Abstract
Gastroenteritis is a clinical illness of humans and other animals that is characterized by vomiting and diarrhea and caused by a variety of pathogens, including viruses. An increasing number of viral species have been associated with gastroenteritis or have been found in stool samples as new molecular tools have been developed. In this work, a DNA microarray capable in theory of parallel detection of more than 100 viral species was developed and tested. Initial validation was done with 10 different virus species, and an additional 5 species were validated using clinical samples. Detection limits of 1 × 103 virus particles of Human adenovirus C (HAdV), Human astrovirus (HAstV), and group A Rotavirus (RV-A) were established. Furthermore, when exogenous RNA was added, the limit for RV-A detection decreased by one log. In a small group of clinical samples from children with gastroenteritis (n = 76), the microarray detected at least one viral species in 92% of the samples. Single infection was identified in 63 samples (83%), and coinfection with more than one virus was identified in 7 samples (9%). The most abundant virus species were RV-A (58%), followed by Anellovirus (15.8%), HAstV (6.6%), HAdV (5.3%), Norwalk virus (6.6%), Human enterovirus (HEV) (9.2%), Human parechovirus (1.3%), Sapporo virus (1.3%), and Human bocavirus (1.3%). To further test the specificity and sensitivity of the microarray, the results were verified by reverse transcription-PCR (RT-PCR) detection of 5 gastrointestinal viruses. The RT-PCR assay detected a virus in 59 samples (78%). The microarray showed good performance for detection of RV-A, HAstV, and calicivirus, while the sensitivity for HAdV and HEV was low. Furthermore, some discrepancies in detection of mixed infections were observed and were addressed by reverse transcription-quantitative PCR (RT-qPCR) of the viruses involved. It was observed that differences in the amount of genetic material favored the detection of the most abundant virus. The microarray described in this work should help in understanding the etiology of gastroenteritis in humans and animals.
INTRODUCTION
Gastroenteritis stands among the five principal causes of mortality by disease and morbidity at all ages worldwide. The most affected population is children under 5 years of age, where it accounts for the second cause of postneonatal death, with approximately 2.6 million deceased per year (1). Although the majority of deaths occur in developing countries, diarrheal disease is among the most common causes of illness worldwide, with approximately 4,620 million episodes annually (1). Besides humans, all vertebrate species suffer from enteric diseases. Infections in farm animals can lead to large economic losses, while household pets, such as dogs and cats, are also affected. On the other hand, wild animals, such as deer, monkeys, bats, foxes, wolves, and boars, among others, can act as potential reservoirs for pathogens (2). Gastrointestinal (GI) infections are caused by a variety of pathogens, including parasites, bacteria, and viruses. The characterization of pathogens causing GI infections of viral etiology has led to the establishment of a main group of pathogens (Rotavirus A [RV-A], Norwalk virus [NV], Human astrovirus [HAstV], and Human adenovirus F [HAdV-F]) (3) for which specific diagnostic tests were developed (4). Tests for secondary or rare viruses are available but are usually restricted to experimental use. Routine diagnostic methods for viral gastroenteritis are nowadays based on the detection of virus components by immunoassays or by molecular methods (5, 6, 7, 8), with the majority of these tests designed to evaluate only a single pathogen at a time.
The use of two or more specific primer sets (multiplexing) in PCR allows the amplification of several targets in one test. Although multiplex tests are available for diverse viruses (9, 10, 11, 12, 13), facilitating rapid and sensitive detection of the main GI disease agents, these assays are still limited in the number of viruses detected, and the results can be affected by mutations at primer binding sites. On the other hand, DNA microarrays represent an alternative to detect hundreds to thousands of potential pathogens in a single assay. Microarray detection is based on solid-phase hybridization, in which specific probes are deposited on a surface and react with a mixture of labeled nucleic acids. So far, different microarrays have been developed to detect causative infectious agents associated with a number of diseases: respiratory (14, 15, 16), hemorrhagic (17), blood borne (18, 19), and central nervous system (20) syndromes. Other broad microarrays have been developed for virus discovery (21); however, a diagnostic microarray specific for viruses found in the GI tract is lacking. Given the recent rise in the number of new viral species (22, 23, 24, 25, 26), diagnostic DNA microarrays represent a possibility for testing their clinical importance and impact on human and animal health.
In this work, the development and validation of a DNA microarray designed to detect in principle more than 100 viral species associated with the GI tract in vertebrates is presented. This microarray was successfully used to identify viruses in a small set of gastroenteritis clinical samples.
MATERIALS AND METHODS
Cells, viruses, and clinical samples.
Viruses were either present in our laboratory or kindly provided by different partner laboratories (Table 1). Clinical samples from children presenting gastroenteritis during the winter season from 2004 to 2005 were obtained in Monterrey, Mexico, with the written consent of a parent or guardian. Analysis of human clinical samples was approved by the Bioethics Committee of the Instituto de Biotecnologia. The initial screening of samples for RV-A was performed in Monterrey by silver staining of RV-A segmented double-stranded RNAs separated by SDS-PAGE. No previous screening for bacterial or parasitic agents was performed on the group of samples. Triple-layered particles of RV-A strain RRV were purified with a cesium chloride density gradient as described previously (27).
TABLE 1.
Reference virus species used in microarray validation
| Family | Genus | Species | Straina | No. of positive probes/totalb |
|---|---|---|---|---|
| Astroviridae | Mammastrovirus | Human astrovirus | Yuc8 | 4/4 |
| Adenoviridae | Mastadenovirus | Human adenovirus C | Adv5 | 10/13 |
| Caliciviridae | Vesivirus | Feline calicivirus | F9 | 14/22 |
| Norovirus | Norwalk virusc | 8/12 | ||
| Sapovirus | Sapporo virusc | 5/14 | ||
| Flaviviridae | Pestivirus | Bovine viral diarrhea virus 1 | NADL | 6/6 |
| Flavivirus | Dengue virus 4 | 9/9 | ||
| Paramyxoviridae | Respirovirus | Bovine parainfluenza virus 3 | SF-4 | 9/9 |
| Reoviridae | Rotavirus | Rotavirus A | RRV | 22/42 |
| TFR-41 | 14/42 | |||
| UK | 19/42 | |||
| Wa | 21/42 | |||
| Orthoreovirus | Mammalian orthoreovirus | T1L | 11/25 | |
| T3D | 19/25 |
Reference strains were provided by Ramon Gonzalez, FC-UAEM (human adenovirus C), Lorena Gutierrez, CINVESTAV-IPN (feline calicivirus, Norwalk virus, and Sapporo virus), Rosa E. Sarmiento, FMVZ-UNAM (bovine viral diarrhea virus 1 and bovine parainfluenza virus 3), Rosa María Del Angel, CINVESTAV-IPN (dengue virus 4), and Terrence S. Dermody, Vanderbilt University School of Medicine (mammalian orthoreovirus).
Number of oligonucleotide probes which recognized virus/total number of oligonucleotide probes designed to bind viral species.
Clinical reference samples.
Microarray probe design.
All virus species that have been either associated with gastroenteritis or found in the gastrointestinal tract were identified by an extensive review of published literature and selected to be included in microarray. All available full-length genomes or complete gene sequences of the selected virus species were obtained from GenBank (up to February 2009), and the proper databases were created. For each virus species, sequence redundancy was eliminated according to sequence similarity with cutoff values of 95 to 99% using CD-HIT software (28). One sequence for each species was selected as a source for probe production and was processed as described previously (29). Specifically, sequences were consecutively split into 70-mers with a shifting window of 3 nucleotides, with each 70-mer corresponding to a potential probe. The 70-mer-length probes have sufficient size to allow for stringent hybridization conditions while allowing for a certain degree of mismatches, but they are small enough to maintain species specificity (30, 31, 32). Target probes were selected to be included in the microarray by analysis of BLAST results and calculation of hybridization thermodynamics (ΔG) calculated by the nearest-neighbor method (33). For the probe to be considered a good candidate for the microarray, the ΔG was required to be at least −70 kcal/mol for homologous sequences and higher than −40 kcal/mol for heterologous sequences. A minimum of 6 nonoverlapping probes from conserved regions in virus genomes were selected for each virus, and each available genome sequence in the target database for given species was recognized by at least two probes. When necessary, due to variability within a species, two or more source sequences were chosen and each single sequence was processed as described above.
