Abstract
We describe the integration of an actively controlled programmable microfluidic sample processor with on-chip optical fluorescence detection to create a single, hybrid sensor system. An array of lifting gate microvalves (automaton) is fabricated with soft lithography, which is reconfigurably joined to a liquid-core, anti-resonant reflecting optical waveguide (ARROW) silicon chip fabricated with conventional microfabrication. In the automaton, various sample handling steps such as mixing, transporting, splitting, isolating, and storing are achieved rapidly and precisely to detect viral nucleic acid targets, while the optofluidic chip provides single particle detection sensitivity using integrated optics. Specifically, an assay for detection of viral nucleic acid targets is implemented. Labeled target nucleic acids are first captured and isolated on magnetic microbeads in the automaton, followed by optical detection of single beads on the ARROW chip. The combination of automated microfluidic sample preparation and highly sensitive optical detection opens possibilities for portable instruments for point-of-use analysis of minute, low concentration biological samples.
I. INTRODUCTION
Chip-scale biosensors have attracted a lot of attention because they allow for highly sensitive and specific determination of a target substance by way of a miniaturized electrical or optical transduction mechanism.1–3 This approach is the foundation of a new generation of instruments that are optimized for analysis at low concentrations and/or small volumes; capabilities that become increasingly important for molecular analysis of nucleic acids and proteins, single-cell studies, and point-of-care or point-of-use approaches to personalize medicine.4–6 The development of labs-on-chip originally focused predominantly on integration of the fluidic aspects and handling of small analyte volumes, while optical readouts remained bulky. Recently, optofluidics has emerged as a field that combines microfluidics and integrated optics in a single chip-scale system.7–9 In particular, the incorporation of on-chip excitation and collection of the optical signal via the use of optical waveguides can provide a path towards a more completely integrated analysis system. Unfortunately, the requirements and approaches for materials and designs that optimize biological sample preparation and optical detection, respectively, are to a large extent incompatible. This makes it difficult to carry out both tasks on a single chip.
Recent works have focused on optimizing particular portions of a microfluidic/optofluidic device or have developed sensitive assays for detection of biomolecules. For example, groups have worked on integrating on-chip micro-lenses—either through laser ablation10 or by incorporation of microspheres11—for easier excitation and parallelization of measurements. Multiple detection modalities have been integrated into Microsystems for diverse applications. Examples include label-free RNA profiling with plasmonic hot spots in gold nanoparticles,12 waveguide-based glucose sensing via evanescent absorption,13 and fluorescence-based optofluidic flow cytometry.14,15 In parallel to microfluidic optimization, studies have also focused on using microfluidic technologies for nucleic acid isolation and detection. Microfluidic assays include nonspecific silica based nucleic acid adsorption,16 antibody mediated nucleic acid isolation,17 and bead-based sequence specific hybridization extraction.18
Optimization of assays and microfluidic/optofluidic functionality simultaneously is possible through hybrid integration in which a sample handling and a sensing layer are designed independently in terms of materials and functionality. Recently, hybrid systems combining PDMS (polydimethylsiloxane)-based microfluidic layers with a liquid-core waveguide optical detection layer have been reported.19,20 Optofluidic chips based on antiresonant reflecting optical waveguides (ARROWs) are ideal for such integration due to their high fluorescence sensitivity and planar optical layout. These devices are based on a series of alternating dielectric layers that allow for interference based guidance of leaky modes in low refractive index, liquid-core waveguides.21–23 While these devices illustrated the basic principle of hybrid integration, they were significantly limited in their functionality as they were operated purely passively, i.e., fluid was moved continuously through the device using a single, fixed pressure provided by an external syringe pump.
