Abstract
HCF-1 is a cellular protein required by VP16 to activate the herpes simplex virus (HSV) immediate-early genes. VP16 is a component of the viral tegument and, after release into the cell, binds to HCF-1 and translocates to the nucleus to form a complex with the POU domain protein Oct-1 and a VP16-responsive DNA sequence. This VP16-induced complex boosts transcription of the viral immediate-early genes and initiates lytic replication. In uninfected cells, HCF-1 functions as a coactivator for the cellular transcription factors LZIP and GABP and also plays an essential role in cell proliferation. VP16 and LZIP share a tetrapeptide HCF-binding motif recognized by the β-propeller domain of HCF-1. Here we describe a new cellular HCF-1 β-propeller domain binding protein, termed HPIP, which contains a functional HCF-binding motif and a leucine-rich nuclear export sequence. We show that HPIP shuttles between the nucleus and cytoplasm in a CRM1-dependent manner and that overexpression of HPIP leads to accumulation of HCF-1 in the cytoplasm. These data suggest that HPIP regulates HCF-1 activity by modulating its subcellular localization. Furthermore, HPIP-mediated export may provide the pool of cytoplasmic HCF-1 required for import of virion-derived VP16 into the nucleus.
Lytic replication in herpes simplex virus begins with the expression of the five viral immediate-early (IE)1 genes that encode multifunctional regulatory proteins necessary for expression of the early genes and creation of an optimal environment for viral replication (reviewed in Ref. 1). Transcription of the IE genes is stimulated by VP16, a component of virion tegument layer (for reviews see Refs. 2–4). Biochemical and genetic analyses have shown that transactivation by VP16 is dependent on HCF-1, a ubiquitous cellular transcription factor (5–8). VP16 binds directly to HCF-1, and this allows subsequent association with the cellular POU domain protein Oct-1 and the VP16-responsive sequence found in each IE gene promoter known as the TAATGARAT motif. Once assembled, the VP16-induced complex directs high level IE gene transcription by virtue of the potent activation domain in the C terminus of VP16 (9, 10). VP16 does not possess its own nuclear localization signal (NLS) but instead relies on association of HCF-1, which contains a bipartite basic residue-rich NLS in the C-terminal subunit (11, 12). Thus HCF-1 provides two functions required by VP16, complex assembly and nuclear targeting.
Human HCF-1 is expressed in a wide range of tissue types, including all of the cell lines tested (13–16). In most cells, HCF-1 is exclusively nuclear and tightly associated with chromatin (17). The HCF-1 polypeptide is synthesized as a 2035-amino acid precursor that is subsequently processed into two subunits through proteolytic cleavage at six HCFPRO repeats located near the center of the precursor (18–21). The bulk of HCF-1 protein exists in the processed form, with the N- and C-terminal subunits tightly but noncovalently associated as a heterodimeric complex (12, 20). In addition to sequences required for proteolytic processing and subunit association, HCF-1 contains several domains that mediate interactions with other transcription factors. At the N terminus there are six kelch repeats that fold into a six-blade β-propeller (22–25). The β-propeller domain is sufficient for interaction with VP16 and formation of the VP16-induced complex. The HCF-1 β-propeller also interacts with two cellular bZIP proteins, LZIP and Zhangfei (15, 26). All three proteins recognize the HCF-1 β-propeller using a conserved tetrapeptide motif known as the HCF-binding motif (HBM) (26, 27). The motif ((D/E)HXY) consists of an acidic residue (asparagine or glutamic acid) followed by an invariant histidine, any residue (X), and then an invariant tyrosine. The HBM is an integral part of the LZIP transactivation domain, and recruitment of HCF-1 is required for activation (28). In VP16, the HBM lies N-terminal to the activation domain but is still required for transactivation (8, 29).