Microarray probe in silico analysis.
The hybridization thermodynamics of RV-A selected probes were evaluated in silico with VP1, VP2, and NSP5 segments of RV-A strains representing all full-genome G and P genotypes available. The hybridization ΔG (kcal/mol) between probe and target was calculated by the nearest-neighbor method. The best probe-target ΔG was plotted in a heat map using R. Detection of a target is when the ΔG is <−50 kcal/mol.
Microarray production.
Selected 70-mer probes were synthesized by Illumina Oligator (Illumina Inc., CA, USA). Oligonucleotides were resuspended to 400 pmol in 3× SSC buffer (0.45 M NaCl, 45 mM sodium citrate, pH 7.0) and spotted onto epoxide-coated glass slides in the Microarray Facility of the Prostate Centre at Vancouver General Hospital, Vancouver, British Columbia, Canada. Each spot contained one specific probe to detect one virus species and 4 pmol of spike70 (a 70-mer without a known biological complementary sequence) (34), used to precisely identify probe spot locations on the microarray. Slides were maintained in a humidity-free chamber until their use.
Nucleic acid extraction, amplification, and labeling.
Genetic material from virus lysates (cell culture supernatants from reference strains) was extracted with the PureLink viral RNA/DNA kit according to the manufacturer's instructions (Invitrogen, USA). For clinical samples and Norwalk and Sapporo virus positive controls, 100 μg of stool was added to conical screw-cap tubes containing 100 mg of 150- to 212-μm glass beads (Sigma, USA), chloroform (100 μl), and phosphate-buffered saline (PBS) up to 1 ml. Samples were homogenized in a bead beater (Biospec Products, USA). After 10 min of centrifugation at 2,000 × g, supernatants were recovered and filtered in Spin-X 22-μm-pore filters (Costar, NY) at 5,000 × g for 10 to 20 min. Filtered samples were treated with Turbo DNase (Ambion, USA) and RNase (Sigma, USA) for 30 min at 37°C and immediately chilled on ice. Nucleic acids were then extracted from 200 μl using the PureLink viral RNA/DNA kit according to the manufacturer's instructions (Invitrogen, USA). Nucleic acids from virus lysates or clinical samples were eluted in nuclease-free water, aliquoted, quantified in NanoDrop ND-1000 (NanoDrop Technologies, DE), and stored at −70°C until further use.
Sample processing and random amplification of nucleic acids were performed essentially as described previously (21, 35, 36). Briefly, reverse transcription was done using SuperScript III reverse transcriptase (Invitrogen, USA) and primer A (5′-GTTTCCCAGTAGGTCTCN9-3′). The cDNA strand was generated by two rounds of synthesis with Sequenase 2.0 (USB, USA). The obtained cDNA was then amplified with KlenTaq polymerase (Sigma, USA) or Taq polymerase (New England BioLabs, USA) using primer B (5′-GTTTCCCAGTAGGTCTC-3′) by 30 cycles of the following program: 30 s at 94°C, 1 min at 50°C, and 1 min at 72°C. As a last step, the nucleotide analogue aminoallyl-dUTP (TriLink, USA) in a 7:3 ratio with dTTP was incorporated during an additional 20 cycles of PCR using the same conditions described above and 5 μl of product from the previous PCR as starting material. The amplified products were purified with the DNA Clean & Concentrator-5 kit (Zymo Research, USA). Coupling reactions of sample DNA with Cy3 and probe 70 (70-mer complementary to spike 70) with Cy5 dyes (GE HealthCare, USA) were done as described elsewhere (31). Fluorophore-labeled DNA was purified with the Zymo DNA Clean & Concentrator-5 kit, and label incorporation was quantified with NanoDrop.
Slide preparation, hybridization and scanning.
Microarray slides were treated just before their use with an ethanolamine wash solution (50 mM ethanolamine, 0.1% SDS, 0.1 M Tris, pH 9) for 15 min at 50°C, followed by two washes in distilled water, and they were then dried by centrifugation for 5 min at 500 rpm. Processed slides were loaded with 30 μl of a combination of Cy3- and Cy5-labeled DNA in 3× SSC buffer, and the hybridization was left to proceed in a sealed chamber submerged in a water bath at 65°C for 8 to 12 h. After incubation the slides were washed consecutively in 2× SSC (65°C), 2× SSC, 1× SSC, and 0.2× SSC and dried for 5 min at 500 rpm. Hybridization images were acquired with an Axon GenePix 4000B scanner (Molecular Devices, USA) synchronized with GenePix Pro 6.0 software to detect and measure spot intensities.
Data analysis.
Hybridization spot intensities were first filtered by the following spot quality control parameters: spot size and shape (denoted as good/bad/absent), channel 532 foreground (F532) signal saturation (% F532 saturated, <5), and F532 signal proportion over channel 532 background (B532) signal [(% > B532 + 2 standard deviations) > 50%]. Spots showing good quality were used to generate microarray level background values. Normalization of intensity values was done with the formula (F532i/F532m) − (B532i-B5532m), where F532i and B532i are the foreground and background signals of spot “i,” respectively, and F532m and B532m are the sums of all foreground or background spots, respectively.
The statistical significance of probe intensities in the reference samples was obtained by the rank products algorithm (37) using a minimum of three technical replicates. Rank values from negative-control samples were recorded and used to generate a “spot rank value” included in subsequent spot quality analysis. For clinical samples, z-score transformed intensities and their P values were analyzed with the fdr tool package (38) in R (39). Positive virus species were defined as having at least two probes with P values of <0.05 and false-discovery rates (FDRs) of <0.01.
Limit-of-detection assays.
In order to determine the amount of virus particles detectable by the microarray, three reference viruses with different genome types were assayed: RV-A double-stranded RNA (dsRNA), HAstV positive single-stranded RNA (ssRNA+), and HadV-C double-stranded DNA (dsDNA). RNA was extracted from purified RV-A strain RRV and MA104 cells. The RV-A genome molecular mass was calculated according to the following formula: (genome length [bp] × 325)/6.022 × 1023 (40). Decreasing dilutions of RV-A RNA corresponding to 1 × 108 to 10 particles were analyzed alone or mixed with an excess of MA104 cells RNA (50 ng). Similarly, decreasing dilutions of focus-forming unit-titrated cell lysates of HAstV or HAdV-C, corresponding to 1 × 107 to 100 virus particles, were extracted, amplified, labeled, and processed using the full microarray protocol as described above.
Conventional diagnostic or confirmatory RT-PCR.