We present a significant advance to optofluidic integration by introducing a hybrid system that combines an actively controlled microfluidic layer with an optical sensing chip with single particle fluorescence detection sensitivity. Active control is achieved by employing an interconnected array of nanoliter microvalves called an automaton. This technology allows for complex sample processing steps and reconfigurable operation that can implement multiple functions in sequence via customized software control. The optofluidic chip, on the other hand, is able to detect fluorescence from single biomolecules.19,23,24 We demonstrate the functional integration of the microfluidic automaton with an optofluidic chip and the implementation of sample preparation steps required for nucleic acid analysis used in genomic studies and disease detection based on single particle fluorescence detection in flow. Specifically, we show on-chip mixing of multiple fluorescent agents as well as sequence-specific solid-phase extraction and detection of synthetic viral nucleic acid targets.
II. MATERIALS AND METHODS
A. Microfluidic automaton fabrication and parameters
Fig. 1(a) shows the integration of the active microfluidic fluid handling (automaton) layer and the on-chip optical detection (ARROW chip) layer. Lifting gate microvalves and pumps are the core components of the programmable microfluidic automaton portion of the system.25 Each automaton is composed of three layers: a substrate, fluidic, and pneumatic layer. Both fluidic and pneumatic layers are made of PDMS, which were molded using conventional SU8 lithography.26,27 In the case of the fluidic layer, a deformable membrane containing fluidic patterns was molded at a thickness of 300 μm and a channel depth of 100 μm by spin-coating the PDMS mixture (Sylgard 184, Dow Corning) at 350 rpm on the SU-8 2025 (MicroChem) master.28 For the pneumatic layer, a 3 mm thick PDMS replica was created on the SU8 master mold by pouring and subsequent degassing. Once baked for 2 h at 60 °C, the pneumatic layer was peeled from the master. Inlets/outlets (2 mm diameter) were punched, followed by alignment and bonding to the top of the fluidic layer by oxygen plasma activation (30 W for 30 s). A UV-ozone treatment was used to reversibly bond the assembled two-layer PDMS device to the substrate glass slide (see supplementary material29 for cross-sectional view).30 Prior to use, the automaton was filled with 10 mg/ml bovine serum albumin (BSA) and incubated for 15 min in order to minimize nonspecific adsorption to the channel walls.
FIG. 1.
Hybrid integration of automaton and ARROW chips principle and implementation. (a) Schematic of hybrid optofluidic device and connections and (b) photograph of assembled device. The white scale bar in (b) represents 1 cm.
For external pressure control of the valves, the automaton was connected to a control box via a custom acrylic manifold consisting of two clamping pieces of plastic that made conformal connections to the pneumatic and substrate layers of the automaton. Furthermore, 3 mm diameter air ports (designed to be aligned with pneumatic layer exits) were available for press fit connection to 3 mm outer diameter (OD) tygon tubing (EW-95702-01, Cole Palmer). This tubing was in turn connected to solenoid valves (S070M-6DC-32, ADI), which were electrically controlled via a switchboard (ULN2803, Elexol), data acquisition board (NI USB 6501, National Instruments), and a custom Labview program.31 A simple mixing program has been included as supplementary material to demonstrate automaton actuation.29
B. Optofluidic chip fabrication and parameters
The optofluidic chips are based on liquid-core ARROWs fabricated using established microfabrication techniques.21,32 More specifically, these chips rely on the intersection of solid and liquid core waveguides to form an orthogonal fluorescence detection scenario (see Fig. 1).33 These optofluidic devices have been shown to provide single biomolecule fluorescence sensitivity.19,24,33 Recent correlated optical and electrical detection of single nanospheres and virus particles has proven the single particle nature of ARROW chip detection.34 The planarity of the ARROW chip allows for easy vertical integration of microfluidic processors, a scenario that still allows for the detection of single