflife.org) is developing synthesis tools for harnessing the power of phylogenetic inference and robust taxonomy to develop a synthetic tree of life. We capitalize on this approach to estimate a synthesis tree for the freshwater crayfish. The crayfish make an exceptional group to demonstrate the utility of the synthesis approach, as there recently have been a number of phylogenetic studies on the crayfishes along with a robust underlying taxonomic framework. Importantly, the crayfish have also been extensively assessed by an IUCN Red List team and therefore have accurate and up-to-date area and conservation status data available for analysis within a phylogenetic context. Here, we develop a synthesis phylogeny for the world's freshwater crayfish and examine the phylogenetic distribution of threat. We also estimate a molecular phylogeny based on all available GenBank crayfish sequences and use this tree to estimate divergence times and test for divergence rate variation. Finally, we conduct EDGE and HEDGE analyses and identify a number of species of freshwater crayfish of highest priority in conservation efforts.
Keywords: crayfish, conservation, EDGE, phylogenetic diversity
1. Introduction
The freshwater ecosystems represent only 0.8% of the Earth's surface, but house nearly 6% of all described species and are under severe pressure from multiple impacts, including: overexploitation, water pollution, flow modification, destruction or degradation of habitat, and invasion by exotic species [1]. Freshwater ecosystems in areas such as the southeastern United States house a highly diverse array of fauna that exhibits high levels of endemism [2]. The fragmented nature of these habitats both drives speciation (resulting in exceptional biodiversity) and results in high susceptibility to habitat destruction and limited dispersal capability. The combination of these factors promotes an accelerated extinction rate (on the order of 4% per decade) on par with extinction rates in tropical rainforests [3]. Thus, freshwater habitats are critical to biodiversity, but are at extremely high risk and therefore are in need of conservation efforts.
Crayfish are an important component of these endangered ecosystems and represent an opportunity to capitalize on our knowledge of their diversity (taxonomic and phylogenetic) to help assess relative conservation priorities for freshwater ecosystems as well as conservation priorities for these endangered species themselves [4,5]. Crayfish play a central ecological role in many freshwater ecosystems and provide an important economic and cultural role in many communities [6,7]. Indeed, they have been categorized as keystone species in stream communities both based on consumer activity [8] as well as directly through predation and indirectly through sediment bioturbation and increasing organic matter processing rates [9]. Even in terrestrial systems, the freshwater crayfish play a significant role as ecological engineers [10,11]. Unfortunately, both the freshwater ecosystems and the crayfish from around the world are under severe pressure and should be high priorities for conservation efforts.
Crayfish are a highly endangered component of these freshwater ecosystems with over 30% of the world's described species considered endangered and at risk of extinction [12]. The extant representatives are composed of over 600 species distributed taxonomically across three families (Parastacidae, Cambaridae and Astacidae) and 30 genera [13,14]. They are distributed in temperate areas across the globe and are on all continents except Antarctica and continental Africa (although there is an endemic genus in Madagascar). The centre of diversity for the Southern Hemisphere family, Parastacidae, is in southeast Australia and Tasmania; whereas, the centre of diversity for the species rich Cambaridae is in the southeastern United States [13]. Crayfish represent an excellent candidate for phylogenetic synthesis because of their robust underlying taxonomy [15,16] and extensive phylogenetic work across the group (table 1 and references therein). Importantly, they have also been the focus of a recent International Union for Conservation of Nature (IUCN) Red List assessment and have been thoroughly assessed using Red List criteria for endangerment [12].
Table 1.
Phylogenetic studies included in the synthesis tree. OTU, operational taxonomic unit.
| study | taxonomic level | OTUs |
|---|---|---|
| molecular phylogeny, this study | Astacidea | 387 |
| Bracken-Grissom et al. [17] | Astacidea | 66 |
| Ainscough et al. [18] | Fallicambarus | 119 |
| Breinholt et al. [19] | Cambarus | 93 |
| Pedraza-Lara et al. [20] | Cambarellinae | 77 |
| Toon et al. [21] | Parastacidae | 61 |
| Breinholt et al. [22] | Astacidea | 21 |
| Buhay & Crandall [23] | Cambarus | 47 |
| Schultz et al. [24] | Engaeus | 53 |
| Buhay & Crandall [25] | Orconectes | 69 |
| Buhay et al. [26] | Cambarus | 130 |
| Fratini et al. [27] | Austropotamobius | 61 |
| Rudolph & Crandall [28] | Virilastacus | 31 |
| Schull et al. [29] | Euastacus | 129 |
| Trontelj et al. [30] | Austropotamobius | 72 |
| Munasinghe et al. [31] | Cherax | 58 |
| Rode & Babcock [32] | Astacidea | 37 |
| Taylor & Hardman [33] | Orconectes | 24 |
| Crandall et al. [34] | Parastacidae | in Toon et al. [21] |
| Crandall et al. [35] | Astacidea | in Bracken-Grissom et al. [17] |
In this study, we bring together recent crayfish phylogenetic studies with this underlying taxonomic framework to estimate a synthesis tree for all the freshwater crayfish (taxonomy + phylogeny). With this synthesis tree, we map IUCN endangered species status and test for associations with phylogenetic clades and taxonomic groups. Additionally, we estimate a crayfish phylogram using GenBank sequence data from across the freshwater crayfish to obtain branch length estimates and anchor these with divergence time estimates calibrated with extensive fossil data [17]. Using the resulting chronogram, we estimate divergence and extinction rates across the freshwater crayfish. Finally, by combining this phylogenetic information with phylogenetic diversity (PD) calculations and endangered status, we identify crayfish species that are especially evolutionarily distinct and globally endangered (EDGE analysis) [36]. Therefore, our study effectively demonstrates the power of combining robust taxonomy with synthetic phylogeny to aid conservation assessment based on PD and endangerment assessment.
2. Material and methods
(a). Phylogenetic analyses and synthesis
(i). Synthetic tree estimation
Phylogenetic synthesis is the merging of multiple sources of phylogenetic information with an underlying taxonomy. Thus, the generation of synthetic trees differs from supertree approaches [37] both conceptually as well as practically. Supertrees treat multiple phylogenetic estimates as the ‘data’ for a new phylogenetic analysis resulting in the supertree without consideration of the underlying taxonomy. Synthetic trees are the graphical uniting of multiple estimates of phylogeny without re-estimation and therefore they do not suffer from signal enhancement, where novel relationships can appear in the supertree that are not present in the input source trees [38]. Instead, conflict can be visualized and traced back to the source trees without novel relationships being generated from conflicting source trees [39]. Because the synthetic tree approach uses a taxonomy as the underlying backbone structure, conflicts in taxonomy can also be quickly identified.