Nucleic acids extracted from clinical samples were used to perform diagnostic reverse transcription-PCR (RT-PCR) using Qiagen's one-step RT-PCR kit (Qiagen, USA) or SuperScript III one-step RT-PCR with Platinum Taq polymerase (Invitrogen, USA). For confirmatory RT-PCR, cDNA was generated with SuperScript III reverse transcriptase (Invitrogen, USA), and Taq polymerase (New England BioLabs) was used for PCRs following the manufacturer's instructions. Oligonucleotide primers used in diagnostic or confirmatory RT-PCR are listed in Table S1 in the supplemental material. PCRs for RV-A detection included a 5-min boiling step followed by immediate ice-chilling step just before RT-PCR. Amplification conditions for RV-A, HAstV, and calicivirus (CV) were as follows: 30 min at 50°C; 15 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 50°C, and 1 min at 72°C; and a final extension of 5 min at 72°C. RT-PCR conditions for human adenovirus (HAdV) were as follows: 30 min at 50°C; 15 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. The human enterovirus (HEV) amplification program was as follows: 30 min at 50°C; 15 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C; and final extension for 5 min at 72°C. Human parechovirus (HPeV) amplification was as follows: 30 min at 50°C; 15 min at 95°C; 35 cycles of 1 min at 95°C, 1 min at 48°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Anellovirus (TTV) confirmation was performed as a seminested PCR. Conditions for the first round were as follows: 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and a final extension for 5 min at 72°C. The second round used the same program but with only 30 cycles. Human bocavirus (HBoV) was detected by Seeplex RV15 OneStep ACE detection (Seegene, USA). PCR products were visualized in 2.0% agarose gels, except for HEV, which required 3.5% gels due to a small amplicon size.
Semiquantitative RT-PCR and PCR detection of viruses.
One-step real-time RT-PCR and real-time PCR were performed using primers targeting conserved genomic regions (see Table S1 in the supplemental material). RV-A detection required previous sample boiling for 5 min and immediate ice chilling. For the RNA viruses (RV-A, HAstV, NV, and HEV), detection was performed as a two-step process. First, 3 μl of RNA (5 ng) was reverse transcribed with 0.125 μl (50 U/μl) SuperScript III reverse transcriptase (Invitrogen, USA), 0.25 μl of RNase inhibitor (20 U/μl), 12.5 μl of 2× SYBR green master mix (Applied Biosystems, USA), 1 μl of the primer, and diethyl pyrocarbonate (DEPC)-treated water in a 24-μl final volume. Samples were incubated for 30 min at 48°C, followed by enzyme inactivation for 10 min at 90°C. In the second step 1 μl of second primer was added, and PCR was carried out as follows. The HAstV and RV-A amplification program consisted of 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. The NV amplification program was 5 min at 95°C and 45 cycles of 10 s at 95°C, 20 s at 48°C, and 45 s at 60°C. The HEV program was 10 min at 95°C, 45 cycles of 20 s at 95°C, 20 s at 55°C, and 1 min at 72°C, and final extension of 5 min at 72°C. In the case of HEV, both specific primers were added before PCR, since the RT step was performed using random hexamers. HAdV amplification reaction mixtures consisted of 3 μl (5 ng) of DNA, 12.5 μl of 2× SYBR green master mix, and 1 μl of each corresponding primer in a 25-μl volume. Conditions were 95°C for 8 min, 45 cycles of 30 s at 95°C, 20 s at 55°C, and 20 s at 72°C, and a final extension of 5 min at 72°C. Amplifications were carried out in an ABI Prism 7500 sequence detector system (Applied Biosystems). Dissociation curves were evaluated for nonspecific products. Threshold cycle (CT) values corresponding to detection of specific virus sequences were obtained from triplicates of selected samples presenting coinfections and compared for the viruses detected. PCR primer sets for detection of CV, HAdV, and HEV were designed to recognize the target at the genus level (5, 6, 41).
RESULTS
Selection of viruses related to gastrointestinal infections.
An advantage of the microarray technology is the capacity to test hundreds and even thousands of targets in a single assay. The main goal of this study was to develop an assay for detection of all viruses that have been found in stool samples from vertebrates, associated or not with gastroenteritis, which should facilitate clinical and epidemiological studies in humans and animals. A deep search of the scientific literature available in public databases resulted in a list of 128 species of viruses reported to be present in the gastrointestinal tract, representing 55 genera that belonged to 17 viral families (see Table S2 in the supplemental material). The list of virus species includes the well-known human gastroenteritis viruses (calicivirus group, rotaviruses, human astroviruses, and enteric adenoviruses), together with some recently described human viruses (Human adenovirus G [23], Human bocavirus [42], Cosavirus [24], Saffold virus [43], and Salivirus A [25, 44]). Classical, nonhuman gastrointestinal viruses (coronavirus, circovirus, and pestiviruses) and other new discovered viral agents (at the time of the microarray design) from different animal species, such as animal anelloviruses (45, 46), bat astroviruses (47), and bovine kobuviruses (48), whose participation as pathogens is not well understood, are also included in the microarray. Thus, the virus species of interest encompassed a variety of viruses with different characteristics, such as RNA and DNA genomes, enveloped/nonenveloped virions, segmented or nonsegmented genomes, and single- or double-stranded genomes. All available complete gene or genome sequences were retrieved from a public database (GenBank) and were organized in a taxonomic hierarchical database following the ICTV classification at the date the microarray probes were designed (ICTV, 2009) or, for novel species, as suggested by the discoverer.
Probe selection and microarray validation.
A set of 1,256 70-mer microarray probes were selected from conserved regions and designed to identify 128 viral species associated with the GI tract, with at least 6 probes designed for each viral species and at least 2 probes corresponding to each sequenced viral genome. To maintain stringent experimental conditions (hybridization at 65°C) while allowing a certain amount of sequence variability, the probes were designed as 70-mers. The highest number of probes covered RV-A (42 probes), Alphacoronavirus (28 probes), and mammalian Orthoreovirus (25 probes) (see Tables S2 and S3 in the supplemental material). For some viruses, the design of a complete set of 6 oligonucleotides was not possible due to the lack of enough complete sequences; nevertheless, available probes were included for each viral species.
Reference strains for 10 viral species were available for probe validation. These species represent 6 viral families and include 4 main human pathogens (HAstV, NV, SV, and RV-A), other human viruses (mammalian Orthoreovirus, HAdV-C, and Dengue virus 4), and three nonhuman viruses (Feline calicivirus, Bovine viral diarrhea virus 1, and Bovine parainfluenza virus 3) (Table 1). All reference strains tested were detected as expected, including four different RV-A strains (human strain Wa, simian strain RRV, porcine virus TFR-41, and bovine strain UK) and two different mammalian Orthoreovirus strains (T1L and T3D) (Table 1). To test the in silico capacity of probes to recognize different and variable strains, 42 probes specific for rotavirus were analyzed with a panel of all available G and P genotypes (see Fig. S1 in the supplemental material). The only genotype that the microarray probably would not detect was G22P[35], belonging to a turkey rotavirus strain.
Sensitivity and specificity of the assay.
To determine the sensitivity limits of the DNA microarray, the virus genetic material was extracted from lysates of HAstV- or HAdV-C-infected cells or from CsCl-purified simian strain RRV particles. In a series of cell lysate dilutions (corresponding from 102 to 107 viral particles), the microarray was able to detect as few as 103 HAdV-C or HAstV virus particles. Similarly, RV-A RNA (corresponding to 10 to 108 viral particles) was amplified before or after addition of a constant amount of cellular RNA (50 ng). In the absence of cellular RNA, the detection limit for viral RNA was 103 genome copies; however, when the complexity of the sample was augmented by adding cellular RNA, the detection limit was one logarithm lower, detecting 104 genome copies.
To evaluate the probe specificity, a rank products algorithm (37) was applied to the results obtained from technical replicates of reference viruses and mock-infected cell controls (MA104 cells, A549 cells, and C6/36 cells). Based on the false-discovery rate (FDR) test included in the software, 16 probes were identified as presenting nonspecific behavior (marked with asterisks in Table S3 in the supplemental material). When analyzed, these nonspecific probes did not show any common feature, although some presented a high GC content (>70%). In the following experiments, the results obtained with these probes were excluded from analysis.
Analysis of clinical samples.