nucleic acids.19 The present device was created by depositing alternating dielectric layers of silicon dioxide (SiO2, refractive index n = 1.47) and tantalum oxide (n = 2.107) on a silicon wafer to ensure optical guiding in the low-index liquid core. The thicknesses of the cladding layers, starting with SiO2 in order of deposition, were 265/102/265/102/265/102 nm, designed for broadband low-loss transmission between 400 and 800 nm.35 Liquid-core waveguides were formed on top of the layers using SU8 sacrificial layers with dimensions of 5 × 12 μm (height × width) using self-aligned pedestals36 and a single layer over coating oxide with transmission optimization.37,38 After the single 6 μm thick over coating SiO2 film was deposited, solid-core ridge waveguides were formed using reactive ion etching. Solid-core waveguides were etched to 4 μm widths and 3 μm ridge heights (see supplementary material29 for cross-sectional view). The orthogonal excitation solid-core waveguide and liquid-core waveguide define an optical excitation volume of 100 fl. Finally, the SU8 sacrificial core was removed using optimized piranha etch procedures to create the hollow liquid core waveguide channel.39
C. Integration of microfluidic and optofluidic layers
The ARROW optofluidic chip and microfluidic automaton were connected as follows: first, a cylindrical brass connector of 2 mm OD was glued to the ARROW liquid-core waveguide chip inlet. A stretchable 1.5 mm inner diameter (ID) piece of tygon tubing was then placed around the connector. Finally, a 2 mm OD, 130 μm ID tygon tubing was press fit into the tygon adapting tubing at one end and the 2 mm punched PDMS outlet of the automaton at the other, creating a perfect seal, as seen in Fig. 1(b). Direct integration by attaching the automaton to ARROW chip connectors is also feasible.19 This setup provides full reconfigurability (i.e., fluidic and optical chips can be swapped out for different experimental tasks) while adding only a small amount of dead volume on the order of 5 μl.
D. Optical detection setups
Fluorescence detection was performed using orthogonal excitation and collection in the chip plane (see Fig. 1(a)).33 In detail, fiber coupled laser light from a 632.8 nm HeNe and 488 nm Ar-ion laser was butt coupled to the ARROW solid-core excitation waveguide using single mode fiber. Upon the generation of fluorescence inside the liquid-core ARROW waveguide, light propagated orthogonally through the liquid-core waveguide to the collection solid-core waveguide. The light was collected from the waveguide by a microscope objective, spectrally separated by a dichroic mirror (3RD640LP, 3rd Millenium), filtered for background (blue and red fluorescence filtered by 513BP17, Semrock and 660BP20, Omega, respectively), and sent to single photon avalanche photodiodes (SPAPDs).
III. RESULTS AND DISCUSSION
We first demonstrate functional connections and fluidic control between these two components with very dissimilar fluidic resistances (see Fig. 1). To this end, an inlet of the automaton was filled with a solution containing 1 μm Tetraspeck fluorescent spheres (diluted to 1 × 108 particles/ml), which was in turn introduced into the hybrid system via automaton pump actuation.40,41 Briefly, any three connected microvalves in the array formed a standard microfluidic pump, where sequential activation moves fluid along the channel, creating pressure. Adding a fourth microvalve creates a loop, enabling circular mixing (for detailed pumping explanation, see supplementary material29). Alternating forward and reverse automaton pumping programs (20 s in length, 600 ms/pump cycle) repeatedly moved fluorescent microbeads through the optical excitation volume as illustrated in Fig. 2(a).
FIG. 2.
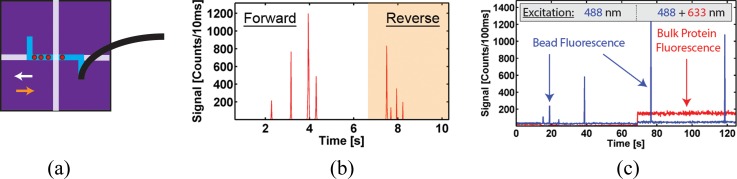
Basic functionality: sealed connection and particle mixing. (a) Bead movement inside the ARROW chip as actuated by automaton with color coded directions of forward (white) and reverse (orange) pumping; (b) corresponding fluorescence signal showing four microbeads repeatedly being moved past excitation spot; and (c) simultaneous detection of fluorescent beads and dye-labeled proteins that were mixed in automaton layer. The different laser excitation wavelengths are indicated; the HeNe laser used to excite the proteins is turned on at t = 70 s.