Published phylogenies representing 20 studies were uploaded as rooted Newick [40] files and stored in The Open Tree of Life Study Curator (http://tree.opentreeoflife.org/curator) (table 1). The Study Curator is a database that provides infrastructure to store phylogenies and all metadata from phylogeny studies (doi, title, year, etc.). It also provides a graphical user interface to map taxon names from uploaded source trees to a user-curated taxonomy. The Open Tree of Life project (opentreeoflife.org) has generated a new user-curated taxonomy for Arthropoda (OTT2.2) by combining and hand-curating public taxonomy databases such as the World Register of Marine Species (WoRMS) [41], GenBank and the Global Biodiversity Information Facility (GBIF). This includes removing species from the taxonomy that are the result of spelling errors. This taxonomy provides flexibility for taxon mapping, including mapping to higher taxonomic ranks if the species designation is missing or in conflict with the OTT. Once studies are uploaded to phylografter and curated, they are exported from phylografter as NeXML files [42]. The NeXML files are loaded into treemachine (https://github.com/OpenTreeOfLife/treemachine) to generate a graph database for all studies that were included in the synthesis, which also includes a NeXML of the taxonomy. The source trees and taxonomy are then merged into a tree alignment graph [39], according to a user pre-defined order, to generate a synthetic tree. Those taxa not represented by a source tree are represented by the taxonomy graph in the synthetic tree.
(ii). Crayfish phylogeny with branch lengths
In order to calculate PD measures, we require a tree with branch lengths. Phylogeny branch lengths are estimates of genetic change along the branch of a phylogeny, usually specified in units of expected number of substitutions per site. The synthesis tree does not include branch length information currently; therefore, we generated a crayfish phylogram using PHLAWD [43] to obtain a phylogeny with branch lengths that contains all species with sequence data from GenBank. We removed intraspecific sequences and tried to retain only species because the PHLAWD method does not distinguish between species and intraspecific variants. This includes those new species that authors of previous crayfish studies identified as potential species, but may not be formally described. The loci assembled included portions of the three mitochondrial genes (12S, 16S and cytochrome c oxidase subunit I (COI)) and portions of two nuclear genes (18S and 28S). Sequences for COI were checked for complete open reading frame and GenBank sequence identifiers that included ‘-like’ to prevent nuclear copies of the COI gene from being included [44]. All loci were aligned using MAFFT 7.130b [45], while poorly aligned regions were identified and removed using gBlocks [46] according to the least stringent settings. Two lobsters, Homarus americanus and Enoplometopus occidentalis, from two superfamilies within the Astacidea were used as outgroup taxa [17]. An optimal partitioning scheme and models of sequence evolution were determined using PartitionFinder v. 1.1.1 [47] according to the Bayesian Information Criterion (BIC) with the nucleotide alignment divided into seven subsets: 12S, 16S, 18S, 28S, COI 1st pos., COI 2nd pos. and COI 3rd pos. A maximum-likelihood (ML) phylogeny was estimated in GARLI 2.0 [48] according to the models and partitions identified in the PartitionFinder analysis. Multiple searches from random starting trees were conducted to ensure searches were not being trapped in local optima. Branch support was assessed using 100 non-parametric bootstrap replicates [49].
(iii). Crayfish chronogram
A chronogram was estimated using penalized likelihood in r8s [50] with the best ML tree. Node calibrations included six fossils used in previous crayfish chronogram estimation studies [21,22]. These six calibration points spanned from Mid Triassic (approx. 225 million years ago (Ma)) [51], through Late Jurassic (approx. 145 Ma) [52], Early Cretaceous (approx. 135 Ma) [53], to the Eocene (approx. 40 Ma) [54], providing a variety of calibration points throughout the phylogeny [55]. The optimal smoothing parameter was chosen based on a cross-validation procedure [56].
(b). Diversification rates through time
To determine whether this group radiated through time at a constant rate, we first assessed whether branching times fit a pure-birth Yule model using the Monte Carlo Constant Rates Test [57]. Studies have shown that this test is sensitive to non-random sampling and missing taxa [58–60]; therefore, we chose to account for missing taxa using birth–death chronogram simulations [58]. We simulated constant rate birth–death chronograms using CorSim [58] assuming 40% missing taxa based on those described species missing from the phylogeny. Furthermore, we assumed the youngest genus was 9.95 Ma old based on the chronogram estimate and the taxonomy used for the synthetic tree. The observed gamma statistic was compared with the null distribution of 1000 simulated birth–death trees using the APE module [61] in R [62].
To assess whether time-dependent speciation and extinction rates varied throughout the history of crayfish, we estimated diversification rate shifts using ML in TreePar [63]. This method moves across a chronogram in intervals and for each interval calculates birth and death, while accounting for missing taxa. At the end of a cycle, the largest change in rate is recorded with the likelihood of the model. This process is continued for additional cycles with each cycle adding one more diversification rate change to the model, while conditioning on the rate changes previously identified in earlier iterations. A χ2-test was used to compare alternative models with different numbers of rate changes with three d.f. We set the maximum number of diversification rate changes to four, while estimating a birth–death model every 1 Myr.
(c). Conservation status, phylogenetic diversity, and EDGE and HEDGE analyses
Crayfish conservation priorities were assigned first by designating IUCN Red List status to each species retrieved from the IUCN Red List of Threatened Species v. 2013.2 [12,64]. Next, we conducted an Evolutionarily Distinct, Globally Endangered (EDGE) analysis [36] and Heightened Evolutionary Distinctiveness and Globally Endangered (HEDGE) analysis [65]. The EDGE analysis ranks species according to their evolutionary distinctness by measuring the length of the branches leading to the tip taxa weighted by the number of descendants from each node, thereby calculating a probability that a species may go extinct. This calculation requires a probability of extinction which we assigned using a numerical designation associated with the IUCN Red List category [36,66]: Least Concern = 0.025, Near Threatened = 0.05, Vulnerable = 0.1, Endangered = 0.2 and Critically Endangered = 0.4. The HEDGE analysis ranks species based on their expected contribution to PD; therefore, the metric aims to preserve species that contribute the most PD. The HEDGE calculation is an extension of probabilistic PD where the probability of extinction can reach 0 [67,68]. Both EDGE and HEDGE metrics were estimated in Mesquite [69] using the Tuatara module [70].