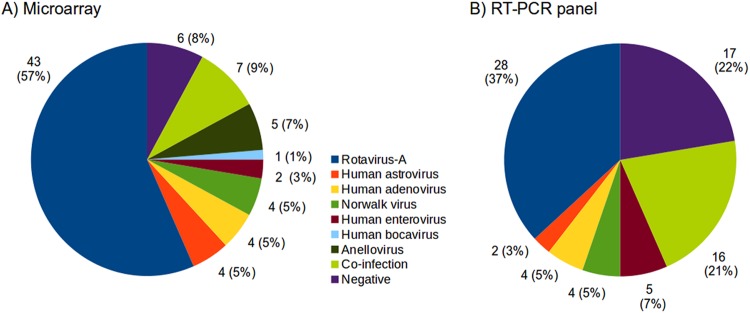
To further test the capacity of the microarray to detect viruses, 76 samples from children under 5 years of age, collected during the winter season from 2004 to 2005 in Monterrey, Mexico, were analyzed. The collection of samples was originally screened for RV-A by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stored at −70°C. Using the microarray developed in this study, a viral agent was detected in 70 out of 76 (92%) samples tested; a single virus was found in 63 (83%) samples, while two or more viral species were detected in 7 (9%) samples (Fig. 1). Among the viruses detected, the most common was RV-A (44 samples), followed by TTV (12 positives), HEV (7), caliciviruses (6 [5 NV and 1 SV]), HAstV (5), HAdV (4 [3 HAdV-F and 1 HAdV-A]), HPeV (2), and HBoV (1) (Fig. 1). It is important to mention that only 6 (8%) samples remained negative after microarray detection and that not all viruses found are known to be pathogenic. As mentioned above, after collection all samples were screened for the presence of RV-A by SDS-PAGE. Additionally, as described below, all samples tested with the microarray were tested for selected viruses, including RV-A, by diagnostic RT-PCR. In 34 samples RV-A was identified by the three methods tested; 5 additional samples were found positive by microarray and RT-PCR tests (Fig. 2). Another 8 were found positive either by microarray (n = 5) or by RT-PCR (n = 3) (Fig. 2). Notably, the 3 samples that were positive only for RV-A by RT-PCR were mixed-infection samples.
FIG 1.
Prevalence of viruses in clinical samples. A group of 76 clinical samples from children presenting gastroenteritis was analyzed by the described microarray (A) or by diagnostic RT-PCR for the 5 most common gastrointestinal pathogens (B). Samples with coinfections are shown. Negative, no virus identified.
FIG 2.
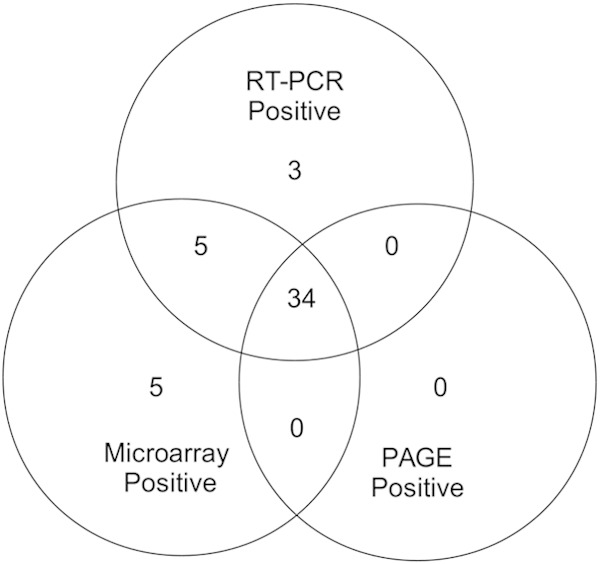
Identification of rotavirus group A. A group of 76 gastroenteritis samples was analyzed by three methods for the presence of rotavirus. These were visualization of rotavirus dsRNA by SDS-PAGE, RT-PCR, and the microarray designed in this work. The circles represent numbers of rotavirus-positive samples identified by one, two, or three of the methods used.
To compare the results of the microarray method with those of a routine diagnostic method for viral gastroenteritis, RT-PCR detection for a panel of 5 viruses (RV-A, HAstV, HAdV, CV [NV and SV], and HEV) was performed in all clinical samples. It is important to point out that the primer sets for HAdV, CV, and HEV are designed to recognize their target at the genus level (5, 6, 41).
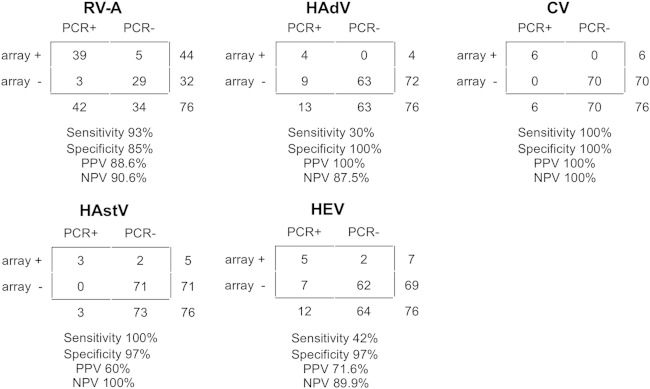
The RT-PCR panel detected at least one virus in 59 samples (78%) (Fig. 1B), a lower detection rate than that with the DNA microarray when analyzing only these 5 viruses (n = 65, 85%). At the individual virus level, the RT-PCR panel confirmed the microarray results in all HAdV-positive samples (1 HAdV-A and 3 HAdV-F), having a positive predictive value (PPV) of 100%, in all CV (5 NV and 1 SV)-positive samples (PPV, 100%), and in 39 of 44 RV-A-positive samples (PPV, 89%), while PPVs were lower for HAstV, with 3 of 5 positive samples identified by microarray (PPV 60%), and HEV, with 5 of 7 positive samples identified by microarray (PPV 71%) (Fig. 3).
FIG 3.
Microarray diagnostic sensitivity and specificity. A panel of 5 virus groups (rotavirus group A [RV-A], human astrovirus [HAstV], human adenovirus [HAdV], calicivirus [CV], and human enterovirus [HEV]) was tested by RT-PCR in all 76 samples. Results were compared to those obtained by microarray analysis. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the microarray (array), compared to RT-PCR (PCR) for detection of particular pathogens are shown.
Detection of viruses in MI.
The RT-PCR screening resulted in the identification of 16 mixed infections (MI), while the microarray identified only 7 MI (Fig. 1). The microarray detected up to 4 different viruses within one sample, with TTV found in all MI samples. The following viral combinations were found by microarray: 3 samples with HEV B/TTV and one sample each with NV/TTV, HEV-B/HAstV/TTV, RV-A/HPeV/TTV, and SV/HEV-B/HPeV/TTV (Fig. 1). Of interest, Human parechovirus and Sapporo virus were detected only in coinfection. The MI combinations observed in RT-PCR were RV-A/HAdV (8 samples), RV-A/HEV (5), HAstV/HEV (1), RV-A/CV (1), and HAdV/CV/HEV (1) (Fig. 1B). Examining these 16 samples, we observed that RV-A was the only virus identified by microarray in all samples with RV-A/HAdV coinfection (n = 8) and in 4 out of 5 RV-A/HEV samples, while HAstV was the only virus identified in samples with HAstV/HEV coinfection (Table 2). In one sample, NV was identified as the sole species by microarray, while RT-PCR results showed CV/HAdV/HEV triple coinfection (Table 2). Thus, in all of these 16 samples, a single virus was identified by the microarray, while at least two viral species were detected by RT-PCR.
TABLE 2.
