Abstract
The etiology of an outbreak of gastroenteritis in humans cannot always be determined, and ∼25% of outbreaks remain unsolved in New Zealand. It is hypothesized that novel viruses may account for a proportion of unsolved cases, and new unbiased high-throughput sequencing methods hold promise for their detection. Analysis of the fecal metagenome can reveal the presence of viruses, bacteria, and parasites which may have evaded routine diagnostic testing. Thirty-one fecal samples from 26 gastroenteritis outbreaks of unknown etiology occurring in New Zealand between 2011 and 2012 were selected for de novo metagenomic analysis. A total data set of 193 million sequence reads of 150 bp in length was produced on an Illumina MiSeq. The metagenomic data set was searched for virus and parasite sequences, with no evidence of novel pathogens found. Eight viruses and one parasite were detected, each already known to be associated with gastroenteritis, including adenovirus, rotavirus, sapovirus, and Dientamoeba fragilis. In addition, we also describe the first detection of human parechovirus 3 (HPeV3) in Australasia. Metagenomics may thus provide a useful audit tool when applied retrospectively to determine where routine diagnostic processes may have failed to detect a pathogen.
INTRODUCTION
Outbreaks of acute gastroenteritis are associated with significant global morbidity and mortality, particularly in pediatric populations (1). Bacterial pathogens are well-described causes of gastroenteritis, e.g., Escherichia coli O157:H7, Salmonella, Campylobacter, and Shigella (2, 3), but are known to occur less frequently than viral pathogens as the cause of acute gastroenteritis (4). Noroviruses, sapoviruses, group A rotaviruses, enteric adenoviruses, and astroviruses have been recognized as the main etiological agents in viral gastroenteritis, although it has been suggested that a number of other viruses may also be involved (5, 6). Parasites such as Dientamoeba fragilis have also been implicated in gastroenteritis; however, there is still debate surrounding their pathogenicity (2, 7).
In New Zealand, no causative agent is identified in approximately 25% of reported gastroenteritis outbreaks at the conclusions of public health investigations (8). A proportion of unsolved cases may be due to the presence of a novel pathogen. Viral pathogens are particularly problematic to discover because well-established methods such as electron microscopy, PCR, or viral culture are not always effective; they may be too specific, lack high-throughput capability, or not be sensitive enough to detect low numbers of organisms within a sample (9). High-throughput sequencing holds promise for resolving the etiology of unsolved gastroenteritis outbreaks, as large volumes of unbiased metagenomic data are produced, allowing for the sequences from all viruses, bacteria, parasites, and fungi present in the sample to be revealed (10). Any knowledge gained from identifying previously unknown pathogens causing outbreaks of gastroenteritis will facilitate public health investigations, allowing for possible identification of the source of infection and informing measures for intervention (6).
Most metagenomic studies of undiagnosed gastroenteritis have focused on identifying the cause of sporadic gastrointestinal disease in individual patients (11, 12) rather than examining outbreaks. To date, there are only three reports of outbreaks that have been investigated using a metagenomic approach (13–15). A large proportion of viruses are yet to be discovered (16), and the number of documented human viruses is incomplete, with at least one new human virus per year expected to be discovered (17). Accordingly, the aim of the present study was to apply metagenomic analysis to unsolved gastrointestinal outbreaks in New Zealand to reveal points where conventional diagnostic algorithms may have failed to detect known viral pathogens and to seek potentially novel viruses associated with gastroenteritis.
MATERIALS AND METHODS
Sample selection and preparation.
Fecal samples from gastroenteritis cases in New Zealand can be tested by diagnostic laboratories for noroviruses, rotaviruses, sapoviruses, astroviruses, or adenovirus types 40 and 41. Bacteria for which samples can be tested include E. coli, Campylobacter spp., Salmonella spp., Vibrio spp., Shigella spp., Listeria monocytogenes, Clostridium perfringens, Staphylococcus aureus, and Bacillus cereus; bacterial enterotoxins may also be sought. Samples may also be tested for the parasites Giardia and Cryptosporidium. Thirty-one fecal samples were randomly selected from a reference laboratory collection of samples from 26 gastroenteritis outbreaks occurring between 2011 and 2012 (8), where no causative agents had been previously identified at the time of sample selection. Continued public health investigations subsequently identified causative agents in five of these outbreaks from linked samples that were examined and reported by other laboratories after the conclusion of the selection process. Campylobacter species were implicated in three outbreaks. Two of the outbreaks were shown to have cases of norovirus present, but the norovirus-positive samples were not available and therefore would not have been included in the group of 31 anonymous samples.