In addition to its role in VP16-induced complex formation, HCF-1 is required for cellular proliferation (30). Analysis of the hamster tsBN67 cell line revealed a temperature-sensitive mutation in HCF-1 that results from a missense mutation in the β-propeller domain that changes proline 134 to a serine. At the nonpermissive temperature, tsBN67 cells undergo a stable arrest in G1/G0 but will reinitiate the cell cycle if returned to the permissive temperature. The mutation prevents recognition of the HBM, and thus at the restrictive temperature, transactivation by both VP16 and LZIP is severely reduced (22, 28). This implies, but does not prove, that the cell cycle arrest arises from a defect in cellular transcription.
To better understand the cellular function of HCF-1, we initiated a screen to identify cellular interacting proteins. Here we describe isolation of cDNAs from a human brain library, encoding a small cellular polypeptide, termed HPIP, which binds to the β-propeller domain. HPIP contains a consensus HBM, which is essential for association with HCF-1. HPIP also contains a leucine-rich nuclear export signal and shuttles between the nucleus and cytoplasm using the CRM1-mediated nuclear export pathway. These properties suggest that HPIP functions as a chaperone for HCF-1, mediating export out of the nucleus. Overexpression of HPIP leads to the accumulation of HCF-1 in the cytoplasm, and this can be blocked using the CRM1 inhibitor leptomycin B. Active shuttling of HCF-1 in and out of the nucleus provides a mechanism for the nuclear import of VP16, whereupon it initiates a cascade of lytic gene expression.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen
The bait plasmid pLexA-HCF-1N380 was constructed by subcloning a fragment encoding the first 380 residues of human HCF-1 into the polylinker of pLexAΔPL (31). Yeast cells expressing LexA-HCF-1N380 were then transformed with a human adult brain cDNA library (Clontech) and scored for lacZ expression by growth on medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). A total of 11 positives were isolated from 6 × 106 independent transformants. Sequence analysis revealed that six of the positives were derived from the same gene and were selected for further analysis.
Northern Blotting
Human multiple tissue and cell line Northern blots (Clontech) were probed according to the manufacturer’s instructions using a 32P-labeled probe corresponding to nucleotides 1–707 of the human HPIP cDNA.
Mammalian Expression Plasmids
Sequences encoding full-length human HPIP were PCR amplified from brain cDNA clone 46.2 using oligonucleotide primers that add an XbaI (5′-GCTCTAGAATCCTGCAGCAGCCCTTGCAGCG-3′) and a BamHI site (5′-GGATCCTCAGAGCTCCATTATGTCCCCAGC-3′) to the 5′- and 3′-terminal ends of the HPIP cDNA, respectively. This fragment was subsequently shuffled into mammalian expression plasmids pCGFLAG2 and pEGFP-C2 (Clontech) generating N-terminal fusions with the FLAG epitope and green fluorescent protein (GFP), respectively. Site-directed mutagenesis was performed by QuikChange™ mutagenesis (Stratagene). Subsequent truncations were generated by PCR and confirmed by DNA sequencing. The nuclear localization signal of HCF-1 was amplified by PCR and subcloned into the unique XbaI site. The plasmids encoding HA-tagged HCF-1N380 has been described previously (22). The cytomegalovirus-driven expression plasmid encoding GFP-IκBα (32) was a kind gift from Dr. Ranjan Sen (Brandeis University).
Transfection and Coimmunoprecipitation
Human 293T cells were transfected with LipofectAMINE 2000 (Invitrogen), using 20 µl of reagent/6-cm dish. The extracts were prepared after 24 h by resuspending the cells in high salt buffer (420 mm KCl, 10 mm Tris-HCl, pH 7.9, 5% glycerol, 0.25% Nonidet P-40, 0.2 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 0.2 mm sodium vanadate, 50 µm sodium fluoride, 1 mm dithiothreitol). Under these conditions, the plasma membrane is ruptured, but the nuclear membrane is maintained, although it becomes porous to soluble nuclear proteins (21). After incubation for 20 min at 4 °C, extracted nuclei and other insoluble debris were removed by centrifugation. For immunoprecipitations, 100 µl of the extract was incubated with 2.4 µl of HA-specific antibody (12CA5)-coupled protein G-agarose beads (Roche Molecular Biochemicals) at 4 °C for 1 h. The beads were washed three times in 1 ml of wash buffer (200 mm KCl, 10 mm Tris-HCl, pH 7.9, 5% glycerol, 0.5 mm EDTA) before separation by SDS-polyacrylamide gel electrophoresis. Immunoblotting was performed with wet transfer and detected by enhanced chemiluminescence (SuperSignal; Pierce). The αHA (Roche Molecular Biochemicals) and αGFP (Molecular Probes) antibodies were diluted 1:5000 and 1:100, respectively. Endogenous HCF-1 was detected using a rabbit polyclonal antibody αrHCF-H12 directed against epitopes in the C terminus (21).