As the particles move through the excitation volume, they produce characteristic fluorescence signals such as those seen in Fig. 2(b). Upon reversing the pumping direction, the same particles are redetected as confirmed by direct, top-down observation using a microscope. The reverse signal has different respective peak heights due to Brownian motion of the beads and the resulting position change in the excitation volume.24,42 This experiment clearly demonstrates the ability of the hybrid optofluidic system to controllably transfer analytes between the interconnected chips with their drastically different fluidic impedances.
The next level of functional complexity is to implement dynamic sample preparation capabilities in the automaton layer, followed by optical detection on the single particle level in the ARROW chip. To this end, mixing functionality—required for important reactions such as labeling, lysing, and dilution—was shown by filling separate inlets of the automaton with Dylight 633 nm labeled antibody (diluted to 50 nM, Thermo) and streptavidin coated Dragon Green (excitation 480 nm) 220 nm diameter polystyrene spheres (diluted to 1 × 106 particles/ml, Bangs Labs). These solutions were then mixed by pre-programmed actuation sequences of the microvalve array, which are described in more detail in the supplementary material.29 The mixing process here is dependent on both convective and diffusive mass transport, whereas straight channels rely only on the latter.31 This enables faster mixing with identical efficiency as that of an off-chip experiment.43 Once the solution was pumped into the ARROW device, an external vacuum—attached to the exit connector of the ARROW chip—was used to drive the flow, resulting in the data traces seen in Fig. 2(c). For approximately 70 s, the sample was excited solely with 488 nm light. In this portion of the trace, only single particle events are detected in the blue fluorescence trace. After 70 s, excitation light was incident from both HeNe and Argon lasers (633 and 488 nm) resulting in simultaneous bulk fluorescence (due to the constant presence of multiple fluorescent antibodies in the excitation volume at any given time) and peaks (representing single nanospheres) in the red and blue fluorescence traces, respectively. Once control, mixing, and distribution had been demonstrated, a more complex biological assay for sequence-specific nucleic acid extraction and detection was developed that takes full advantage of the active fluid control.
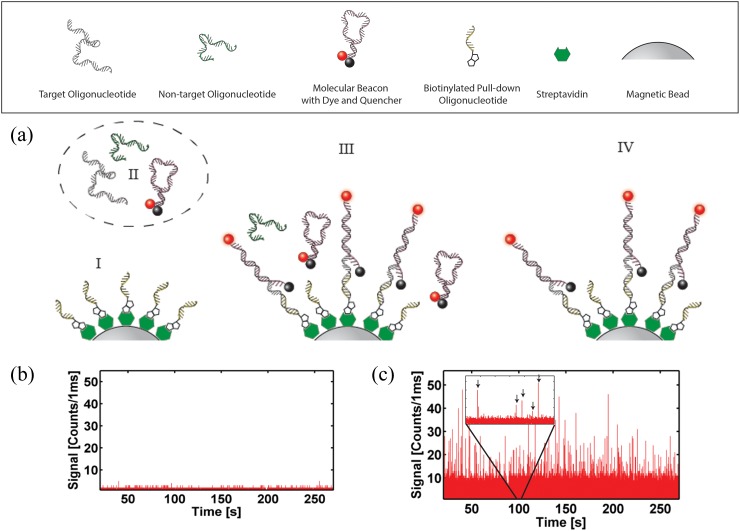
The nucleic acid of interest was chosen here to be a protein-coding region of Zaire Ebola virus (GeneBank ID AY354458.1). This target region was chosen to be specific to the virus and of synthesizable length (nt. 6832–6931, see supplementary material29 for sequence). A hairpin molecular beacon44 was designed to recognize a portion of the target nucleotide (nt. 6900–6923 5′-/TYE665/CGCATGGGCCTTCTGGGAAACTAAAAAATGCG/IAbRQSp/-3′). The beacon was designed to have thermodynamically higher stability when bound to the target oligonucleotide (simulated melting temperature from target Tm,t = 61.6 °C vs. intramolecular hairpin formation of Tm,h = 42.6 °C). Finally, a biotinylated “pull-down” oligonucleotide was designed to hybridize to a different portion of the target sequence (nt. 6832–6846, 5′-/BiotinTEG/AAAGGAGCAATACCA-3′) and function as a means of separating target oligonucleotide from complex sample mixtures. All synthetic oligonucleotides were purchased from Integrated DNA Technologies.