Fecal samples were resuspended in 2 ml of phosphate-buffered saline (PBS), incubated for 1 h at 4°C, and centrifuged at 6,000 × g for 5 min to remove cellular debris and bacteria; the supernatant was then filtered through a 0.45-μm syringe filter (Millipore, Billerica, MA). Nucleic acid was extracted from 400 μl of filtered supernatant and eluted in 50 μl of molecular-grade water using the iPrep PureLink virus kit (Life Technologies, Carlsbad, CA), producing a metagenome representative for each sample.
High-throughput sequencing.
A metagenome of the RNA present in each sample was achieved by removing DNA from the extracted nucleic acid using Ambion DNA-free (Life Technologies) as per the manufacturer's instructions. Following treatment, 8 μl of DNA-free RNA was incorporated into first-strand cDNA synthesis primed by random hexamers (Life Technologies). cDNA was then amplified by multiple-displacement amplification in a whole-transcriptome amplification kit (Qiagen, Valencia, CA), producing more than 1 μg of DNA. DNA libraries were then prepared using the Illumina TruSeq DNA library preparation kit V2 (Illumina, San Diego, CA), followed by sequencing of 150-bp paired-end reads on an Illumina MiSeq instrument (New Zealand Genomics Limited, Massey Genome Service, Massey University, Palmerston North, New Zealand).
This method not only is capable of detecting RNA viruses but also is capable of detecting DNA viruses due to the presence of viral mRNA or of DNA that has evaded digestion due to the inefficiency of DNase enzymes (18).
Bioinformatics.
The quality of the sequence data from a total of 15 MiSeq runs was checked using FastQC (version 0.10.1; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The total read length of 150 bp was trimmed where the average quality score was <30. Duplicate reads were collapsed using FASTX-Toolkit (version 0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/index.html). Velvet (version 1.2.07) (19) was used for de novo assembly of the trimmed sequence data with a k-mer of 75. Contigs were compared to the GenBank nonredundant nucleotide sequence database using BLASTN from BLAST+ (version 2.2.27) (20) and an E value threshold for reporting of 0.001. BLAST outputs were visualized in MEGAN (version 4.7) (21) for taxonomic assignment. A protein-level homology search was also performed by using RAPSearch2 (22) in order to identify highly divergent viruses.
Confirmatory PCR and quantitative PCR (qPCR) assays.
Sequences for human parechovirus type 3 (HPeV3) identified in high-throughput sequencing data were confirmed by customized conventional one-step reverse transcription-PCR (RT-PCR) assay. Primers were designed in Geneious 6.1.3 (Biomatters, New Zealand) targeting a 180-bp region of the HPeV3 VP1 gene (HPeV3_VP1_F2 [5′-GCTGGTGAGCAGATGACACT-3′] and HPeV3_VP1_R [5′-GGCTGGTACGGGGAAAAAGA-3′]). A 25-μl reaction volume for the Invitrogen SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Life Technologies) was used as per the manufacturer's instructions, including 5 μl of RNA-DNA and thermocycling parameters as follows: 50°C for 30 min; 85°C for 5 min; 95°C for 2 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 30 s; and a final extension of 68°C for 5 min. A PCR product of 180 bp was identified by microchip electrophoresis (MultiNA; Shimadzu, Kyoto, Japan) and then sequenced by the Sanger method on a capillary sequencer (model 3100 Avant; Applied Biosystems).
Confirmatory testing for D. fragilis was conducted using a qPCR assay targeting the 5.8S rRNA gene (23), with some modifications. A 25-μl reaction mixture consisted of 10 μl of PerfeCTa qPCR ToughMix Low Rox (Quanta Biosciences, Gaithersburg, MD), 3 pmol of each D. fragilis-specific primer, 5 pmol of D. fragilis-specific probe, and 5 μl of DNA. Thermocycling parameters were 15 min at 95°C followed by 45 cycles of 15 s at 95°C and 60 s at 60°C.
Phylogenetic analysis for human parechovirus type 3 sequence.
Sequences were aligned using ClustalW (24) and phylogenetic trees inferred by MEGA5 (25). The Tamura-Nei model selected was based on the lowest Bayesian informative criterion values from a model test within MEGA5.
Ethical approval.
This study was approved by the New Zealand Health & Disabilities Ethics Committee (approval number LRS/11/EXP/026). An amendment to this study was also approved (approval number LRS/11/EXP/026/AM02).
RESULTS
Identification of candidate pathogens using metagenomics.
A total of 193,505,576 sequence reads were produced for the 31 samples, with an average of 3,121,058 reads per sample. These sequences were assembled into 552,057 contigs; 62% of these could be assigned to a taxonomic group using MEGAN, and bacteria were the most predominant (data not shown). The delineation of bacterial species was not performed, as this requires targeted amplification of a conserved gene region, e.g., 16S rRNA. The de novo metagenomic data set does not target specific genes and was therefore analyzed for the purpose of identifying viruses and/or parasites. Eight viruses and one parasite (D. fragilis) were identified as candidates for gastroenteritis etiology (Table 1). Additional statistics on the metagenomic data are presented in Table 2. The RAPSearch2 results did not reveal any novel viral sequences but were concordant with the BLASTN results (data not shown).