Green Fluorescence and Immunofluorescence Microscopy
Cos-1 cells were seeded onto sterile coverslips in a 24-well plate and transfected with 100 ng of expression plasmids encoding HPIP or HCF-1N380 using LipofectAMINE 2000 (Invitrogen). After 24 h, the cells were fixed in 3.7% formaldehyde for 15 min, washed three times in phosphate-buffered saline (PBS), quenched in 100 mm ammonium chloride, and then permeabilized with 0.1% Triton X-100 diluted in PBS. After washing in PBS, the samples were blocked with 10% fetal bovine serum and 0.25% saponin in PBS for 30 min at 37 °C. The samples were then incubated with αFLAG or αHCF-1 polyclonal antibody (αrHCF (21), diluted 1:100) in blocking buffer for 1 h at 37 °C and washed three times with washing buffer (7% fish gelatin and 0.025% saponin in PBS). The coverslips were incubated with the secondary antibody (Texas Red α-rabbit or Texas Red α-mouse; Molecular Probes) diluted 1:250 in blocking buffer for 1 h at 37 °C. The samples were washed, counterstained for 5 min with Hoechst 33528, washed, and fixed onto slides in fluorescence mounting medium (DAKO Corp.). Fluorescence was observed using a Zeiss Axioplan microscope, and images were captured using Axioplan software and exported into Adobe Photoshop 5.0 for further processing. In addition, 100–150 cells from each sample (up to three coverslips) were scored by visual inspection for nuclear or cytoplasmic accumulation of the tagged proteins, and the cells shown in the photomicrographs were chosen to illustrate the predominant patterns.
RESULTS
Characterization of HPIP
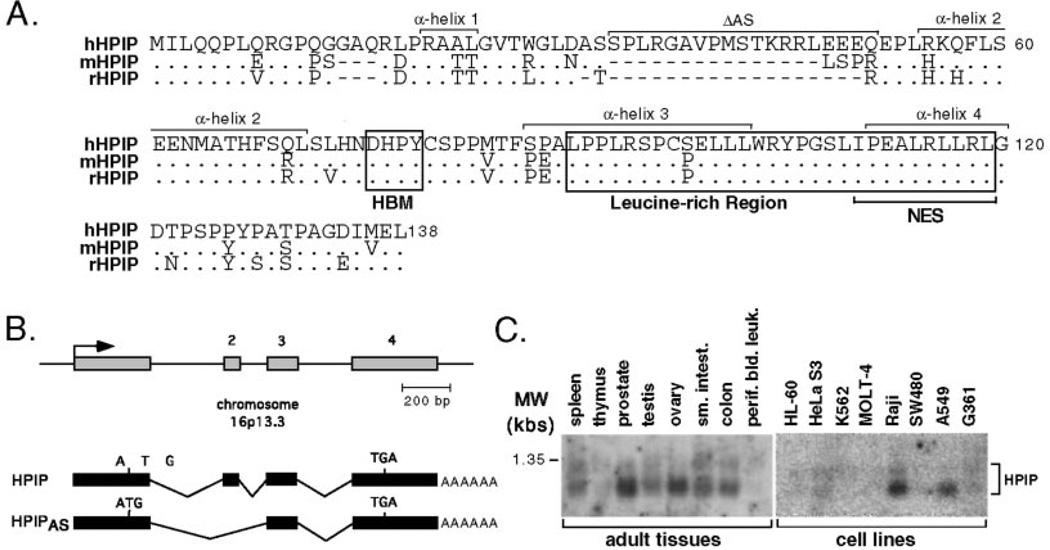
To identify cellular factors that interact with the β-propeller domain of HCF-1, we performed a yeast interaction screen using residues 1–380 (HCF-1N380) of human HCF-1 fused to the LexA DNA-binding domain to screen a human brain cDNA library. We obtained a number of clones that interacted specifically with the bait. Six of these positive clones were independently derived from the same gene and encode a previously undescribed polypeptide that we will refer to as HPIP (HCF β-propeller interacting protein). The predicted HPIP open reading frame encodes a polypeptide of 138 residues with a calculated molecular mass of 15.3 kDa (Fig. 1A). Searches of the expressed sequence tag data base (dbEST) identified a number of additional cDNA clones including those from mouse (AK013438) and rat (AA944494). An alignment of the rodent sequences is given below the human sequence in Fig. 1A and exhibits a relatively high degree of conservation. We were unable to identify counterparts in Drosophila or Caenorhabditis elegans. Searches of the