CT values for viral nucleic acid quantification in samples with coinfection
| Virus identified by: |
CT value determined by real-time RT-PCR fora: |
|||||
|---|---|---|---|---|---|---|
| Diagnostic RT-PCR | Microarray | RV-A | HAdV | HEV | NVb | HAstV |
| RV-A | RV-A | 21.9 | ||||
| HAdV | HAdV | 14.7 | ||||
| HEV | HEV | 23.8 | ||||
| NV | NV | 19.6 | ||||
| HAstV | HAstV | 14.8 | ||||
| RV-A/HAdV | RV-A | 20.5 | 37.6 | |||
| RV-A/HAdV | RV-A | 22.5 | 28.4 | |||
| RV-A/HAdV | RV-A | 28.2 | 41.3 | |||
| RV-A/HAdV | RV-A | 28.6 | 44.5 | |||
| RV-A/HAdV | RV-A | 29.1 | 43.8 | |||
| RV-A/HAdV | RV-A | 29.2 | 43.4 | |||
| RV-A/HAdV | RV-A | 30.4 | 30.6 | |||
| RV-A/HEV | RV-A | 29.2 | 25.4 | |||
| RV-A/HEV | RV-A | 29.2 | 27.3 | |||
| RV-A/HEV | RV-A | 29.6 | 28.5 | |||
| RV-A/HEV | RV-A | 30.8 | 27.8 | |||
| RV-A/HEV | HEV | 38.4 | 28 | |||
| RV-A/NV | NV | 30.1 | 23.6 | |||
| HAstV/HEV | HAstV | 28 | 23.2 | |||
| NV/HEV/HAdV | NV | 34.1 | 28.4 | 20.7 | ||
| NTCc | 36.4 | 44.5 | 33.7 | 33.7 | 29.9 | |
RV-A, rotavirus group A; HAdV, human adenovirus; HEV, human enterovirus; NV, Norwalk virus; HAstV, human astrovirus. Single-infection samples were used as positive controls. Lower CT values are shown in bold.
NV is detected at the genus level as calicivirus.
NTC, nontemplate control.
One possible explanation for the discrepancies in the identification of mixed infections using microarrays and RT-PCR could be the variability in the relative amount of genetic material from each virus in clinical samples, as it has been observed that individuals infected with some viruses, for example RV-A and NV, can shed large amounts of viral particles in the acute stage of infection (49, 50, 51). To explore this possibility, the amount of viral genetic material in selected samples with mixed infection was quantified by real-time RT-PCR. The use of equal quantities of starting material allowed us to compare directly the amplification CTs of two viruses within a sample. The results showed that the single virus detected by microarray had, in most cases, a lower CT value than the second virus detected by quantitative RT-PCR (qRT-PCR), with the only exception being the combination RV-A/HEV, where RV-A was the only virus identified by microarray despite the fact that HEV had lower CT values (Table 2). This indicates that MI presenting large differences in the amounts of the genetic material of the viral agents involved are prone to result in single-virus detection by the microarray (generally detecting the one present more abundantly).
Consequently, when comparing the sensitivity and specificity of the microarray with the panel of individual diagnostic RT-PCRs, the most prevalent or most frequently found viruses in single infections, such as RV-A, HAstV, and CV, showed good sensitivity and specificity (from 85 to 100%), while the sensitivity for viruses such as HAdV and HEV was low, ranging from 30 to 42%, clearly being affected by other viruses present in the sample (Fig. 3; Table 2). For example, 4 samples that presented only HAdV were found positive by both microarray and RT-PCR, while in the remaining 9 samples, which presented HAdV coinfection with RV-A (8 samples) and CV (one sample), only the second virus was identified by microarray (Table 2). It should be pointed out that most of these samples contained a low level of HAdV genetic material, with CT values close to the nontemplate control value (44.5) (Table 2).
Detection of uncommon GI viruses.
Of note, the microarray found 3 viruses that usually are not evaluated in gastroenteritis samples. Two samples presented HPeV, both in coinfection (one with RV-A/TTV and another with SV/HEV B/TTV). An additional sample containing HBoV was identified (RV-A was identified by RT-PCR in this sample), and 12 samples presented TTV, 5 samples as single infection and 7 in coinfection with other viruses. As reference samples for these viruses were not available, confirmation RT-PCR coupled with capillary sequencing was performed, and the viruses detected by the microarray were confirmed in all these samples (results not shown). The fact that single TTV-positive samples were found is not an indicator of causation.
DISCUSSION
Current routine viral testing is designed to detect only the most prevalent viruses, frequently leaving 30 to 50% of cases without an agent identified (52). In recent years, advances in molecular biology and the implementation of next-generation sequencing has allowed the identification of several new viruses in intestinal samples (53, 54, 55, 56, 57). The roles of most of these viruses (Aichi virus, Anellovirus, Human bocavirus, Human parechovirus, Human picobirnavirus, and some enteroviruses, among others) in diarrheal disease remains unclear, raising the need to study in detail their epidemiology. In order to gather information on GI virus diversity, proper tools are required for their monitoring. In this work, a comprehensive and sensitive DNA microarray was developed and tested, which allows in principle the parallel detection of more than 100 gastrointestinal tract-associated virus species.
Implementation of a microarray for detection of viruses is not an easy task. Design of probes and experimental conditions are two important parameters to consider. Resequencing microarrays permit identification of mutations but require high numbers of probes for a single agent, increasing the cost (58). Arrays for subtyping use fewer and shorter probes but are often designed for only one viral species (59, 60, 61, 62, 63). Microarrays used for virus discovery have proven to be very useful when usual suspects are discarded or in cases of rare diseases, but identification is not clear and requires complex analysis (34).
Several DNA microarrays have been previously reported for identification of the main known gastrointestinal pathogenic viruses (59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75); however, they were oriented mostly to the identification or subtyping of one viral species, and none had specifically addressed the list of viruses that can be found in stool samples.
The microarray platform described in this work has been validated with 14 reference viral strains, representing 10 different virus species. Importantly, 5 other viral species were identified using the microarray when analyzing clinical samples: HAdV-F, HAdV-A, HPeV, HBoV, and several TTVs. The capacity of the microarray to correctly identify viruses whose probes were not validated in this work with cultured reference strains confirms that the methodology used to design probes is adequate and increases the probability that the remaining probes will be also capable to identify their target viruses, and this is additionally supported by in silico detection of a wide variety of RV-A strains using probes obtained from conserved genes; however, testing with other reference strains would be necessary. During the validation experiments, some probes were found to react nonspecifically with the amplified labeled DNA, regardless of its origin; in other words, they were found to be “sticky,” and they were excluded from further analysis. No common characteristic was found between these probes that could account for their nonspecific binding behavior.
One of the critical parameters in virus detection is the sensitivity of the assay. There are several factors that can affect the sensitivity. In the case of a microarray, sample nucleic acids are generally processed by random-primed amplification prior to hybridization to ensure amplification of a wide variety of viruses. The product of random PCR could be lower than that of specific PCR, decreasing the sensitivity of the assay, as all genetic material is amplified, diluting the positive signal (76). The limit of detection for three viruses with different genome types (dsDNA, dsRNA, and ssRNA+) was established at 103 virus particles, suggesting that the nature of the genome does not affect the sensitivity of the assay. Moreover, testing the sensitivity of the microarray with purified RV-A RNA, we observed that addition of 50 ng of cellular RNA as a nonspecific diluting RNA decreased the sensitivity of detection 10-fold. To try to solve the sensitivity problem in complex clinical samples, agent-specific primers have been included in previous studies, together with random primers during amplification of the genetic material (14, 15), with the disadvantage of narrowing the scope of targets for the microarray assay.
We subsequently analyzed a group of clinical samples collected from children with diarrhea. Initially, the clinical samples were screened by SDS-PAGE, which led to the identification of 34 RV-A-positive samples, while the microarray presented in this work identified 44, suggesting that the microarray platform has a higher sensitivity than traditional methods. A similar sensitivity was obtained by RT-PCR, as 42 samples were found to be RV-A positive. Even though our results indicate that the limit of detection of purified virus (1 × 103 viral particles) is similar to that reached with PCR assays (8), the microarray had a higher number of positive results when clinical samples were tested, possibly due to the natural genetic variation in primer binding regions of viruses found in sample viruses.