In preparation for the on-chip assay, pull-down magnetic beads were created.45,46 It is noted that off-chip, amplification free imaging of captured nucleic acids has been shown on magnetic beads.47 An aliquot (0.1 mg) of MyOne T1 streptavidin coated magnetic beads (Invitrogen) was washed three times with 1 × T50 (10 mM Tris-HCl pH 8.0, 50 mM NaCl) and incubated with 100 pmol of pull-down oligonucleotide for 2 h at room temperature on a rotary mixer. Next, a magnet was used to retain the magnetic beads during a 4× washing cycle with 1 × T50, after which the sample was resuspended in 100 μl of 1 × T50 (with 2 mM sodium azide) and stored for later use. Fig. 3(a) shows the complete sequence of the strain-specific solid-phase extraction protocol to label and isolate the target viral nucleic acids. Part I of the figure illustrates the magnetic beads coated with the pull-down sequences. In a second preparation step, beacon and target oligonucleotides were hybridized at a concentration of 1 μM by heating to 95 °C for 5 min in a dry bath (MyBlock), followed by subsequent cooling to room temperature (Fig. 3(a), part II).
FIG. 3.
Advanced functionality: viral nucleic acid detection using on-chip solid-phase extraction and detection. (a) Solid phase nucleic acid pull-down assay—(I) prepared “pull-down” magnetic beads and (II) nucleic acid target are mixed on automaton (III); washed magnetic particles are sent to the ARROW chip (IV). Particle detection for (b) negative (pull-down non-cognate)—see I—and (c) positive (pull-down cognate) nucleic acid samples—see IV. Single bead signals are marked by arrows in the figure inset.
On-chip, the assay was conducted as illustrated in Fig. 3(a) (steps III and IV). First, three inlets of the automaton were filled with the pull-down bead solution (∼1 × 107 beads/ml), oligonucleotide mixture (target or non-target oligonucleotide with molecular beacon, each at a concentration of 1 μM), and 1 × T50 buffer, respectively. Each valve actuation (∼125 nl) then corresponds to dispensation of about 1250 microspheres or 125 fmol of each nucleic acid. Single valve volumes of the pull-down and beacon-target solutions were circularly mixed for 30 s in the microvalve array (step III, see supplementary material29 for details). The solution was then pumped to a separate valve under which a magnet was fixed and was allowed to rest for 10 s in order to pull down the beads along with any bead-hybridized beacon-target complexes. Buffer was then used to wash the automaton to remove any unbound and potential carryover materials, sending them to the waste outlet. The retained solution containing only labeled target nucleic acids is shown in part IV of Fig. 3(a). This entire cycle takes 60 s for processing. It is possible to detect bead signal from a single program cycle—using only 100 nl of nucleic acid sample; however, here steps III and IV (see Fig. 3(a)) were repeated 50 times (without removal of the magnet) in order to accrue larger concentrations of magnetic beads for analysis. After completion of these cycles, the magnet was removed and the beads were pumped to the ARROW chip for optical analysis. Signal traces for experiments containing non-target oligonucleotide plus beacon (negative control) and beacon-target hybrids are shown in Figs. 3(b) and 3(c), respectively. It is clear that the magnetic beads, under negative control conditions (Figure 3(b))—no target or completely non-complementary contaminating oligos, in this case a 100 nt. H1N1 synthetic oligonucleotide—yield on average 3 counts/ms per particle. In contrast, beads that are subjected to mix with beacon bound targets have an average signal of 15 counts/ms per particle (up to a maximum of 50 counts/ms per particle), corresponding to a signal-to-noise ratio of 5 and allowing for specific detection of the viral target. As ARROW devices have single nucleic acid sensitivity,19 the limit of detection (LOD) of this devices is dependent only on the counting time of the assay. These results demonstrate the ability of the hybrid optofluidic system to process and extract small volumes (about 5 μl) of target nucleic acid material with sequence specificity. It is notable that the signal for target-beacon bound beads is high for each individual bead. Therefore, it would be possible to further reduce the sample volume, and thus the number of fluorescent targets per bead.