nearly complete human genome sequence revealed that the human gene encoding HPIP is located on chromosome 16p13.3 (GenBank™ Hs16_10709). From comparison of the genomic and cDNA sequences, we predict a total of four exons (Fig. 1B). One of the six clones isolated in the screen (clone 47.7) represents an alternatively spliced variant in which exon 2, encoding residues 33–51, is skipped, creating an internal deletion of 19 amino acids.
Fig. 1. Characterization of HPIP cDNA clones.
A, predicted amino acid sequences of human (h), mouse (m), and rat (r) HPIP. Identical residues are indicated by a dot, whereas introduced gaps are indicated with dashes. The HCF-binding motif or HBM (DHPY) and leucine-rich region are boxed. In human cells, alternative mRNA splicing removes 19 residues (indicated as ΔAS) that are not conserved in the rodent counterparts. The nucleotide and predicted protein sequences reported here have been deposited in the GenBank™ data base with accession number AY116892. B, the deduced exon-intron structure of human HPIP, indicating the origins of the alternative splice variant (HPIPAS), which skips exon 2. C, Northern blot analysis showing the distribution of HPIP mRNA expression in a variety of human primary tissues and cells lines. MW, molecular mass.
The predicted HPIP polypeptide reveals limited similarity to other characterized proteins; however, it does possess a number of interesting features. The most notable is a potential HCF-binding motif or HBM (DHPY, residues 76–79) located near the center of the polypeptide (Fig. 1A) and is perfectly conserved in the rodent sequences. This tetrapeptide motif is found in other HCF-1 interacting proteins, notably VP16 and LZIP, and is the primary determinant for association with the HCF-1 β-propeller domain (26–28). Toward the C terminus of HPIP, there is a leucine-rich region (residues 90–119) that is also well conserved between the three mammalian HPIP sequences. Secondary structure analysis of HPIP predicts four α-helices separated by unstructured loops (indicated in Fig. 1A).
Using commercially obtained filter sets, we performed Northern blot analysis to examine the tissue distribution of HPIP mRNA (Fig. 1C). Two major transcripts of ~950 and 1100 nucleotides in length were detected in poly(A)+ selected mRNA derived from a variety of primary human tissues. In general, HPIP mRNA appeared less abundant in cultured cell lines, although unambiguous signals could be detected in mRNA derived from Raji and A549 cells. These results are consistent with the analysis of expressed sequence tags, which indicate a relatively broad distribution of HPIP mRNA expression.