Although multiplexed assays are being developed, their use in routine testing is not generally implemented, and most studies use single-pathogen tests. When RT-PCR screening for the most common viruses is performed, the percentage of clinical samples without a virus identified remains around 30 to 50% (13, 77, 78), while the microarray presented in this study detected a virus in 92% of the samples. This high detection rate could have been influenced by the time of sampling, since winter is a high season for viral gastroenteritis in the region and no preselection for pathogens was performed. An additional advantage of the microarray test compared to a set of different RT-PCR assays is the capacity to identify viruses that are not commonly tested for, such as those previously associated with diarrhea (like HPeV) and those of unclear clinical significance in GI disease (HBoV and TTV). In this work we found a wide range of circulating atypical viruses among children, similarly as observed in other studies (79, 80), and their continuous surveillance should be considered. To our knowledge, this is first report of HPeV, HBoV, and TTV in Mexican children.
As a consequence of the limited number of virus species routinely tested, the prevalence of coinfections is a poorly explored issue. Usually, when a panel of up to 5 viruses is used, coinfection rates of between 4 and 18% are observed, with the most common combination being RV-A/NV (2, 13, 77, 81, 82, 83). More recently, wide-ranging metagenomic studies have shown that mixed infections are more common than previously thought (4, 80), even in healthy individuals (79). The analysis of the small set of clinical samples in this work showed that 30% (23 out of 76) contained more than 1 gastrointestinal virus. The identification of individual viruses in coinfections presented some discrepancies when comparing the results from microarray and RT-PCR tests. Of seven samples with mixed infections identified by the microarray, five were confirmed by RT-PCR, while in 16 mixed infections identified by RT-PCR, a single virus was identified by the microarray, suggesting that the microarray may be less sensitive than RT-PCR for detection of mixed infections. To address this inconsistency, real-time RT-PCR was implemented for the principal combinations of viruses that were missed by the microarray. This platform showed a certain advantage for detection of RV-A over HAdV and HEV, as RV-A was identified even when the HEV genome was present in larger amounts. HEV was identified by the microarray in samples coinfected with RV-A only when RV-A RNA was present in small amounts, close to negative-control levels (Table 2). Preferential identification of RV-A by the microarray could be due to the large amount of virus particles excreted during the acute phase of infection and to the large number of probes selected (42 oligonucleotides, compared to 5 and 17 probes for HEV A and HEV C, respectively, and 17 probes for HAdV). On the other hand, the two HEV samples positive by microarray that were missed by RT-PCR correspond to mixed infections with HAstV/TTV and SV/HpeV/TTV, respectively. Several attempts to identify HEV in these samples by RT-PCR resulted in negative results, and thus the possibility of a microarray false-positive result cannot be discarded.
The number of virus species identified has increased considerably in the last decade with the application of emerging genomic technologies such as microarrays and unbiased next-generation sequencing in studies of fatal or rare cases of disease in humans and in wild and domestic animals (25, 56, 84, 85, 86). Adequate tools that allow detection of well-known pathogenic viruses while being capable of detecting the new or rare viruses in a single assay will contribute useful epidemiological information about both kinds of viruses. This microarray includes viruses of different host origins in order to extend the range of use to veterinary studies. The oligonucleotide probes selected should allow the identification of target viruses despite the sequence variations that will occur in the future; however, it will be important to update the microarray design on a regular basis to maintain the capacity to broadly detect pathogenic viruses and to include newly found viral species.
Parallel detection of gastroenteric viruses beyond the most common viruses should facilitate a better understanding of virus etiology, as it increases the rate of positive cases, closing the diagnostic gap, and allows inspection for mixed infections where secondary viral agents could represent an important factor. Adding data from case-control studies and inclusion of other host parameters, such as serological data, will help to provide evidence of virus pathogenicity. Furthermore, adequate and comprehensive epidemiological studies in wild and domestic animals should be considered.
Supplementary Material
ACKNOWLEDGMENTS
The computational analysis was performed using the cluster of the Instituto de Biotecnología, UNAM, with the assistance of M. C. Jerome Verleyen. We thank Paul Gaytán Colín, Eugenio López-Bustos, and Santiago Becerra Ramírez from Unidad de Síntesis y Secuenciación de DNA, Instituto de Biotecnología, UNAM, for synthesis of the oligonucleotides used in RT-PCR assays.
This work was supported by grant S0008-111593 from CONACYT. Miguel A. Martínez and María de los Dolores Soto-del Río were supported by a scholarship from CONACYT-Mexico.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01317-14.
REFERENCES
- 1.WHO. 2011. World health statistics 2011. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Simpson VR. 2002. Wild animals as reservoirs of infectious diseases in the UK. Vet J 163:128–146. doi: 10.1053/tvjl.2001.0662. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelmi I, Roman E, Sanchez-Fauquier A. 2003. Viruses causing gastroenteritis. Clin Microbiol Infect 9:247–262. doi: 10.1046/j.1469-0691.2003.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rovida F, Campanini G, Sarasini A, Adzasehoun KM, Piralla A, Baldanti F. 2013. Comparison of immunologic and molecular assays for the diagnosis of gastrointestinal viral infections. Diagn Microbiol Infect Dis 75:110–111. doi: 10.1016/j.diagmicrobio.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Allard A, Albinsson B, Wadell G. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol 39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X. 2004. Genetic diversity among sapoviruses. Arch Virol 149:1309–1323. doi: 10.1007/s00705-004-0296-9. [DOI] [PubMed] [Google Scholar]
- 7.Royuela E, Negredo A, Sanchez-Fauquier A. 2006. Development of a one step real-time RT-PCR method for sensitive detection of human astrovirus. J Virol Methods 133:14–19. doi: 10.1016/j.jviromet.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz BA, Bange R, Vahlenkamp TW, Johne R, Muller H. 2002. Detection and quantitation of group A rotaviruses by competitive and real-time reverse transcription-polymerase chain reaction. J Virol Methods 105:277–285. doi: 10.1016/S0166-0934(02)00118-0. [DOI] [PubMed] [Google Scholar]