In addition to estimating EDGE and HEDGE, we measured the PD of crayfish in each of eight terrestrial Freshwater Animal Diversity Assessment (FADA: http://fada.biodiversity.be) recognized ecozones [71] to examine broad geographical patterns of diversity and endangerment within the context of pre-established freshwater ecozones. We used the IUCN Red List [64] to gather range data and code the presence or absence of FADA ecozones for each of the 382 species in the chronogram. This information was used to estimate the phylogenetic species variability (PSV), which is a measure of PD that is independent of species richness [72]. PSV is a metric that evolves a hypothetical independent neutral trait forward in time along a phylogeny to quantify how shared evolutionary history decreases the variance of the hypothetical trait [72]. As applied here, we are examining the variance in branch lengths as a proxy of relatedness within FADA ecozones. It is important to note that we also estimate PSV variance because our phylogeny is not clocklike; therefore, there is variation around the single PSV value. We chose to use PSV as measure of diversity and relatedness due to the variability in the number of species across regions. PSV estimates can range from 0 to 1 with 0 indicating species within a FADA ecozone are closely related and 1 indicating species within a FADA ecozone are distantly related. We calculated PSV and Faith's PD [73] in the R package picante [74] using the chronogram without the outgroup; picante calculates PD for each FADA ecozone by summing the total length of all branches connecting species within a FADA zone on the unrooted phylogeny.
3. Results
(a). Phylogenetic synthesis
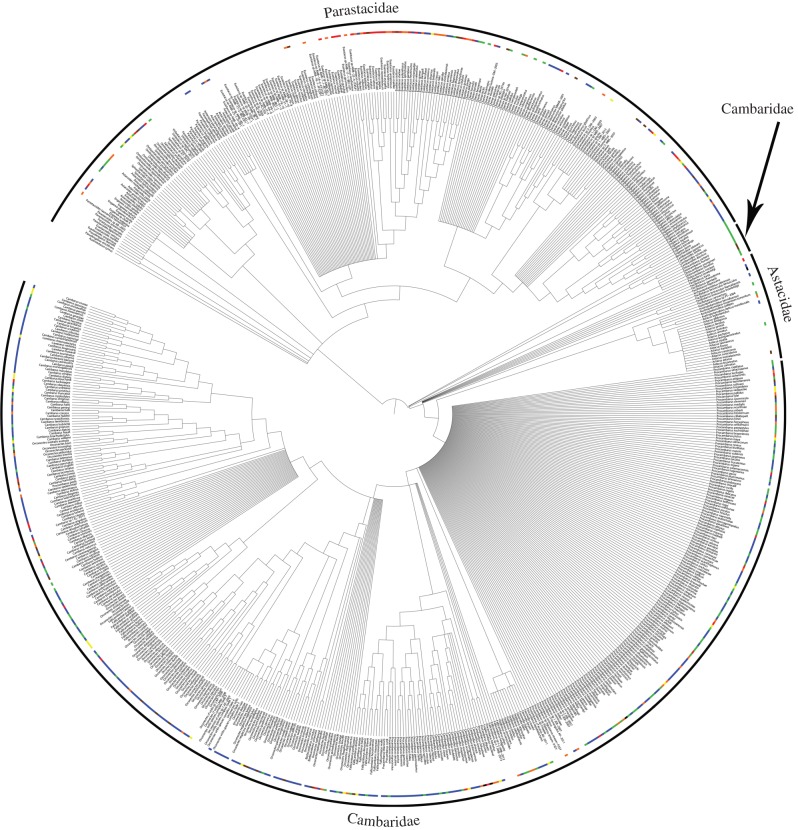
Construction of the synthetic tree and curation of the taxonomy yielded 719 terminal taxa (figure 1). The terminal taxa consist of all 590 described species of freshwater crayfish [12] and multiple representatives of species with unique GenBank identifiers from population level data. Of the 719 taxa, 387 (60% of all described species) are unique species represented with publicly available sequence data. The genus Procambarus is the largest genus with the lowest number of species with available sequence data (31 available/178 species described).
Figure 1.
Synthetic tree consisting of the studies in table 1 and a combined taxonomy including WoRMS, GBIF and National Center for Biotechnology Information taxonomies. Colours indicate those species assigned an IUCN Red List status: black, Extinct; red, Critically Endangered; orange, Endangered; brown, Vulnerable; yellow, Near Threatened; blue, Least Concern; and green, Data Deficient.
(b). Molecular phylogeny
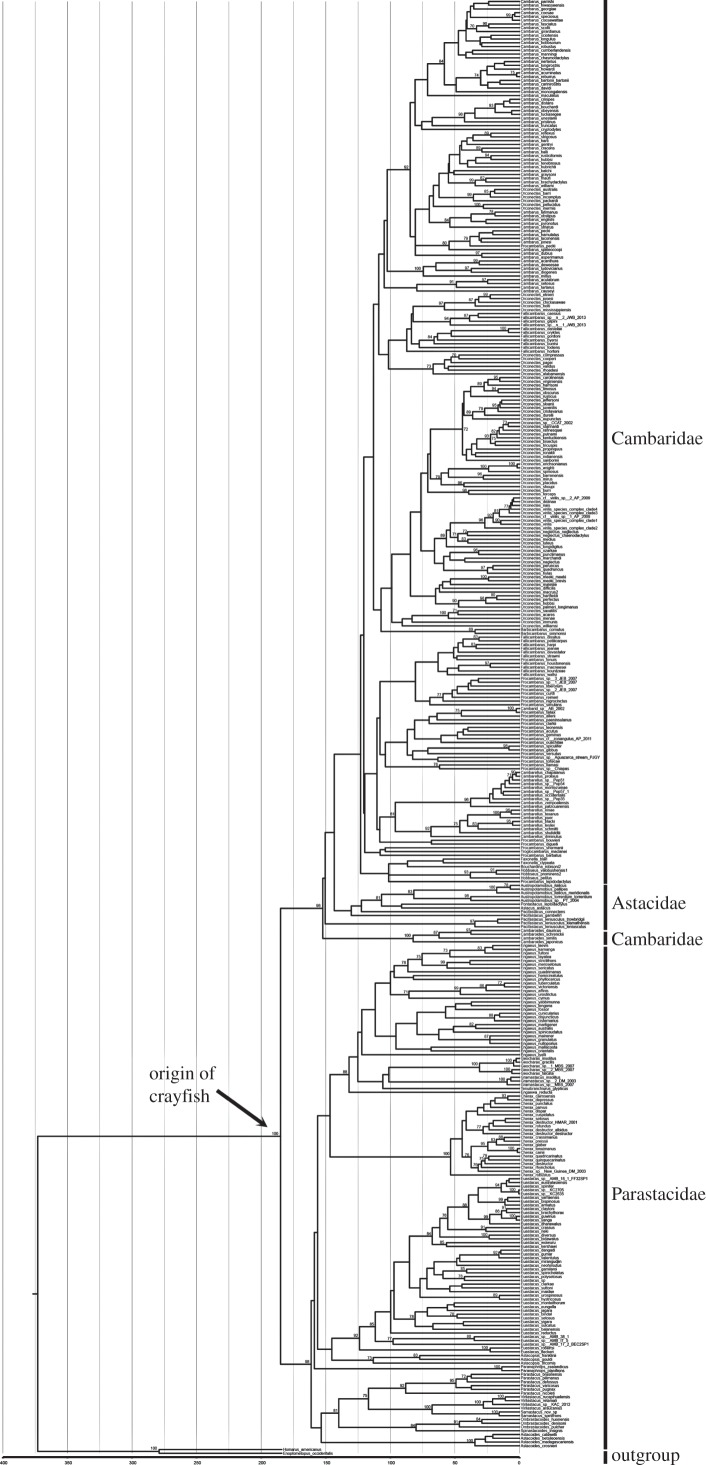
The final nucleotide alignment contained 5259 bp of nucleotides after gBlocks trimming. The BIC favoured each partition subset as the best partitioning scheme. The ML tree strongly supported sister clades consisting of the Northern Hemisphere and Southern Hemisphere taxa (bootstrap value (BS) = 100); however, branch support only favoured monophyly for one family (figure 2). Parastacidae is strongly supported (BS = 98), while Cambaridae and Astacidae are paraphyletic (figure 2). The monophyly of Southern Hemisphere genera is mostly supported, but no support is present for the relationships among the genera (figure 2). This is similar to the Northern Hemisphere genera, but in the Northern Hemisphere the genera containing many species (e.g. Orconectes, Procambarus, Cambarus) are not supported as being monophyletic (figure 2).