The HBM Is Required for Specific Association with the HCF-1 β-Propeller HCF-1
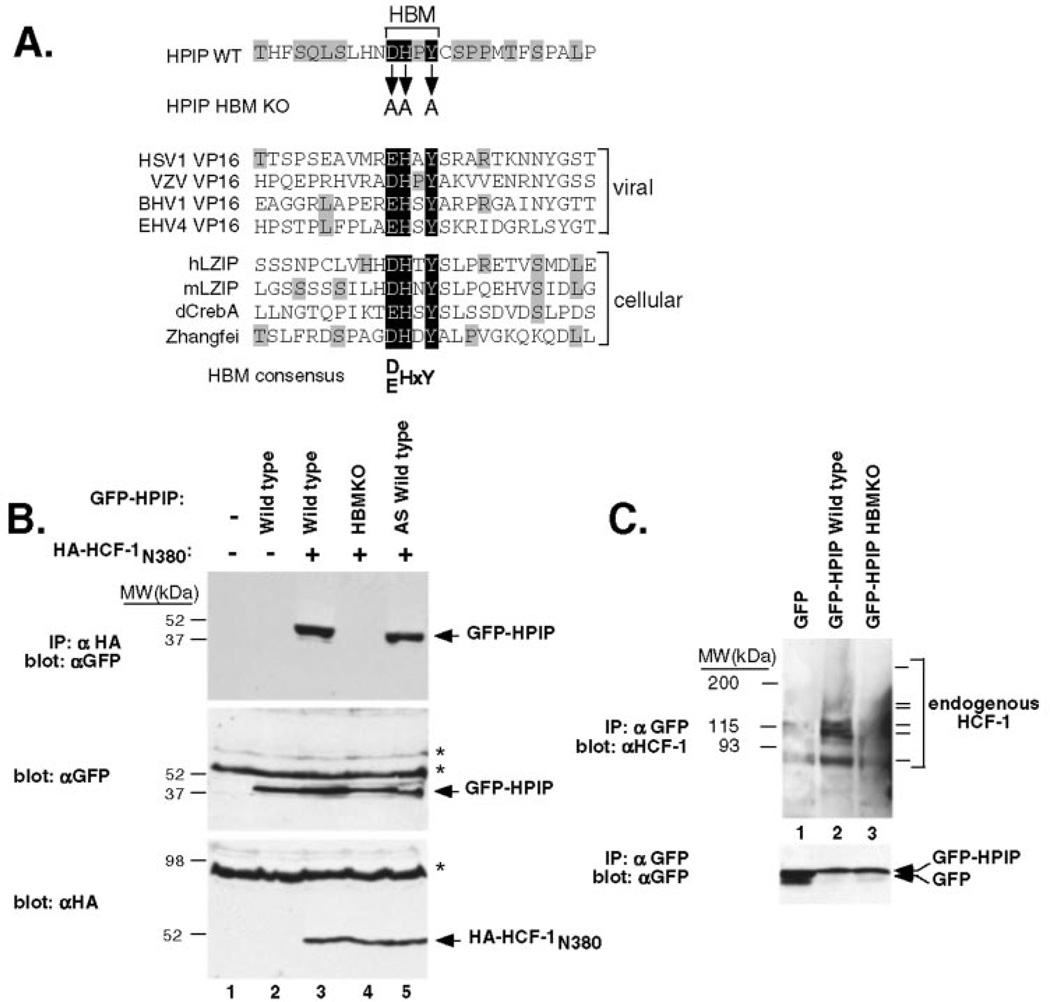
The presence of a candidate HBM near the center of the predicted HPIP sequence suggests a mechanism for association with the β-propeller domain of HCF-1. Fig. 2A shows an alignment of the HPIP HBM and surrounding sequences with the corresponding peptides from several VP16-like proteins encoded by herpesviruses as well as members of the cellular bZIP family. The core tetrapeptide sequence from HPIP (76DHPY79) is identical to that of the varicella-zoster virus (HHV3) VP16 homologue (also known as the ORF10 protein), which is known to interact with HCF-1 (33). As noted previously, there is almost no sequence conservation outside of the core tetrapeptide; however, five of the eight sequences have a hydrophobic residue (valine or isoleucine) at position −3 relative to the acidic residue of the HBM consensus. In HPIP this position corresponds to the hydrophobic residue leucine. These similarities support, but do not prove, the notion that HPIP contains a genuine HBM.
Fig. 2. HPIP and HCF-1 associate in mammalian cells.