TABLE 1.
Candidate pathogenic organisms detected in metagenomic data from unsolved outbreaks of gastroenteritis, and results of independent confirmatory testing
| Metagenomic data |
Independent test |
Disease(s) already known to be associated with the candidate pathogen | |||
|---|---|---|---|---|---|
| Candidate pathogen | No. of samples | % of total no. of samples | Method(s) | Result | |
| Adenovirus | 1 | 3 | qPCR | Confirmed | Gastroenteritis, respiratory illness |
| Human enterovirus B | 2 | 7 | qPCR | Confirmeda | Paralysis; gastrointestinal symptomsb; hand, foot and mouth disease |
| Human parechovirus type 3 | 1 | 3 | qPCR | Confirmed | Neonatal sepsis, encephalitis, paralysis |
| Picobirnavirus | 14 | 45 | Gastroenteritis | ||
| Influenza A virus | 1 | 3 | qPCR | Not confirmed | Respiratory illness, gastrointestinal symptoms |
| Pepper mild mottle virus | 3 | 10 | Plant virus; link to abdominal pain in humans (44) | ||
| Rotavirus | 1 | 3 | qPCR, ICAc | Confirmed | Gastroenteritis |
| Sapovirus | 1 | 3 | qPCR | Confirmed | Gastroenteritis |
| Dientamoeba fragilis | 5 | 16 | qPCR | Confirmed | Possible link to gastroenteritis |
| None | 10 | 32 | |||
Confirmed in one sample only.
Gastrointestinal symptoms are classified as vomiting and diarrhea.
ICA, immunochromatographic assay (SD Bioline).
TABLE 2.
BLASTN output statistics for the longest contig matching a virus/parasite from each of the fecal samplesa
| Anonymous sample no. | Virus or parasite | Longest contig matching a virus/parasite (bp)b | BLASTN output statistics for the best hit recorded for the longest contig |
||||
|---|---|---|---|---|---|---|---|
| E value | Score | % identity | % query coverage | Best hitc | |||
| 4 | Picobirnavirus | 1,196 | 3.5E−126 | 512 | 96 | 23 | GQ915049 |
| 4 | D. fragilis | 198 | 8.0E−55 | 360 | 98 | 100 | JQ677148 |
| 5 | Influenza virus A | 151 | 2.0E−31 | 234 | 100 | 85 | CY188670 |
| 5 | D. fragilis | 151 | 1.0E−68 | 268 | 99 | 100 | AY730405 |
| 7 | Picobirnavirus | 540 | 3.0E−60 | 242 | 70 | 84 | GQ915026 |
| 7 | D. fragilis | 214 | 2.0E−43 | 407 | 98 | 100 | JQ677168 |
| 8 | Picobirnavirus | 796 | 6.0E−58 | 388 | 82 | 62 | AB517736 |
| 10 | Picobirnavirus | 592 | 3.5E−21 | 124 | 67 | 77 | KF861773 |
| 11 | Picobirnavirus | 1,738 | 0 | 2950 | 95 | 100 | KJ663816 |
| 12 | Picobirnavirus | 375 | 1.0E−04 | 57 | 82 | 14 | KF861769 |
| 12 | D. fragilis | 343 | 4.0E−112 | 642 | 100 | 100 | JQ677148 |
| 13 | Picobirnavirus | 579 | 4.0E−08 | 70 | 77 | 17 | AB517737 |
| 13 | Tobamovirus | 560 | 1.4E−171 | 678 | 99 | 62 | AB000709 |
| 13 | D. fragilis | 218 | 5.0E−64 | 253 | 99 | 100 | JQ677149 |
| 15 | Parechovirus | 268 | 1.0E−30 | 475 | 99 | 99 | AB759205 |
| 16 | Tobamovirus | 286 | 3.9E−143 | 572 | 100 | 100 | AB126003 |
| 17 | Picobirnavirus | 585 | 2.0E−10 | 77 | 79 | 15 | KC692366 |
| 18 | Rotavirus | 573 | 0 | 928 | 95 | 100 | JX567765 |
| 21 | Picobirnavirus | 257 | 3.0E−43 | 185 | 80 | 82 | AB517731 |
| 22 | Picobirnavirus | 288 | 1.0E−47 | 199 | 78 | 89 | AF246939 |
| 23 | Adenovirus | 238 | 1.0E−65 | 455 | 100 | 100 | AB901016 |
| 23 | Picobirnavirus | 562 | 3.0E−21 | 113 | 68 | 60 | AF246941 |
| 26 | Sapovirus | 341 | 5.0E−162 | 580 | 98 | 100 | AY237420 |
| 29 | Enterovirus | 822 | 0 | 969 | 86 | 99 | FJ460595 |
| 33 | Enterovirus | 233 | 1.2E−21 | 124 | 82 | 96 | GQ141875 |
| 35 | Picobirnavirus | 324 | 1.0E−137 | 499 | 94 | 100 | GQ915028 |
| 38 | Picobirnavirus | 1,603 | 0 | 750 | 70 | 92 | KJ663814 |
| 40 | Picobirnavirus | 1,195 | 1.0E−94 | 358 | 69 | 82 | AB517733 |
| 40 | Tobamovirus | 403 | 0.0E + 00 | 806 | 100 | 95 | AB000709 |
Samples that showed no evidence for any candidate virus were omitted.