Figure 2.
Chronogram estimated in r8s using the ML phylogeny. Node calibrations identical to Breinholt et al. [22] and Toon et al. [21]. Bootstrap support greater than 70% from 100 non-parametric bootstrap replicates shown.
(c). Diversification rates through time
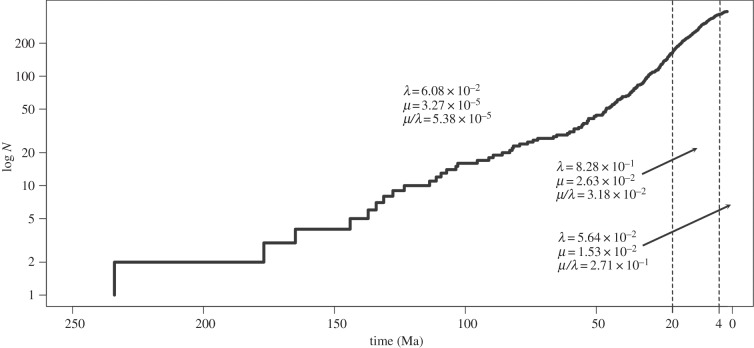
The Monte Carlo Constant Rates test rejected the Yule model with a single rate of diversification through time. A three-rate birth–death model fitted the branching times the best according to the χ2-test with changes in diversification rate occurring at 20 Ma and 4 Ma (figure 3). The death parameter increased towards the present, while the birth parameter decreases and then increases at the present. Finally, the turnover rate (death/birth) is the greatest for the rate nearest the present.
Figure 3.
Log-lineage through time plot of the ML chronogram. Vertical dashed lines indicate timing of the χ2 favoured changes in diversification rate. ML parameter estimates are given for each birth–death model.
(d). Conservation status, phylogenetic diversity, and EDGE and HEDGE analyses
The PD and PSV calculations of FADA ecozones differed when compared with one another owing to variation in species richness (table 2). The Nearctic region had the highest PD, while the Neotropical region had the highest PSV. The Australasian region, on the other hand, has a high PSV owing to the relatively long branch lengths. This indicates that the species in this region have fewer close relatives and represent a greater amount of evolutionary history and more distinct lineages. Although both measures predict different regions with the highest diversity, both estimate the Afrotropical region to be the least diverse (there is only the Malagasy genus with seven species). The low PSV can be attributed to shorter branches, thus being more closely related, ultimately lowering the PSV value. It should be noted that the PSV variance is the greatest for this group, shedding light on the fact that PSV is a mean and although they are closely related, there is branch length heterogeneity (non-clocklike) among the seven taxa.
Table 2.
Estimates of phylogenetic species variability (PSV), PSV variance and phylogenetic diversity (PD) using the molecular phylogeny with species grouped according to FADA ecozone.
| ecozone | PSV | PSV variance | PD |
|---|---|---|---|
| Australasian | 0.72 | 1.07 × 10−5 | 6025.27 |
| Afrotropical | 0.16 | 7.40 × 10−3 | 266.54 |
| Nearctic | 0.59 | 2.49 × 10−6 | 11026.91 |
| Neotropical | 0.74 | 2.19 × 10−4 | 1407.77 |
| Palearctic | 0.63 | 8.53 × 10−4 | 861.7 |
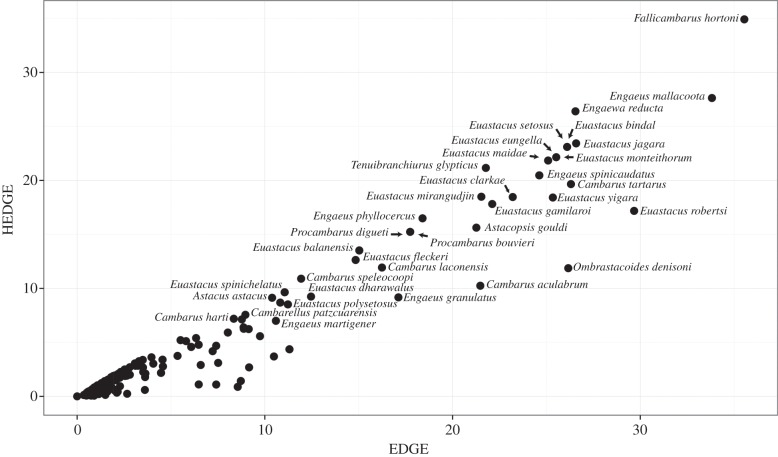
The EDGE and HEDGE scores were calculated for all species with IUCN values in our molecular phylogeny (figure 4). Calculations of EDGE ranged from 35.54 to 0.36, while calculations for HEDGE ranged from 34.91 to 0.12. Fallicambarus hortoni had the largest EDGE and HEDGE scores of all species. The 10 species with the highest EDGE and HEDGE scores include species that are critically endangered and endangered, with the bulk of these species being located in Australia (e.g. Engaeus sp., Engaewa sp., Euastacus sp.) plus a few North American cave species (e.g. Cambarus tartarus, C. laconensis, C. aculabrum).
Figure 4.
Bivariate plot of the largest EDGE and HEDGE scores for taxa included in the molecular phylogeny. Calculations were performed in the Mesquite module Tuatara.
4. Discussion
Recently, phylogenies have been used in combination with IUCN values and associated statistics to address conservation questions in order to focus species-based conservation efforts on preserving the greatest amount of PD [36,65,66,75]. Species that have few close relatives have a greater responsibility for evolutionary history [73]. Thus, the combination of phylogenetic information with endangered status allows for conservation biologists to make informed decisions about conservation priorities that incorporate threat and evolutionary history and processes [76]. Despite its predictive value, no comprehensive phylogeny has been proposed for the crayfish. Crayfish suffer from severe habitat loss, and as a result a large percentage of the species are endangered [12]. A phylogenetically informed conservation plan would benefit this group of organisms immensely to prioritize species-based conservation efforts and provide overarching protection for diverse freshwater ecosystems. Here we synthesize all taxonomic and phylogenetic information to define priorities given the current available information. Finally, we describe the limitations of findings and what is needed to achieve a robust framework to inform conservation priorities in the future.