- 9.Coupland LJ, McElarney I, Meader E, Cowley K, Alcock L, Naunton J, Gray J. 2013. Simultaneous detection of viral and bacterial enteric pathogens using the Seeplex(R) Diarrhea ACE detection system. Epidemiol Infect 141:2111–2121. doi: 10.1017/S0950268812002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khamrin P, Okame M, Thongprachum A, Nantachit N, Nishimura S, Okitsu S, Maneekarn N, Ushijima H. 2011. A single-tube multiplex PCR for rapid detection in feces of 10 viruses causing diarrhea. J Virol Methods 173:390–393. doi: 10.1016/j.jviromet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Rohayem J, Berger S, Juretzek T, Herchenroder O, Mogel M, Poppe M, Henker J, Rethwilm A. 2004. A simple and rapid single-step multiplex RT-PCR to detect norovirus, astrovirus and adenovirus in clinical stool samples. J Virol Methods 118:49–59. doi: 10.1016/j.jviromet.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Svraka S, van der Veer B, Duizer E, Dekkers J, Koopmans M, Vennema H. 2009. Novel approach for detection of enteric viruses to enable syndrome surveillance of acute viral gastroenteritis. J Clin Microbiol 47:1674–1679. doi: 10.1128/JCM.00307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Maarseveen NM, Wessels E, de Brouwer CS, Vossen AC, Claas EC. 2010. Diagnosis of viral gastroenteritis by simultaneous detection of adenovirus group F, astrovirus, rotavirus group A, norovirus genogroups I and II, and sapovirus in two internally controlled multiplex real-time PCR assays. J Clin Virol 49:205–210. doi: 10.1016/j.jcv.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Palacios G, Quan PL, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Zhai J, Renwick N, Hui J, Hegyi H, Grolla A, Strong JE, Towner JS, Geisbert TW, Jahrling PB, Buchen-Osmond C, Ellerbrok H, Sanchez-Seco MP, Lussier Y, Formenty P, Nichol MS, Feldmann H, Briese T, Lipkin WI. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan PL, Palacios G, Jabado OJ, Conlan S, Hirschberg DL, Pozo F, Jack PJ, Cisterna D, Renwick N, Hui J, Drysdale A, Amos-Ritchie R, Baumeister E, Savy V, Langer KM, Rucht JA, Boyle DB, Garcia-Sastre A, Casas I, Perez-Brena P, Briese T, Lipkin WI. 2007. Detection of respiratory viruses and subtype identification of influenza A viruses by GreeneChipResp oligonucleotide microarray. J Clin Microbiol 45:2359–2364. doi: 10.1128/JCM.00737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sengupta S, Onodera K, Lai A, Melcher U. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J Clin Microbiol 41:4542–4550. doi: 10.1128/JCM.41.10.4542-4550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao-Ping K, Yong-Qiang L, Qing-Ge S, Hong L, Qing-Yu Z, Yin-Hui Y. 2009. Development of a consensus microarray method for identification of some highly pathogenic viruses. J Med Virol 81:1945–1950. doi: 10.1002/jmv.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Striebel HM, Birch-Hirschfeld E, Egerer R, Foldes-Papp Z, Tilz GP, Stelzner A. 2004. Enhancing sensitivity of human herpes virus diagnosis with DNA microarrays using dendrimers. Exp Mol Pathol 77:89–97. doi: 10.1016/j.yexmp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhaohui S, Wenling Z, Bao Z, Rong S, Wenli M. 2004. Microarrays for the detection of HBV and HDV. J Biochem Mol Biol 37:546–551. doi: 10.5483/BMBRep.2004.37.5.546. [DOI] [PubMed] [Google Scholar]
- 20.Boriskin YS, Rice PS, Stabler RA, Hinds J, Al-Ghusein H, Vass K, Butcher PD. 2004. DNA microarrays for virus detection in cases of central nervous system infection. J Clin Microbiol 42:5811–5818. doi: 10.1128/JCM.42.12.5811-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. 2002. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A 99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtz LR, Finkbeiner SR, Zhao G, Kirkwood CD, Girones R, Pipas JM, Wang D. 2009. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol J 6:86. doi: 10.1186/1743-422X-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones MS II, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J Virol 81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor A, Victoria J, Simmonds P, Wang C, Shafer RW, Nims R, Nielsen O, Delwart E. 2008. A highly divergent picornavirus in a marine mammal. J Virol 82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, Mason CJ, Pandey P, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besses JM, Bartkus JM, Delwart EL. 2009. A novel picornavirus associated with gastroenteritis. J Virol 83:12002–12006. doi: 10.1128/JVI.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rockett RJ, Sloots TP, Bowes S, O'Neill N, Ye S, Robson J, Whiley DM, Lambert SB, Wang D, Nissen MD, Bialasiewicz S. 2013. Detection of novel polyomaviruses, TSPyV, HPyV6, HPyV7, HPyV9 and MWPyV in feces, urine, blood, respiratory swabs and cerebrospinal fluid. PLoS One 8:e62764. doi: 10.1371/journal.pone.0062764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarate S, Espinosa R, Romero P, Guerrero CA, Arias CF, Lopez S. 2000. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50–54. doi: 10.1006/viro.2000.0660. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 29.Paulin LF, Soto-Del Rio Mde L, Sanchez I, Hernandez J, Gutierrez-Rios RM, Lopez-Martinez I, Wong-Chew RM, Parissi-Crivelli A, Isa P, Lopez S, Arias CF. 2014. PhyloFlu, a DNA microarray for determining the phylogenetic origin of influenza A virus gene segments and the genomic fingerprint of viral strains. J Clin Microbiol 52:803–813. doi: 10.1128/JCM.03134-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Z, Wu L, Fields MW, Zhou J. 2005. Use of microarrays with different probe sizes for monitoring gene expression. Appl Environ Microbiol 71:5154–5162. doi: 10.1128/AEM.71.9.5154-5162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Cofrey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsey PS. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 32.Wang HY, Malek RL, Kwitek AE, Greene AS, Luu TV, Behbahani B, Frank B, Quackenbush J, Lee NH. 2003. Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol 4:R5. doi: 10.1186/gb-2003-4-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SantaLucia J., Jr 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urisman A, Fischer KF, Chiu CY, Kistler AL, Beck S, Wang D, DeRisi JL. 2005. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol 6:R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohlander SK, Espinosa R III, Le Beau MM, Rowley JD, Diaz MO. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322–1324. doi: 10.1016/0888-7543(92)90057-Y. [DOI] [PubMed] [Google Scholar]
- 36.Chen EC, Miller SA, DeRisi JL, Chiu CY. 27 April 2011. Using a pan-viral microarray assay (Virochip) to screen clinical samples for viral pathogens. J Vis Exp doi: 10.3791/2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 38.Strimmer K. 2008. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 40.Teo J, Di Pietro P, San Biagio F, Capozzoli M, Deng YM, Barr I, Caldwell N, Ong KL, Sato M, Tan R, Lin R. 2011. VereFlu: an integrated multiplex RT-PCR and microarray assay for rapid detection and identification of human influenza A and B viruses using lab-on-chip technology. Arch Virol 156:1371–1378. doi: 10.1007/s00705-011-0999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotbart HA. 1990. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol 28:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen AC, Gyhrs ML, Nielsen LP, Pedersen C, Bottiger B. 2013. Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J Clin Virol 57:239–242. doi: 10.1016/j.jcv.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, DeRisi JL. 2009. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol J 6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biagini P, Uch R, Belhouchet M, Attoui H, Cantaloube JF, Brisbarre N, de Micco P. 2007. Circular genomes related to anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequence-independent single primer amplification approach. J Gen Virol 88:2696–2701. doi: 10.1099/vir.0.83071-0. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto H, Takahashi M, Nishizawa T, Tawara A, Fukai K, Muramatsu U, Naito Y, Yoshikawa A. 2002. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J Gen Virol 83:1291–1297. [DOI] [PubMed] [Google Scholar]
- 47.Chu DK, Poon LL, Guan Y, Peiris JS. 2008. Novel astroviruses in insectivorous bats. J Virol 82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khamrin P, Maneekarn N, Peerakome S, Okitsu S, Mizuguchi M, Ushijima H. 2008. Bovine kobuviruses from cattle with diarrhea. Emerg Infect Dis 14:985–986. doi: 10.3201/eid1406.070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerg Infect Dis 14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee N, Chan MC, Wong B, Choi KW, Sin W, Lui G, Chan PK, Lai RW, Cockram CS, Sung JJ, Leung WK. 2007. Fecal viral concentration and diarrhea in norovirus gastroenteritis. Emerg Infect Dis 13:1399–1401. doi: 10.3201/eid1309.061535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramani S, Sankaran P, Arumugam R, Sarkar R, Banerjee I, Mohanty I, Jana AK, Kuruvilla KA, Kang G. 2010. Comparison of viral load and duration of virus shedding in symptomatic and asymptomatic neonatal rotavirus infections. J Med Virol 82:1803–1807. doi: 10.1002/jmv.21872. [DOI] [PubMed] [Google Scholar]