Only matches to candidate vertebrate viruses, pepper mild mottle virus (PMMV), or Dientamoeba fragilis were considered. Plant viruses (except PMMV) and bacteriophage were excluded from the analysis.
GenBank accession number for the first sequence found to match the query in the BLASTN result.
Known pathogenic human viruses.
The common enteric viral pathogens human adenovirus, rotavirus, and sapovirus were detected in the metagenomic data. Human enterovirus B and influenza A virus sequences were also present (Table 1).
Fecal samples were resubmitted to the reference laboratory for repeat testing by qPCR (26–29) (http://www.who.int/influenza/resources/documents/molecular_diagnosis_influenza_virus_humans_update_201108.pdf). The presence of sapovirus and adenovirus in the original samples was confirmed, human enterovirus B could be detected in only one sample, and influenza A virus could not be detected in any samples. A rapid antigen immunoassay (SD Bioline Rota/Adeno rapid kit; Standard Diagnostics Inc., Yongin-si, South Korea) and qPCR (28) confirmed the presence of group A rotavirus.
Detection of D. fragilis and HPeV3.
Of particular interest was the detection of D. fragilis and HPeV3 sequences (Table 1). Permission was sought by ethical review (New Zealand Health & Disabilities Ethics Committee: approval number LRS/11/EXP/026/AM02) to return to the original reference collection of 64 identifiable samples from the 26 outbreaks to specifically test for HPeV3 and D. fragilis; epidemiological data were therefore also available.
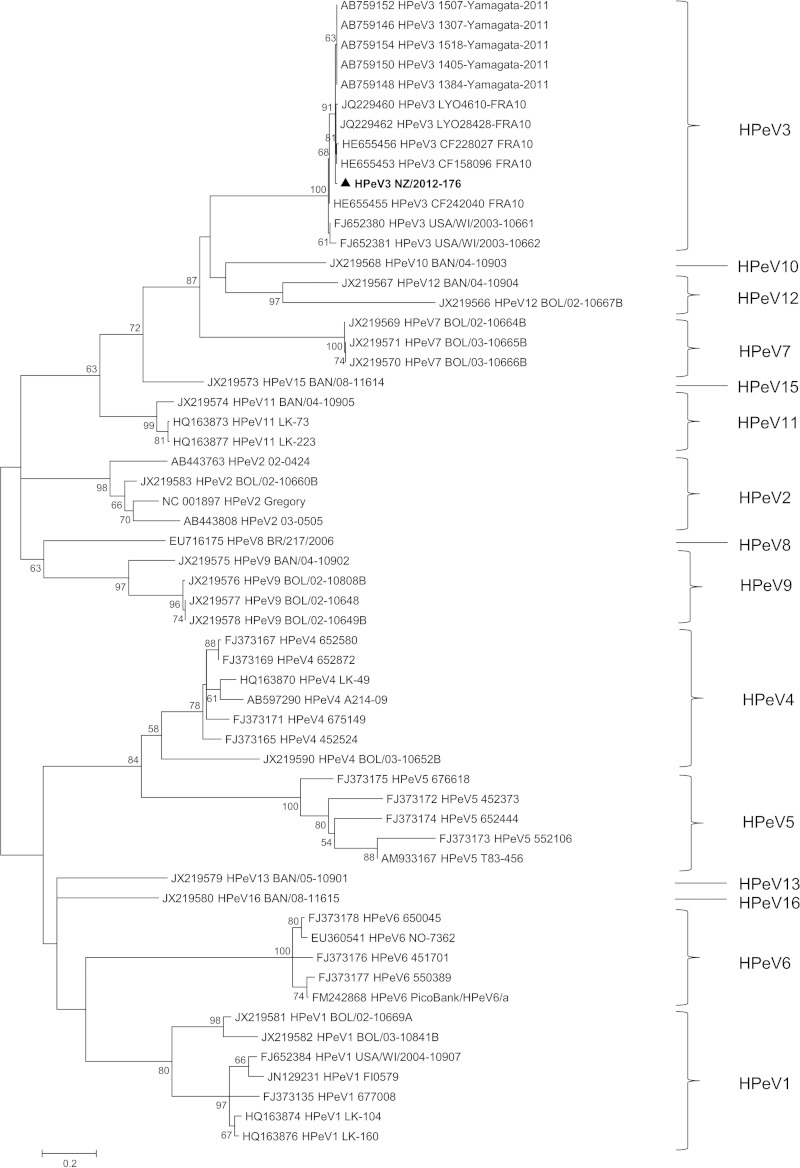
A customized one-step RT-PCR assay revealed HPeV3 in a fecal sample collected from a 2-year-old child as part of a gastroenteritis outbreak investigation at a child care facility; eight individuals were affected over a 17-day period. No other samples were available from linked cases. Phylogenetic analysis confirmed the identity of the virus as HPeV3 (Fig. 1), designated HPeV3 NZ/2012-176.