(a). Taxonomy
Taxonomy is the foundation of conservation, and without sound taxonomy conservation priorities can become misleading [76–78]. Our phylogenetic analyses, in addition to previous published phylogenies, show crayfish taxonomy is in need of review and curation. For example, both our molecular phylogeny and synthetic tree confirm multiple genera are paraphyletic (figures 1 and 2). This is concordant with previous molecular studies of North American fauna where species are not monophyletic [20] and genera are not monophyletic [18]. In addition, our dataset only supports one of three families as monophyletic (figures 1 and 2).
Two issues may be causing the paraphyly and polyphyly of families and genera. First, traditional molecular loci used in crayfish systematics may not be the best choice to resolve the relationships being estimated (discussed in the molecular data section below). The second potential cause of conflict between taxonomy and molecular phylogeny may be that the morphological characters used to diagnose species are not informative about species relationships at higher taxonomic levels. The majority of taxonomic conflict occurs among the North American fauna. This group of crayfish forms a clade that most likely resulted from a recent rapid radiation, which is portrayed on a phylogeny as short branches near the terminal nodes (figure 2). Quick bursts of cladogenesis leave few morphological synapomorphies, making it difficult to establish relationships among species due to the short amount of time sister taxa were in isolation or gene flow was limited. We suggest a major reappraisal of North American crayfish taxonomy with the aid of a molecular phylogeny that capitalizes on loci from throughout the genome (see suggestions below).
The Southern Hemisphere taxonomy is not exempt from problems. The centre of diversity for the Southern Hemisphere is Australia, where the phylogeny consists of long terminal branches relative to the North American fauna [21]. The long terminal branches reflect low diversification rates in recent years, which may have resulted from the desiccation of Australia starting with the formation of the Antarctic Circumpolar Current during the Miocene and the formation of the Antarctic ice sheets and glacial cycles [79]. With the long terminal branches and increased time since speciation, the genera form monophyletic groups, unlike the Northern Hemisphere crayfish. However, published phylogenies have shown paraphyletic relationships for genera such as Euastacus [29] and Engaeus [24]. Although there is limited conflict between phylogeny and taxonomy of Southern Hemisphere taxa, most of these discrepancies represent undescribed species rather than true taxonomy conflict.
As part of this study, we revised the existing taxonomy that forms the basis of the synthetic tree. Although we removed non-recognized species names from the list of all species in use in morphological and molecular studies, a larger effort is needed to describe the undescribed material and formally re-describe higher taxa according to new molecular, highly supported, phylogenetic hypotheses. This level of curation will be difficult as the number of alpha taxonomists has declined greatly in the molecular and genomics era [80], but it is desperately needed for conservation efforts.
(b). Crayfish phylogeny
Our effort here to reconstruct the crayfish phylogeny with publically available sequence data shows the immense historical effort needed to reconstruct the evolutionary relationships. Despite this great effort, many generic and intergeneric relationships remain uncertain. This can be seen in our synthetic tree and supermatrix we assembled for this study (figures 1 and 2, respectively). We have sampled approximately 60% of all crayfish species. Unfortunately, from a conservation priority standpoint, our confidence in conservation priorities estimated in this study is limited by the percentage of sampled species. The EDGE and HEDGE calculations all rely on a fully resolved and sampled phylogeny with branch lengths. Our phylogeny contains 60% of the known species; therefore, missing 40% of the known species may drastically overestimate EDGE and HEDGE calculations owing to the missing taxa not shortening the edge lengths of terminal taxa. EDGE and HEDGE analyses have primarily focused on the mammals, where there are estimates of a complete phylogeny. Although the mammal phylogeny is nearly complete, researchers have dealt with missing taxa by placing them as best they can on the phylogeny [81]. While we can place these taxa according to taxonomy (and have done so in our synthetic tree, figure 1), we do not have branch length information for these data which is critical to the EDGE/HEDGE calculations. Furthermore, many of the genera in the Northern Hemisphere crayfish are paraphyletic, making the placement of species based on taxonomy suspect. Therefore, our EDGE and HEDGE estimates should be taken lightly until a more robust phylogeny and taxonomy are assembled (work in progress).
Traditionally in crayfish phylogenetics, five core genes are sequenced: COI, 12S, 16S, 18S and 28S [82]. Although providing a well-supported backbone for many groups (figure 2), this set of loci in combination has failed here to resolve many of the relationships within and among genera and families. We are at a time in molecular systematics where genomic data are being generated at substantial pace and systematics can begin to look across the genome for loci with a modest amount of work. Three techniques are currently being used to target single-copy nuclear genes across the nuclear genome: highly conserved regions [83], ultraconserved regions [84] and transcriptomes [85]. Each of these methods produces hundreds to thousands of loci informative across time scales. The crayfish phylogeny would benefit greatly from a study that targets one of these sources for loci to estimate and confirm the phylogenetic backbone and add support to the relationships within and among genera.
(c). Diversification rates and extinction
Recent advances in comparative methods have allowed researchers to estimate extinction from phylogenies [86], although some are sceptical [87]. Our diversification analyses show diversification rates were not constant through time (figure 3). In fact, the best-fit birth–death model for our data supports three different time-dependent divergence rates. The timing of changes in diversification are fairly congruent with the timing of the formation of the Antarctic Circumpolar Current and the formation of ice sheets [88] approximately 20 Ma, while the most recent change in diversification rate corresponds to recent glaciation in the Northern Hemisphere [89]. Future crayfish divergence time estimation should attempt estimation in a Bayesian framework. Unfortunately, getting convergence of parameter estimates for a large tree is difficult; therefore, we have relied on point estimates here as an approximation and recognize the variance associated with these dates.
Although we give a general picture of historical diversification and extinction here on a geological time scale, future studies can leverage a more complete tree and other comparative methods to obtain diversification and extinction rates on a local time scale. For example, new binary and multi-state character models of diversification rates exist [90,91]. We envision using these models in conjunction with environmental data layers to look at diversification and extinction rates of taxa associated with certain types of habitat and natural history characteristics. Analyses that combine ecology, morphology, geography and phylogeny can provide powerful correlative evidence with high extinction rates that are associated with particular characters.
Acknowledgements
We thank the Royal Society and the bioGENESIS working group for organizing and supporting the ‘Phylogeny, extinction risks and conservation’ workshop at the Royal Society. We are grateful to the George Washington University Colonial One high performance computing cluster for providing analysis capacity for this work. We thank Joseph Brown for his help with the synthesis analyses. We thank two anonymous reviewers and the associate editor for providing helpful comments to improve our manuscript.