IV. CONCLUSION
In summary, we have introduced a hybrid chip-scale system combining automated microfluidic processing with highly sensitive on-chip optical detection. Actively controlled manipulation of fluidic volumes is necessary to carry out the complex sample preparation tasks required for realistic molecular diagnostic tests. We implemented this active control using programmable microvalve arrays and demonstrated that these can be fluidically coupled to a liquid-core waveguide analysis layer. In addition to basic functions such as reversible pumping, valving, and mixing processes, we implemented an assay for molecular diagnosis of viral nucleic acids from complex solutions. The target nucleic acids were extracted and isolated in the microfluidic layer and optically detected in the optofluidic layer. The simultaneous miniaturization of both fluidic and optical components thus allows for highly sensitive and specific detection of minute quantities of target molecules in complex biological fluids.
ACKNOWLEDGMENTS
We thank T. Yuzvinsky at the W. M. Keck Center for Nanoscale Optofluidics at UCSC for assistance with sample imaging and fruitful discussions. This work was supported by the NIH/NIAID under Grant Nos. 1R21AI100229 and 4R33AI100229-03, the NSF under Grant Nos. CBET-1159423 and CBET-1159453, and an NIH laboratory construction Grant No. CO6 RR12087. J.W.P. acknowledges support by the Eugene Cota-Robles Fellowship and the National Science Foundation Graduate Fellowship Research Program under Grant No. DGE 0809125.
References
- 1.Fan X., Advanced Photonic Structures for Biological and Chemical Detection ( Springer Verlag, 2009). [Google Scholar]
- 2.Hoa X. D., Kirk A. G., and Tabrizian M., Biosens. Bioelectron. 23, 151 (2007). 10.1016/j.bios.2007.07.001 [DOI] [PubMed] [Google Scholar]
- 3.Länge K., Rapp B. E., and Rapp M., Anal. Bioanal. Chem. 391, 1509 (2008). 10.1007/s00216-008-1911-5 [DOI] [PubMed] [Google Scholar]
- 4.Craighead H., Nature 442, 387 (2006). 10.1038/nature05061 [DOI] [PubMed] [Google Scholar]
- 5.Hughes A. J. and Herr A. E., Proc. Natl. Acad. Sci. U.S.A. 109, 21450 (2012). 10.1073/pnas.1207754110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher S., Nestler J., Otto T., Wegener M., Ehrentreich-Förster E., Michel D., Wunderlich K., Palzer S., Sohn K., Weber A., Burgard M., Grzesiak A., Teichert A., Brandenburg A., Koger B., Albers J., Nebling E., and Bier F. F., Lab Chip 12, 464 (2012). 10.1039/c1lc20693a [DOI] [PubMed] [Google Scholar]
- 7.Fan X. and White I. M., Nat. Photonics 5, 591 (2011). 10.1038/nphoton.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt H. and Hawkins A. R., Nat. Photonics 5, 598 (2011). 10.1038/nphoton.2011.163 [DOI] [Google Scholar]
- 9.Psaltis D., Quake S. R., and Yang C., Nature 442, 381 (2006). 10.1038/nature05060 [DOI] [PubMed] [Google Scholar]