FIG 1.
Phylogenetic analysis showing the genetic relatedness of partial viral protein 1 (VP1) nucleotide sequences for human parechovirus type 3 identified in New Zealand (shown in boldface and indicated by a triangle) with known human parechoviruses. Evolutionary history was inferred for a total of 479 nucleotide sites by using a maximum likelihood method based on the Tamura-Nei model, with gamma distribution and invariant sites in MEGA5 software. Bootstrap values were calculated from 1,000 trees (only bootstrap values of >50% are shown). Scale bar indicates nucleotide substitutions per site.
Specific testing using qPCR for D. fragilis in the reference collection showed that this organism was present in seven samples from six outbreaks (Table 3).
TABLE 3.
Dientamoeba fragilis testing by qPCR in 64 unsolved outbreak samples
| Outbreak identifier | No. of samples testing positive for D. fragilis in outbreaka | Total no. of probable cases in outbreak | Outbreak setting | Symptom(s) |
|---|---|---|---|---|
| 11–1 | 1 | 8 | Child care center | Diarrhea, vomiting |
| 11–2 | 1 | 43 | Child care center | Fever, nausea and vomiting |
| 12–1 | 1 | 8 | Child care center | None given |
| 12–2 | 1 | 22 | Commercial food operator | Diarrhea |
| 12–3 | 1 | 8 | Hostel/institution | Diarrhea, vomiting |
| 12–4 | 2 | 28 | Child care center | Diarrhea, abdominal pain |
Denominator cannot be calculated for each outbreak, as the total number of samples available for testing is unknown.
DISCUSSION
In this study, metagenomic analysis using high-throughput sequencing was applied to reveal viruses present in human feces from unsolved outbreaks of gastroenteritis occurring in New Zealand between 2011 and 2012. No evidence for the presence of any novel viral agents was obtained, but there were numerous instances where known viral pathogens were detected, including well-known enteric viruses. The utility of metagenomics may therefore extend beyond the detection of novel organisms by providing a useful audit tool when applied retrospectively, thus highlighting any potential inadequacies in the routine diagnostic process.
Until recently, only two unsolved outbreaks of human gastroenteritis have been investigated using metagenomics and implicated novel viruses in the etiology (13, 14). A more recent study investigated seven outbreaks of gastroenteritis (and sporadic cases) and found the presence of novel viruses; however, none of these could be implicated in disease (15). The present study is one of the first to apply de novo metagenomics to a cohort of multiple unsolved outbreaks of gastroenteritis on a larger scale than has been previously attempted, with a total of 26 outbreaks included. It was hypothesized that novel viral agents would account for at least a portion of the etiology, but none were detected even when a protein-level homology search was applied (RapSearch2). Instead, the detection of known pathogens rendered the application of metagenomics akin to the use of a sophisticated multiplex PCR. These data do not support the hypothesis that there is a large unknown quotient of viruses accounting for the majority of unsolved outbreaks. However, completely novel or highly divergent viruses may be so different that they will escape detection regardless of whether nucleotide or protein searches are used. A more tenable explanation could be a “diagnostic gap,” a theory also suggested by others (5, 13, 30). The present study reveals instances where routine testing failed. For example, common enteric viruses were present in the metagenomic data; the most likely explanation for this is that the pathogens were not tested for in the first instance. The successful reapplication of routine diagnostic methods in the present study excludes the possibility that genomic changes would account for evasion of testing. There are two main reasons why a pathogen may not have been initially tested for: (i) there is an economic imperative to test only a selected number of cases from an outbreak, or (ii) the actual request for testing does not always include every agent; e.g., norovirus may be requested without consideration for rotavirus. However, definitive conclusions cannot be drawn as to how known viruses were missed, because anonymous samples in this study (as required by ethics) prevent access to previous test results or epidemiological information.