Funding statement
This work was supported by the US NSF grant no. DEB 13-01820.
References
- 1.Dudgeon D, et al. 2006. Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. Camb. Philos. Soc. 81, 163–182. ( 10.1017/S1464793105006950) [DOI] [PubMed] [Google Scholar]
- 2.Lydeard C, Mayden RL. 1995. A diverse and endangered aquatic ecosystem of the Southeast United States. Conserv. Biol. 9, 800–805. ( 10.1046/j.1523-1739.1995.09040800.x) [DOI] [Google Scholar]
- 3.Ricciardi A, Rasmussen JB. 1999. Extinction rates in North American freshwater fauna. Conserv. Biol. 13, 1220–1222. ( 10.1046/j.1523-1739.1999.98380.x) [DOI] [Google Scholar]
- 4.Crandall KA. 1998. Conservation phylogenetics of Ozark crayfishes: assigning priorities for aquatic habitat protection. Biol. Conserv. 84, 107–117. ( 10.1016/S0006-3207(97)00112-2) [DOI] [Google Scholar]
- 5.Whiting AS, Lawler SH, Horwitz P, Crandall KA. 2000. Biogeographic regionalisation of Australia: assigning conservation priorities based on endemic freshwater crayfish phylogenetics. Anim. Conserv. 3, 155–163. ( 10.1111/j.1469-1795.2000.tb00240.x) [DOI] [Google Scholar]
- 6.Jones JPG, Andriahajaina FB, Hockley NJ, Crandall KA, Ravoahangimalala OR. 2007. The ecology and conservation status of Madagascar's endemic freshwater crayfish (Parastacidae: Astacoides). Freshw. Biol. 52, 1820–1833. ( 10.1111/j.1365-2427.2007.01766.x) [DOI] [Google Scholar]
- 7.Jones JPG, Andriahajaina FB, Hockley NJ, Balmford A, Ravoahangimalala OR. 2005. A multidisciplinary approach to assessing the sustainability of freshwater crayfish harvesting in Madagascar. Conserv. Biol. 19, 1863–1871. [Google Scholar]
- 8.Robert P, Creed J. 1994. Direct and indirect effects of crayfish grazing in a stream community. Ecology 75, 2091 ( 10.2307/1941613) [DOI] [Google Scholar]
- 9.Parkyn SM, Rabeni CF, Collier KJ. 1997. Effects of crayfish (Paranephrops planifrons: Parastacidae) on in-stream processes and benthic faunas: a density manipulation experiment. N Z J. Mar. Freshw. Res. 31, 685–692. ( 10.1080/00288330.1997.9516798) [DOI] [Google Scholar]
- 10.Richardson AMM. 1983. The effect of the burrows of a crayfish on the respiration of the surrounding soil. Soil Biol. Biochem. 15, 239–242. ( 10.1016/0038-0717(83)90065-2) [DOI] [Google Scholar]
- 11.Reynolds J, Souty-Grosset C, Richardson A. 2013. Ecological roles of crayfish in freshwater and terrestrial habitats. Freshw. Crayfish 19, 197–218. ( 10.5869/fc.2013.v19-2.197) [DOI] [Google Scholar]
- 12.Richman NI, et al. 2015. Multiple drivers of decline in the global status of freshwater crayfish (Decapoda: Astacidea). Phil. Trans. R. Soc. B 370, 20140060 ( 10.1098/rstb.2014.0060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crandall KA, Buhay JE. 2008. Global diversity of crayfish (Astacidae, Cambaridae, and Parastacidae—Decapoda) in freshwater. Hydrobiologia 595, 295–301. ( 10.1007/s10750-007-9120-3) [DOI] [Google Scholar]
- 14.De Grave S, et al. 2009. Classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zool. Suppl. 21, 1–109. [Google Scholar]
- 15.Hobbs HH., Jr 1989. An illustrated checklist of the American crayfishes (Decapoda: Astacidae, Cambaridae, and Parastacidae). Smithson. Contrib. Zool. 480, 1–236. ( 10.5479/si.00810282.480) [DOI] [Google Scholar]
- 16.Horwitz P. 1995. A preliminary key to the species of Decapoda (Crustacea: Malacostraca) found in Australian inland waters, pp. 1–69. Albury, Australia: Co-operative Research Centre for Freshwater Ecology. [Google Scholar]
- 17.Bracken-Grissom HD, et al. 2014. The emergence of the lobsters: phylogenetic relationships, morphological evolution and divergence time comparisons of an ancient group (Decapoda: Achelata, Astacidea, Glypheidea, Polychelida). Syst. Biol. 63, 457–479. ( 10.1093/sysbio/syu008) [DOI] [PubMed] [Google Scholar]
- 18.Ainscough BJ, Breinholt JW, Robison HW, Crandall KA. 2013. Molecular phylogenetics of the burrowing crayfish genus Fallicambarus (Decapoda: Cambaridae). Zool. Scr. 42, 306–316. ( 10.1111/zsc.12006) [DOI] [Google Scholar]
- 19.Breinholt JW, Porter ML, Crandall KA. 2012. Testing phylogenetic hypotheses of the subgenus of the freshwater crayfish genus Cambarus (Decapoda: Cambaridae). PLoS ONE 7, e46105 ( 10.1371/journal.pone.0046105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedraza-Lara C, Doadrio I, Breinholt JW, Crandall KA. 2012. Phylogeny and evolutionary patterns in the dwarf crayfish subfamily (Decapoda: Cambarellinae). PLoS ONE 7, e48233 ( 10.1371/journal.pone.0048233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toon A, Pérez-Losada M, Schweitzer CE, Feldmann RM, Carlson M, Crandall KA. 2010. Gondwanan radiation of the Southern Hemisphere crayfishes (Decapoda: Parastacidae): evidence from fossils and molecules. J. Biogeogr. 37, 2275–2290. ( 10.1111/j.1365-2699.2010.02374.x) [DOI] [Google Scholar]
- 22.Breinholt J, Perez-Losada M, Crandall KA. 2009. The timing of the diversification of the freshwater crayfishes. In Decapod crustacean phylogenetics (eds Martin JW, Crandall KA, Felder DL.), pp. 343–356. New York, NY: CRC Press, Taylor and Francis. [Google Scholar]
- 23.Buhay JE, Crandall KA. 2009. Taxonomic revision of cave crayfish in the genus Cambarus, subgenus Aviticambarus (Decapoda: Cambaridae) with descriptions of two new species, C. speleocoopi and C. laconensis, endemic to Alabama, USA. J. Crustacean Biol. 29, 121–134. ( 10.1651/08-3089.1) [DOI] [Google Scholar]
- 24.Schultz MB, Smith SA, Horwitz P, Richardson AMM, Crandall KA, Austin CM. 2009. Evolution underground: a molecular phylogenetic investigation of Australian burrowing freshwater crayfish (Decapoda: Parastacidae) with particular focus on Engaeus Erichson. Mol. Phylogenet. Evol. 50, 580–598. ( 10.1016/j.ympev.2008.11.025) [DOI] [PubMed] [Google Scholar]