- 10.Mohammed M.-I. and Desmulliez M. P. Y., Biomicrofluidics 7, 64112 (2013). 10.1063/1.4837755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y. J., Wu Y. C., Chen Y., Kung Y. C., Wu T. H., Huang K. W., Sheen H. J., and Chiou P. Y., Biomicrofluidics 7, 44121 (2013). 10.1063/1.4818944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S., Yan Y., Wang Y., Senapati S., and Chang H.-C., Biomicrofluidics 7, 61102 (2013). 10.1063/1.4832095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryckeboer E., Bockstaele R., Vanslembrouck M., and Baets R., Biomed. Opt. Express 5, 1636 (2014). 10.1364/BOE.5.001636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan Z., Zou Y., Zhang M., Lv J., Shen H., Yang P., Zhang H., Zhu Z., and Yang C. James, Biomicrofluidics 8, 014110 (2014). 10.1063/1.4866766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho S. H., Godin J. M., Chen C.-H., Qiao W., Lee H., and Lo Y.-H., Biomicrofluidics 4, 43001 (2010). 10.1063/1.3511706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J., Johnson M., Hill P., Sonkul R. S., Kim J., and Gale B. K., J. Micromech. Microeng. 22, 015007 (2012). 10.1088/0960-1317/22/1/015007 [DOI] [Google Scholar]
- 17.Cho Y.-K., Lee J.-G., Park J.-M., Lee B.-S., Lee Y., and Ko C., Lab Chip 7, 565 (2007). 10.1039/b616115d [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Morabito K., Tang J. X., and Tripathi A., Biomicrofluidics 7, 44107 (2013). 10.1063/1.4816943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parks J. W., Cai H., Zempoaltecatl L., Yuzvinsky T. D., Leake K., Hawkins A. R., and Schmidt H., Lab Chip 13, 4118 (2013). 10.1039/c3lc50818h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Testa G., Persichetti G., Sarro P. M., and Bernini R., Biomed. Opt. Express 5, 417 (2014). 10.1364/BOE.5.000417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duguay M., Kokubun Y., Koch T., and Pfeiffer L., Appl. Phys. Lett. 49, 13 (1986). 10.1063/1.97085 [DOI] [Google Scholar]
- 22.Yin D., Deamer D. W., Schmidt H., Barber J. P., and Hawkins A. R., Appl. Phys. Lett. 85, 3477 (2004). 10.1063/1.1807966 [DOI] [Google Scholar]
- 23.Yin D., Lunt E. J., Rudenko M. I., Deamer D. W., Hawkins A. R., and Schmidt H., Lab Chip 7, 1171 (2007). 10.1039/b708861b [DOI] [PubMed] [Google Scholar]
- 24.Rudenko M. I., Kühn S., Lunt E. J., Deamer D. W., Hawkins A. R., and Schmidt H., Biosens. Bioelectron. 24, 3258 (2009). 10.1016/j.bios.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J., Kang M., Jensen E. C., and Mathies R. A., Anal. Chem. 84, 2067 (2012). 10.1021/ac202934x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duffy D. C., McDonald J. C., Schueller O. J., and Whitesides G. M., Anal. Chem. 70, 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 27.Xia Y. and Whitesides G. M., “Soft lithography,” Annu. Rev. Mater. Sci. 28, 153–184 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- 28.Kim J., Jensen E., Stockton A., and Mathies R., Anal. Chem. 85, 7682 (2013). 10.1021/ac303767m [DOI] [PubMed] [Google Scholar]
- 29.See supplementary material at http://dx.doi.org/10.1063/1.4897226E-BIOMGB-8-016405 for automaton assay and nucleic acid sequence information.