The most interesting clinical finding for this study is the detection of HPeV3 in New Zealand. HPeV3 was discovered in 2004 (31) and is now thought to be prevalent worldwide. It has yet to be reported in the Southern Hemisphere, with the exception of Bolivia (32). HPeV3 is mainly reported as a pediatric pathogen, particularly for children under 1 year of age. It is capable of causing more severe disease, such as neonatal sepsis, encephalitis, sudden unexpected death in infancy (SUDI), and paralysis (33–35), likely due in part to central nervous system involvement. Cases of HPeV3 infection may present with gastrointestinal and respiratory symptoms, but this virus has also been detected in healthy individuals (36, 37). The relevance of HPeV3 to the specific gastroenteritis outbreak in this study cannot be determined, especially given that no samples from linked cases were available. However, it is noted that no testing for HPeV3 is currently performed in New Zealand, and given that HPeV3 can cause severe disease, it is recommended that clinicians now consider testing for HPeV3.
Although the focus of this study was primarily on revealing viruses present in unsolved outbreaks of gastroenteritis, it was also possible to examine higher organisms within the metagenomic data, where genes that are specific for such organisms, e.g., parasites, allow for unequivocal identification. A strong etiological candidate was identified, the parasite D. fragilis. While the role of this parasite in gastroenteritis still remains unclear, it has been suggested that in the absence of another pathogen, D. fragilis may be considered a cause (7). In the present study, D. fragilis was detected in 7/64 (10.9%) outbreak samples, but interestingly, it showed a higher frequency in 4/9 (44%) outbreaks occurring in pediatric settings. Only one previous study has reported an estimate of D. fragilis prevalence in New Zealand, at ∼2% in one city in 1987 (38). Current routine testing in community pathology laboratories uses light microscopy, which has been shown to provide a sensitivity inferior to that of qPCR (7), and therefore may underestimate the prevalence of D. fragilis. Further testing for D. fragilis in a larger number of outbreak samples than of healthy control samples is therefore warranted to clarify any involvement of this pathogen in outbreaks of gastroenteritis.
Viruses with more circumspect involvement in gastroenteritis were also detected in the metagenomic data, e.g., pepper mild mottle virus, picobirnaviruses, human enterovirus B, and influenza A virus (Table 1). Pepper mild mottle virus is often found at high titers in human fecal samples due to its dietary origin (39), while picobirnavirus has been detected in 20% of human diarrheal samples of unknown etiology (40). Human enterovirus B and influenza A virus are respiratory pathogens, and while there is some evidence of gastrointestinal involvement (41, 42), it is unclear if they are involved in these outbreaks.
The aim of this study was to search for viruses, and thus, virus enrichment steps were incorporated into the methodology, such as the use of centrifugation and nuclease treatment. These methods decrease the amount of host and bacterial material, thus increasing the sensitivity of detection for viruses (18, 43). As previously reported, it was still possible to detect bacterial DNA, DNA viruses (e.g., adenovirus), and parasites (i.e., D. fragilis) in the metagenome. This is not unexpected; DNase treatment is not 100% efficient, allowing the detection of DNA viruses and bacteria when using RNA-targeted methods (18, 43). It has also been suggested that the identification of a DNA virus in data from an RNA-targeted approach may reflect the expression of viral mRNA transcribed from DNA genomes during infection (18).
There are still major issues to be resolved before metagenomics can be transitioned from the research setting into a clinical diagnostic laboratory. First, metagenomic studies, like many other diagnostic assays, are only able to detect the presence or absence of a pathogen; they do not provide any information on functional interactions or prove causality in the disease or outbreak. In the current study, while candidate viral pathogens were identified, causality could not be proven for their involvement in the outbreak, especially given that only one or two samples were available from each outbreak. Second, viruses with unique genomes that have no known relatives in the GenBank database could have been missed due to the absence of any homology to known sequences. As more viral genomes become available and bioinformatic tools are further developed, metagenomics will become more effective in the discovery of novel viruses. Third, it will be necessary for both clinicians and patients to consider issues that may attend unexpected findings and incidental detection of pathogens with significant health consequences. Randomized anonymous samples and ethical review as a specific research project avoided such an issue in this study. However, the use of anonymous samples also limits interpretations, as no clinical or epidemiological information relating to the outbreaks could be obtained. Lastly, even though sequencing hardware is well developed, the bioinformatic software is presently suited only to an expert user and is often ad hoc or not specifically designed for unbiased metagenomic data. These ethical and analytical aspects will need significant consideration, along with financial costs, before metagenomic detection of viruses becomes commonplace in clinical diagnostic laboratories.
This metagenomic study revealed a list of viruses and a parasite occurring in unsolved gastrointestinal outbreaks in New Zealand, providing a starting point for further investigations into outbreak etiology and an audit process for routine diagnostic algorithms. This study also provides the first report of HPeV3 in Australasia, and given the more serious illness associated with this pathogen, it is recommended that clinicians and health authorities consider routine testing for this pathogen in instances of severe disease such as encephalitis, neonatal sepsis, and paralysis, as already occurs in the United States, Japan, and Europe.