- 25.Buhay JE, Crandall KA. 2008. Taxonomic revision of cave crayfishes in the genus Orconectes, subgenus Orconectes (Decapoda: Cambaridae) along the Cumberland Plateau, including a description of a new species, Orconectes barri. J. Crustacean Biol. 28, 57–67. ( 10.1651/07-2827R.1) [DOI] [Google Scholar]
- 26.Buhay JE, Moni G, Mann N, Crandall KA. 2007. Molecular taxonomy in the dark: evolutionary history, phylogeography, and diversity of cave crayfish in the subgenus Aviticambarus, genus Cambarus. Mol. Phylogenet. Evol. 42, 435–448. ( 10.1016/j.ympev.2006.07.014) [DOI] [PubMed] [Google Scholar]
- 27.Fratini S, Zaccara S, Barbaresi S, Grandjean F, Souty-Grosset C, Crosa G, Gherardi F. 2005. Phylogeography of the threatened crayfish (genus Austropotamobius) in Italy: implications for its taxonomy and conservation. Heredity 94, 108–118. ( 10.1038/sj.hdy.6800581) [DOI] [PubMed] [Google Scholar]
- 28.Rudolph EH, Crandall KA. 2012. A new species of burrowing crayfish, Virilastacus jarai (Crustacea, Decapoda, Parastacidae) from central-southern Chile. Proc. Biol. Soc. Wash. 125, 258–275. ( 10.2988/11-39.1) [DOI] [Google Scholar]
- 29.Shull HC, Pérez-Losada M, Blair D, Sewell K, Sinclair EA, Lawler S, Ponniah M, Crandall KA. 2005. Phylogeny and biogeography of the freshwater crayfish Euastacus (Decapoda: Parastacidae) based on nuclear and mitochondrial DNA. Mol. Phylogenet. Evol. 37, 249–263. ( 10.1016/j.ympev.2005.04.034) [DOI] [PubMed] [Google Scholar]
- 30.Trontelj P, Machino Y, Sket B. 2005. Phylogenetic and phylogeographic relationships in the crayfish genus Austropotamobius inferred from mitochondrial COI gene sequences. Mol. Phylogenet. Evol. 34, 212–226. ( 10.1016/j.ympev.2004.09.010) [DOI] [PubMed] [Google Scholar]
- 31.Munasinghe DHN, Murphy NP, Austin CM. 2003. Utility of mitochondrial DNA sequences from four gene regions for systematic studies of Australian freshwater crayfish of the genus Cherax (Decapoda: Parastacidae). J. Crustacean Biol. 23, 402–417. ( 10.1163/20021975-99990350) [DOI] [Google Scholar]
- 32.Rode AL, Babcock LE. 2003. Phylogeny of fossil and extant freshwater crayfish and some closely related Nephropid lobsters. J. Crustacean Biol. 23, 418–435. ( 10.1163/20021975-99990351) [DOI] [Google Scholar]
- 33.Taylor CA, Hardman M. 2002. Phylogenetics of the crayfish subgenus Crockerinus, genus Orconectes (Decapoda: Cambaridae), based on cytochrome oxidase I. J. Crustacean Biol. 22, 874–881. ( 10.1163/20021975-99990299) [DOI] [Google Scholar]
- 34.Crandall KA, Fetzner JW, Jr, Jara CG, Buckup L. 2000. On the phylogenetic positioning of the South American freshwater crayfish genera (Decapoda: Parastacidae). J. Crustacean Biol. 20, 530–540. ( 10.1163/20021975-99990069) [DOI] [Google Scholar]
- 35.Crandall KA, Harris DJ, Fetzner JW. 2000. The monophyletic origin of freshwater crayfishes estimated from nuclear and mitochondrial DNA sequences. Proc. R. Soc. Lond. B 267, 1679–1686. ( 10.1098/rspb.2000.1195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 ( 10.1371/journal.pone.0000296.g001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson MJ, Purvis A, Henze C. 1998. Phylogenetic supertrees: assembling the trees of life. Trends Ecol. Evol. 13, 105–109. ( 10.1016/S0169-5347(97)01242-1) [DOI] [PubMed] [Google Scholar]
- 38.Bininda-Emonds ORP. 2004. The evolution of supertrees. Trends Ecol. Evol. 19, 315–322. ( 10.1016/j.tree.2004.03.015) [DOI] [PubMed] [Google Scholar]
- 39.Smith SA, Brown JW, Hinchliff CE. 2013. Analyzing and synthesizing phylogenies using tree alignment graphs. PLoS Comput. Biol. 9, e1003223 ( 10.1371/journal.pcbi.1003223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardona G, Rosselló F, Valiente G. 2008. Extended Newick: it is time for a standard representation of phylogenetic networks. BMC Bioinform. 9, 532 ( 10.1186/1471-2105-9-532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Board WE. 2014. World Register of Marine Species. http://www.marinespecies.org (accessed 27 May 2014). [Google Scholar]
- 42.Vos RA, et al. 2012. NeXML: rich, extensible, and verifiable representation of comparative data and metadata. Syst. Biol. 61, 675–689. ( 10.1093/sysbio/sys025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SA, Beaulieu J, Donoghue MJ. 2009. Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol. Biol. 9, 37 ( 10.1186/1471-2148-9-37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song H, Buhay JE, Whiting MF, Crandall KA. 2008. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl Acad. Sci. USA 105, 13 486–13 491. ( 10.1073/pnas.0803076105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. ( 10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 47.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 48.Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, The University of Texas, Austin, TX. [Google Scholar]
- 49.Felsenstein J. 1985. Confidence limits on phylogenies with a molecular clock. Syst. Zool. 34, 152–161. ( 10.2307/2413323) [DOI] [Google Scholar]
- 50.Sanderson MJ. 2003. R8S: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302. ( 10.1093/bioinformatics/19.2.301) [DOI] [PubMed] [Google Scholar]
- 51.Amati L, Feldmann RM, Zonneveld J-P. 2004. A new family of Triassic lobsters (Decapoda: Astacidea) from British Columbia and its phylogenetic context. J. Paleontol. 78, 150–168. () [DOI] [Google Scholar]
- 52.Van Straelen V. 1928. On a fossil freshwater crayfish from eastern Mongolia. Bull. Geol. Soc. China 7, 173–178. [Google Scholar]
- 53.Martin AJ, Rich TH, Poore GCB, Schultz MB, Austin CM, Kool L, Vickers-Rich P. 2008. Fossil evidence in Australia for oldest known freshwater crayfish of Gondwana. Gondwana Res. 14, 287–296. ( 10.1016/j.gr.2008.01.002) [DOI] [Google Scholar]
- 54.Feldmann RM, Grande L, Birkhimer CP, Hannibal JT, McCoy DL. 1981. Decapod fauna of the Green River formation (Eocene) of Wyoming. J. Paleontol. 55, 788–799. [Google Scholar]