- 30.Bhattacharyya A. and Klapperich C. M., Lab Chip 7, 876 (2007). 10.1039/b700442g [DOI] [PubMed] [Google Scholar]
- 31.Jensen E. C., Bhat B. P., and Mathies R. A., Lab Chip 10, 685 (2010). 10.1039/b920124f [DOI] [PubMed] [Google Scholar]
- 32.Yin D., Schmidt H., Barber J., and Hawkins A. R., Opt. Express 12, 2710 (2004). 10.1364/OPEX.12.002710 [DOI] [PubMed] [Google Scholar]
- 33.Yin D., Deamer D. W., Schmidt H., Barber J. P., and Hawkins A. R., Opt. Lett. 31, 2136 (2006). 10.1364/OL.31.002136 [DOI] [PubMed] [Google Scholar]
- 34.Liu S., Zhao Y., Parks J. W., Deamer D. W., Hawkins A. R., and Schmidt H., “Correlated electrical and optical analysis of single nanoparticles and biomolecules on a nanopore-gated optofluidic chip,” Nano Lett. 14, 4816–4820 (2014). 10.1021/nl502400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh P., Optical Waves in Layered Media ( John Wiley & Sons, Inc., Hoboken, NJ, 1998). [Google Scholar]
- 36.Lunt E. J., Wu B., Keeley J. M., Measor P., Schmidt H., and Hawkins A. R., IEEE Photonics Technol. Lett. 22, 1147 (2010). 10.1109/LPT.2010.2051145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lunt E. J., Measor P., Phillips B. S., Kühn S., Schmidt H., and Hawkins A. R., Opt. Express 16, 20981 (2008). 10.1364/OE.16.020981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y., Phillips B., Ozcelik D., Parks J., Measor P., Gulbransen D., Schmidt H., and Hawkins A. R., “ Tailoring the spectral response of liquid waveguide diagnostic platforms,” J. Biophotonics 5, 703–711 (2012). 10.1002/jbio.201200049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmes M., Keeley J., Hurd K., Schmidt H., and Hawkins A. R., J. Micromech. Microeng. 20, 115008 (2010). 10.1088/0960-1317/20/11/115008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlinsey J. M., Monahan J., Marchiarullo D. J., Ferrance J. P., and Landers J. P., Anal. Chem. 77, 3637 (2005). 10.1021/ac048115z [DOI] [PubMed] [Google Scholar]
- 41.Jensen E. C., Zeng Y., Kim J., and Mathies R. A., JALA 15, 455 (2010). 10.1016/j.jala.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt H. and Hawkins A. R., Microfluid. Nanofluid. 4, 3 (2008). 10.1007/s10404-007-0199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen E. C., Stockton A. M., Chiesl T. N., Kim J., Bera A., and Mathies R. A., Lab Chip 13, 288 (2013). 10.1039/c2lc40861a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi S. and Kramer F. R., “Molecular beacons: Probes that fluoresce upon hybridization,” Nat. Biotechnol. 14, 303–8 (1996). 10.1038/nbt0396-303 [DOI] [PubMed] [Google Scholar]
- 45.Jungell-Nortamo A., Syvänen A. C., Luoma P., and Söderlund H., Mol. Cell. Probes 2, 281 (1988). 10.1016/0890-8508(88)90012-6 [DOI] [PubMed] [Google Scholar]
- 46.Day P. J., Flora P. S., Fox J. E., and Walker M. R., Biochem. J. 278(Pt 3), 735 (1991); available at http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1151408/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klamp T., Camps M., Nieto B., Guasch F., Ranasinghe R. T., Wiedemann J., Petrášek Z., Schwille P., Klenerman D., and Sauer M., Sci. Rep. 3, 1852 (2013). 10.1038/srep01852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4897226E-BIOMGB-8-016405 for automaton assay and nucleic acid sequence information.