- 52.Dennehy PH. 2011. Viral gastroenteritis in children. Pediatr Infect Dis J 30:63–64. doi: 10.1097/INF.0b013e3182059102. [DOI] [PubMed] [Google Scholar]
- 53.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. 2003. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. 2009. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin B, Blaney KM, Malanoski AP, Ligler AG, Schnur JM, Metzgar D, Russell KL, Stenger DA. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J Clin Microbiol 45:443–452. doi: 10.1128/JCM.01870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brinkman NE, Fout GS. 2009. Development and evaluation of a generic tag array to detect and genotype noroviruses in water. J Virol Methods 156:8–18. doi: 10.1016/j.jviromet.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Brown DW, Gunning KB, Henry DM, Awdeh ZL, Brinker JP, Tzipori S, Herrmann JE. 2008. A DNA oligonucleotide microarray for detecting human astrovirus serotypes. J Virol Methods 147:86–92. doi: 10.1016/j.jviromet.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chizhikov V, Wagner M, Ivshina A, Hoshino Y, Kapikian AZ, Chumakov K. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J Clin Microbiol 40:2398–2407. doi: 10.1128/JCM.40.7.2398-2407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lovmar L, Fock C, Espinoza F, Bucardo F, Syvanen AC, Bondeson K. 2003. Microarrays for genotyping human group a rotavirus by multiplex capture and type-specific primer extension. J Clin Microbiol 41:5153–5158. doi: 10.1128/JCM.41.11.5153-5158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinje J, Koopmans MP. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J Clin Microbiol 38:2595–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayodeji M, Kulka M, Jackson SA, Patel I, Mammel M, Cebula TA, Goswami BB. 2009. A microarray based approach for the identification of common foodborne viruses. Open Virol J 3:7–20. doi: 10.2174/1874357900903010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H, Mammel M, Kulka M, Patel I, Jackson S, Goswami BB. 2011. Detection and identification of common food-borne viruses with a tiling microarray. Open Virol J 5:52–59. doi: 10.2174/1874357901105010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu YQ. 2010. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology 53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honma S, Chizhikov V, Santos N, Tatsumi M, Timenetsky Mdo C, Linhares AC, Mascarenhas JD, Ushijima H, Armah GE, Gentsch JR, Hoshino Y. 2007. Development and validation of DNA microarray for genotyping group A rotavirus VP4 (P[4], P[6], P[8], P[9], and P[14]) and VP7 (G1 to G6, G8 to G10, and G12) genes. J Clin Microbiol 45:2641–2648 doi: 10.1128/JCM.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaaskelainen AJ, Piiparinen H, Lappalainen M, Koskiniemi M, Vaheri A. 2006. Multiplex-PCR and oligonucleotide microarray for detection of eight different herpesviruses from clinical specimens. J Clin Virol 37:83–90. doi: 10.1016/j.jcv.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Kim JM, Kim SY, Park YB, Kim HJ, Min BS, Cho JC, Yang JM, Cho YH, Ko G. 2012. Simultaneous detection of major enteric viruses using a combimatrix microarray. J Microbiol 50:970–977. doi: 10.1007/s12275-012-2228-9. [DOI] [PubMed] [Google Scholar]
- 70.Leblanc N, Gantelius J, Schwenk JM, Stahl K, Blomberg J, Andersson-Svahn H, Belak S. 2009. Development of a magnetic bead microarray for simultaneous and simple detection of four pestiviruses. J Virol Methods 155:1–9. doi: 10.1016/j.jviromet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Lin B, Vora GJ, Thach D, Walter E, Metzgar D, Tibbetts C, Stenger DA. 2004. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J Clin Microbiol 42:3232–3239. doi: 10.1128/JCM.42.7.3232-3239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lopez-Campos G, Coiras M, Sanchez-Merino JP, Lopez-Huertas MR, Spiteri I, Martin-Sanchez F, Perez-Brena P. 2007. Oligonucleotide microarray design for detection and serotyping of human respiratory adenoviruses by using a virtual amplicon retrieval software. J Virol Methods 145:127–136. doi: 10.1016/j.jviromet.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 73.Mattison K, Corneau N, Berg I, Bosch A, Duizer E, Gutierrez-Aguirre I, L'Homme Y, Lucero Y, Luo Z, Martyres A, Myrmel M, O'Ryan M, Pagotto F, Sano D, Svraka S, Urzua U, Bidawid S. 2011. Development and validation of a microarray for the confirmation and typing of norovirus RT-PCR products. J Virol Methods 173:233–250. doi: 10.1016/j.jviromet.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 74.Shih SR, Wang YW, Chen GW, Chang LY, Lin TY, Tseng MC, Chiang C, Tsao KC, Huang CG, Shio MR, Tai JH, Wang SH, Kuo RL, Liu WT. 2003. Serotype-specific detection of enterovirus 71 in clinical specimens by DNA microchip array. J Virol Methods 111:55–60. doi: 10.1016/S0166-0934(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 75.Susi P, Hattara L, Waris M, Luoma-Aho T, Siitari H, Hyypia T, Saviranta P. 2009. Typing of enteroviruses by use of microwell oligonucleotide arrays. J Clin Microbiol 47:1863–1870. doi: 10.1128/JCM.02226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vora GJ, Meador CE, Stenger DA, Andreadis JD. 2004. Nucleic acid amplification strategies for DNA microarray-based pathogen detection. Appl Environ Microbiol 70:3047–3054. doi: 10.1128/AEM.70.5.3047-3054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pang XL, Preiksaitis JK, Lee BE. 2014. Enhanced enteric virus detection in sporadic gastroenteritis using a multi-target real-time PCR panel: a one-year study. J Med Virol 86:1594–1601. doi: 10.1002/jmv.23851. [DOI] [PubMed] [Google Scholar]
- 78.Simpson R, Aliyu S, Iturriza-Gomara M, Desselberger U, Gray J. 2003. Infantile viral gastroenteritis: on the way to closing the diagnostic gap. J Med Virol 70:258–262. doi: 10.1002/jmv.10386. [DOI] [PubMed] [Google Scholar]
- 79.Kapusinszky B, Minor P, Delwart E. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitui MT, Bozdayi G, Ahmed S, Matsumoto T, Nishizono A, Ahmed K. 2014. Detection and molecular characterization of diarrhea causing viruses in single and mixed infections in children: a comparative study between Bangladesh and Turkey. J Med Virol 86:1159–1168. doi: 10.1002/jmv.23744. [DOI] [PubMed] [Google Scholar]
- 81.Koh H, Baek SY, Shin JI, Chung KS, Jee YM. 2008. Coinfection of viral agents in Korean children with acute watery diarrhea. J Korean Med Sci 23:937–940. doi: 10.3346/jkms.2008.23.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roman E, Wilhelmi I, Colomina J, Villar J, Cilleruelo ML, Nebreda V, Del Alamo M, Sanchez-Fauquier A. 2003. Acute viral gastroenteritis: proportion and clinical relevance of multiple infections in Spanish children. J Med Microbiol 52:435–440. doi: 10.1099/jmm.0.05079-0. [DOI] [PubMed] [Google Scholar]
- 83.Tran A, Talmud D, Lejeune B, Jovenin N, Renois F, Payan C, Leveque N, Andreoletti L. 2010. Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in northern France. J Clin Microbiol 48:1943–1946. doi: 10.1128/JCM.02181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belak S, Karlsson OE, Blomstrom AL, Berg M, Granberg F. 2013. New viruses in veterinary medicine, detected by metagenomic approaches. Vet Microbiol 165:95–101. doi: 10.1016/j.vetmic.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 85.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 86.Bodewes R, van de Bildt MW, Schapendonk CM, van Leeuwen M, van Boheemen S, de Jong AA, Osterhaus AD, Smits SL, Kuiken T. 2013. Identification and characterization of a novel adenovirus in the cloacal bursa of gulls. Virology 440:84–88. doi: 10.1016/j.virol.2013.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.