- 55.Parham JF, et al. 2012. Best practices for justifying fossil calibrations. Syst. Biol. 61, 346–359. ( 10.1093/sysbio/syr107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109. ( 10.1093/oxfordjournals.molbev.a003974) [DOI] [PubMed] [Google Scholar]
- 57.Pybus OG, Harvey PH. 2000. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc. R. Soc. Lond. B 267, 2267–2272. ( 10.1098/rspb.2000.1278) [DOI] [Google Scholar]
- 58.Cusimano N, Stadler T, Renner SS. 2012. A new method for handling missing species in diversification analysis applicable to randomly or nonrandomly sampled phylogenies. Syst. Biol. 61, 785–792. ( 10.1093/sysbio/sys031) [DOI] [PubMed] [Google Scholar]
- 59.Brock CD, Harmon LJ, Alfaro ME. 2011. Testing for temporal variation in diversification rates when sampling is incomplete and nonrandom. Syst. Biol. 60, 410–419. ( 10.1093/sysbio/syr007) [DOI] [PubMed] [Google Scholar]
- 60.Hohna S, Stadler T, Ronquist F, Britton T. 2011. Inferring speciation and extinction rates under different sampling schemes. Mol. Biol. Evol. 28, 2577–2589. ( 10.1093/molbev/msr095) [DOI] [PubMed] [Google Scholar]
- 61.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 62.R Core Team. 2013. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 63.Stadler T. 2011. Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl Acad. Sci. USA 108, 6187–6192. ( 10.1073/pnas.1016876108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.IUCN. 2013. IUCN Red List of Threatened Species. v. 2013.2 Gland, Switzerland: IUCN. [Google Scholar]
- 65.Steel M, Mimoto A, Mooers AO. 2007. Hedging our bets: the expected contribution of species to future phylogenetic diversity. Evol. Bioinf. 2, 237–244. [PMC free article] [PubMed] [Google Scholar]
- 66.Mooers AO, Faith DP, Maddison WP. 2008. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE 3, e3700 ( 10.1371/journal.pone.0003700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faith DP. 2008. Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 22, 1461–1470. ( 10.1111/j.1523-1739.2008.01068.x) [DOI] [PubMed] [Google Scholar]
- 68.Witting L, Loeschcke V. 1995. The optimization of biodiversity conservation. Biol. Conserv. 71, 205–207. ( 10.1016/0006-3207(94)00041-N) [DOI] [Google Scholar]
- 69.Maddison WP, Maddison DR. 2011. Mesquite: a modular system for evolutionary analysis. Version 3.01. http://mesquiteproject.org. [Google Scholar]
- 70.Maddison WP, Mooers AØ. 2007. Tuatara: conservation priority in a phylogenetic context. v. 1.0. http://mesquiteproject.org/packages/tuatara.
- 71.Balian EV, Segers H, Leveque C, Martens K. 2008. An introduction to the freshwater animal diversity assessment (FADA) project. Hydrobiologia 595, 3–8. ( 10.1007/s10750-007-9235-6) [DOI] [Google Scholar]
- 72.Helmus MR, Bland TJ, Williams CK, Ives AR. 2007. Phylogenetic measures of biodiversity. Am. Nat. 163, E68–E83. ( 10.1086/511334) [DOI] [PubMed] [Google Scholar]
- 73.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 74.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 75.Redding DW, Mooers AO. 2006. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678. ( 10.1111/j.1523-1739.2006.00555.x) [DOI] [PubMed] [Google Scholar]
- 76.Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. 2000. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295. ( 10.1016/S0169-5347(00)01876-0) [DOI] [PubMed] [Google Scholar]
- 77.Mace GM. 2004. The role of taxonomy in species conservation. Phil. Trans. R. Soc. Lond. B 359, 711–719. ( 10.1098/rstb.2003.1454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sites JW, Crandall KA. 1997. Testing species boundaries in biodiversity studies. Conserv. Biol. 11, 1289–1297. ( 10.1046/j.1523-1739.1997.96254.x) [DOI] [Google Scholar]
- 79.Crisp MD, Cook LG. 2007. A congruent molecular signature of vicariance across multiple plant lineages. Mol. Phylogenet. Evol. 43, 1106–1117. ( 10.1016/j.ympev.2007.02.030) [DOI] [PubMed] [Google Scholar]
- 80.Hopkins GW, Freckleton RP. 2002. Declines in the numbers of amateur and professional taxonomists: implications for conservation. Anim. Conserv. 5, 245–249. ( 10.1017/S1367943002002299) [DOI] [Google Scholar]
- 81.May-Collado LJ, Agnarsson I. 2011. Phylogenetic analysis of conservation priorities for aquatic mammals and their terrestrial relatives, with a comparison of methods. PLoS ONE 6, e22562 ( 10.1371/journal.pone.0022562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sinclair EA, Fetzner JW, Buhay J, Crandall KA. 2004. Proposal to complete a phylogenetic taxonomy and systematic revision for freshwater crayfish (Astacidea). Freshw. Crayfish 14, 21–29. [Google Scholar]
- 83.Lemmon AR, Emme SA, Lemmon EM. 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61, 727–744. ( 10.1093/sysbio/sys049) [DOI] [PubMed] [Google Scholar]
- 84.Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. ( 10.1093/sysbio/sys004) [DOI] [PubMed] [Google Scholar]
- 85.Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749. ( 10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- 86.Slater GJ, Harmon LJ. 2013. Unifying fossils and phylogenies for comparative analyses of diversification of trait evolution. Methods Ecol. Evol. 4, 699–702. ( 10.1111/2041-210X.12091) [DOI] [Google Scholar]
- 87.Rabosky DL. 2009. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824. ( 10.1111/j.1558-5646.2009.00926.x) [DOI] [PubMed] [Google Scholar]
- 88.McGowran B, Holdgate GR, Li Q, Gallagher SJ. 2004. Cenozoic stratigraphic succession in southeastern Australia. Aust. J. Earth Sci. 51, 459–496. ( 10.1111/j.1400-0952.2004.01078.x) [DOI] [Google Scholar]
- 89.Raymo ME. 1994. The initiation of Northern Hemisphere glaciation. Annu. Rev. Earth Planet. Sci. 22, 353–383. ( 10.1146/annurev.ea.22.050194.002033) [DOI] [Google Scholar]
- 90.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 91.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. ( 10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]