ACKNOWLEDGMENTS
We acknowledge our colleagues at ESR for their assistance, including Maurice Wilson and Chris Graham for providing the background on gastroenteritis testing in New Zealand, Donia Macartney-Coxson for her contribution to formative bioinformatic work that led to the present study, and Steve Pyne and Ned Rajanayagam for the provision of computing resources. We also acknowledge the provision of sequencing by New Zealand Genomics Limited, with special thanks to Lorraine Berry at the Massey Genome Service (Massey University, New Zealand). Finally, we also acknowledge the provision of high-performance computing resources from the New Zealand e-Science Infrastructure.
This study was funded by the Health Research Council of New Zealand (HRC Reference 11/411). The New Zealand Ministry of Health provides funding for the reference laboratory.
REFERENCES
- 1.Elliott EJ. 2007. Acute gastroenteritis in children. BMJ 334:35–40. doi: 10.1136/bmj.39036.406169.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denno DM, Klein EJ, Young VB, Fox JG, Wang D, Tarr PI. 2007. Explaining unexplained diarrhea and associating risks and infections. Anim Health Res Rev 8:69–80. doi: 10.1017/S1466252307001302. [DOI] [PubMed] [Google Scholar]
- 3.Harada S, Okada M, Yahiro S, Nishimura K, Matsuo S, Miyasaka J, Nakashima R, Shimada Y, Ueno T, Ikezawa S, Shinozaki K, Katayama K, Wakita T, Takeda N, Oka T. 2009. Surveillance of pathogens in outpatients with gastroenteritis and characterization of sapovirus strains between 2002 and 2007 in Kumamoto Prefecture, Japan. J Med Virol 81:1117–1127. doi: 10.1002/jmv.21454. [DOI] [PubMed] [Google Scholar]
- 4.Chen SM, Ni YH, Chen HL, Chang MH. 2006. Microbial etiology of acute gastroenteritis in hospitalized children in Taiwan. J Formos Med Assoc 105:964–970. doi: 10.1016/S0929-6646(09)60280-1. [DOI] [PubMed] [Google Scholar]
- 5.Svraka S, Duizer E, Vennema H, de Bruin E, van der Veer B, Dorresteijn B, Koopmans M. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J Clin Microbiol 45:1389–1394. doi: 10.1128/JCM.02305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass RI. 2013. Beyond discovering the viral agents of acute gastroenteritis. Emerg Infect Dis 19:1190–1191. doi: 10.3201/eid1908.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barratt JL, Harkness J, Marriott D, Ellis JT, Stark D. 2011. A review of Dientamoeba fragilis carriage in humans: several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes 2:3–12. [DOI] [PubMed] [Google Scholar]
- 8.The Institute of Environmental Science and Research. 2012. Annual summary of outbreaks in New Zealand, 2012. FW0753 The Institute of Environmental Science and Research, Porirua, New Zealand: https://surv.esr.cri.nz/surveillance/annual_outbreak.php?we_objectID=3576. [Google Scholar]
- 9.Xie G, Yu J, Duan Z. 2013. New strategy for virus discovery: viruses identified in human feces in the last decade. Sci China Life Sci 56:688–696. doi: 10.1007/s11427-013-4516-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipkin WI. 2010. Microbe hunting. Microbiol Mol Biol Rev 74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, DeRisi JL. 2009. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol J 6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinje J, Wang D, Tong S. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol 83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones MS II, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J Virol 81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits SL, Schapendonk CM, van Beek J, Vennema H, Schurch AC, Schipper D, Bodewes R, Haagmans BL, Osterhaus AD, Koopmans MP. July 2014. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis doi: 10.3201/eid2007.140190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA, Daszak P, Lipkin WI. 2013. A strategy to estimate unknown viral diversity in mammals. mBio 4(5):e00598–13. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woolhouse ME, Howey R, Gaunt E, Reilly L, Chase-Topping M, Savill N. 2008. Temporal trends in the discovery of human viruses. Proc Biol Sci 275:2111–2115. doi: 10.1098/rspb.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall RJ, Wang J, Todd AK, Bissielo AB, Yen S, Strydom H, Moore NE, Ren X, Huang QS, Carter PE, Peacey M. 2014. Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J Virol Methods 195:194–204. doi: 10.1016/j.jviromet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 21.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Tang H, Ye Y. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij JJ, Mulder B, Poell B, van Middelkoop D, Brienen EA, van Lieshout L. 2007. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol Cell Probes 21:400–404. doi: 10.1016/j.mcp.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernroth BE, Conden-Hansson AC, Rehnstam-Holm AS, Girones R, Allard AK. 2002. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl Environ Microbiol 68:4523–4533. doi: 10.1128/AEM.68.9.4523-4533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberste MS, Penaranda S, Rogers SL, Henderson E, Nix WA. 2010. Comparative evaluation of Taqman real-time PCR and semi-nested VP1 PCR for detection of enteroviruses in clinical specimens. J Clin Virol 49:73–74. doi: 10.1016/j.jcv.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Pang XL, Lee B, Boroumand N, Leblanc B, Preiksaitis JK, Yu Ip CC. 2004. Increased detection of rotavirus using a real time reverse transcription-polymerase chain reaction (RT-PCR) assay in stool specimens from children with diarrhea. J Med Virol 72:496–501. doi: 10.1002/jmv.20009. [DOI] [PubMed] [Google Scholar]
- 29.Svraka S, van der Veer B, Duizer E, Dekkers J, Koopmans M, Vennema H. 2009. Novel approach for detection of enteric viruses to enable syndrome surveillance of acute viral gastroenteritis. J Clin Microbiol 47:1674–1679. doi: 10.1128/JCM.00307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyman WH, Walsh JF, Kotch JB, Weber DJ, Gunn E, Vinje J. 2009. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J Pediatr 154:253–257. doi: 10.1016/j.jpeds.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 31.Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K. 2004. Isolation and identification of a novel human parechovirus. J Gen Virol 85:391–398. doi: 10.1099/vir.0.19456-0. [DOI] [PubMed] [Google Scholar]
- 32.Nix WA, Khetsuriani N, Penaranda S, Maher K, Venczel L, Cselko Z, Freire MC, Cisterna D, Lema CL, Rosales P, Rodriguez JR, Rodriguez W, Halkyer P, Ronveaux O, Pallansch MA, Oberste MS. 2013. Diversity of picornaviruses in rural Bolivia. J Gen Virol 94:2017–2028. doi: 10.1099/vir.0.053827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuzurihara SS, Ao K, Hara T, Tanaka F, Mori M, Kikuchi N, Kai S, Yokota S. 2013. Human parechovirus-3 infection in nine neonates and infants presenting symptoms of hemophagocytic lymphohistiocytosis. J Infect Chemother 19:144–148. doi: 10.1007/s10156-012-0420-9. [DOI] [PubMed] [Google Scholar]
- 34.Shoji K, Komuro H, Miyata I, Miyairi I, Saitoh A. 2013. Dermatologic manifestations of human parechovirus type 3 infection in neonates and infants. Pediatr Infect Dis J 32:233–236. doi: 10.1097/INF.0b013e31827b1fd0. [DOI] [PubMed] [Google Scholar]
- 35.Harvala H, Wolthers KC, Simmonds P. 2010. Parechoviruses in children: understanding a new infection. Curr Opin Infect Dis 23:224–230. doi: 10.1097/QCO.0b013e32833890ca. [DOI] [PubMed] [Google Scholar]
- 36.Zhang DL, Jin Y, Li DD, Cheng WX, Xu ZQ, Yu JM, Jin M, Yang SH, Zhang Q, Cui SX, Liu N, Duan ZJ. 2011. Prevalence of human parechovirus in Chinese children hospitalized for acute gastroenteritis. Clin Microbiol Infect 17:1563–1569. doi: 10.1111/j.1469-0691.2010.03390.x. [DOI] [PubMed] [Google Scholar]
- 37.Kolehmainen P, Oikarinen S, Koskiniemi M, Simell O, Ilonen J, Knip M, Hyoty H, Tauriainen S. 2012. Human parechoviruses are frequently detected in stool of healthy Finnish children. J Clin Virol 54:156–161. doi: 10.1016/j.jcv.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Oxner RB, Paltridge GP, Chapman BA, Cook HB, Sheppard PF. 1987. Dientamoeba fragilis: a bowel pathogen? N Z Med J 100:64–65. [PubMed] [Google Scholar]
- 39.Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SWL, Hibberd ML, Liu ET, Rohwer F, Ruan Y. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. 2010. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol 48:1787–1794. doi: 10.1128/JCM.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 42.Tebruegge M, Curtis N. 2009. Enterovirus infections in neonates. Semin Fetal Neonatal Med 14:222–227. doi: 10.1016/j.siny.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Cotten M, Oude Munnink B, Canuti M, Deijs M, Watson SJ, Kellam P, van der Hoek L. 2014. Full genome virus detection in fecal samples using sensitive nucleic acid preparation, deep sequencing, and a novel iterative sequence classification algorithm. PLoS One 9:e93269. doi: 10.1371/journal.pone.0093269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colson P, Richet H, Desnues C, Balique F, Moal V, Grob J-J, Berbis P, Lecoq H, Harlé J-R, Berland Y, Raoult D. 2010. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One 5:e10041. doi: 10.1371/journal.pone.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]