A, alignment of the HPIP sequence (black background) with the HBM regions of VP16-like proteins from herpes simplex virus-1 (HSV1), varicella-zoster virus (VZV), bovine herpesvirus-1 (BHV1), and equine herpesvirus-4 (EHV4) as well as the cellular HCF-1 interacting proteins human (hLZIP) and mouse LZIP (mLZIP), Drosophila dCREB-A/BBF2 (dCrebA), and human Zhangfei (15, 26, 54). Identical residues outside the core HBM are indicated with shading. B, 293T cells were cotransfected with expression plasmids encoding wild type or mutant versions of GFP-HPIP (3 µg) and HA-tagged HCF-1N380 (2 µg). The extracts were prepared and subject to coimmunoprecipitation using αHA antibody-coupled beads. Immunoprecipitates were resolved on a SDS-10% polyacrylamide gel and immunoblotted using an αGFP antibody (top panel). To monitor protein expression, extracts were blotted directly using the αGFP (middle panel) or αHA (bottom panel) antibodies. Nonspecific cross-reacting bands are indicated with asterisks. C, coimmunoprecipitation of endogenous HCF-1. The extracts were prepared from transfected 293T cells expressing GFP alone (lane 1), GFP-HPIP (lane 2), and GFP-HPIP HBM KO mutant (lane 3). After immunoprecipitation (IP) with αGFP antibody beads, the extracts were resolved by SDS-10% polyacrylamide gel electrophoresis and probed with αHCF-1 polyclonal sera (upper panel) or αGFP antibody (lower panel). The series of HCF-1 polypeptides detected by the αrHCF-H12 antibody (21) are indicated with bars. As reported previously, the higher molecular mass HCF-1300 and HCF-1150 polypeptides transfer poorly from 10% acrylamide gels and barely detected in this exposure.
To confirm the interaction between HPIP and the HCF-1 β-propeller domain, human 293T cells were cotransfected with expression plasmids encoding HA-tagged HCF-1N380 and full-length HPIP fused to the green fluorescence protein (GFP-HPIP). Protein extracts were prepared and subjected to coimmunoprecipitation using αHA antibody beads. The precipitates were resolved on a SDS-polyacrylamide gel and blotted using an αGFP antibody (Fig. 2B). Coimmunoprecipitation of GFP-HPIP was dependent on coexpression of HA-tagged HCF-1N380 (compare lanes 2 and 3), consistent with the interaction detected in yeast. To examine the role of the putative HBM motif, we generated a substitution mutant in which the critical aspartate, histidine, and tyrosine residues were changed to alanine (Fig. 2A, GFP-HPIP HBM KO). Mutations of these residues in VPI6 and LZIP are sufficient to disrupt the interaction with HCF-1 (26–28). In contrast to wild type, the HBM mutant failed to coimmunoprecipitate with wild type HCF-1N380 (lane 4), indicating that this is a bona fide HBM sequence. Lastly, we tested the alternative splice variant (GFP-HPIPAS), which lacks 19 residues on the N-terminal side of the HBM (Fig. 1A). GFP-HPIPAS polypeptides were recovered with similar efficiency to GFP-HPIP (lane 5), consistent with its identification of both variants in the yeast two-hybrid screen. These results show that HCF-1 and HPIP can form a complex in mammalian cells and that this interaction is dependent on the HBM sequence in HPIP.
To confirm this observation, we asked whether the GFP-HPIP fusion was capable of coimmunoprecipitating endogenous HCF-1 proteins (Fig. 2C). Extracts were prepared from 293T cells transiently transfected with plasmids encoding GFP (lane 1), GFP-HPIP wild type (lane 2), and GFP-HPIP HBM KO (lane 3) and immunoprecipitated using αGFP-coupled beads. Precipitated proteins were resolved on a SDS-10% polyacrylamide gel and probed with antibodies to HCF-1 (upper panel) or GFP (lower panel). Endogenous HCF-1 was readily detected in the sample expressing wild type GFP-HPIP (lane 2). Significantly less HCF-1 was recovered using the HBM mutant (lane 2) and was similar to the background level detected with GFP alone (lane 1). This result indicates that exogenously expressed GFP-HPIP associates with the endogenous HCF-1 protein in an HBM-dependent manner.
HPIP Shuttles between Cytoplasmic and Nuclear Compartments
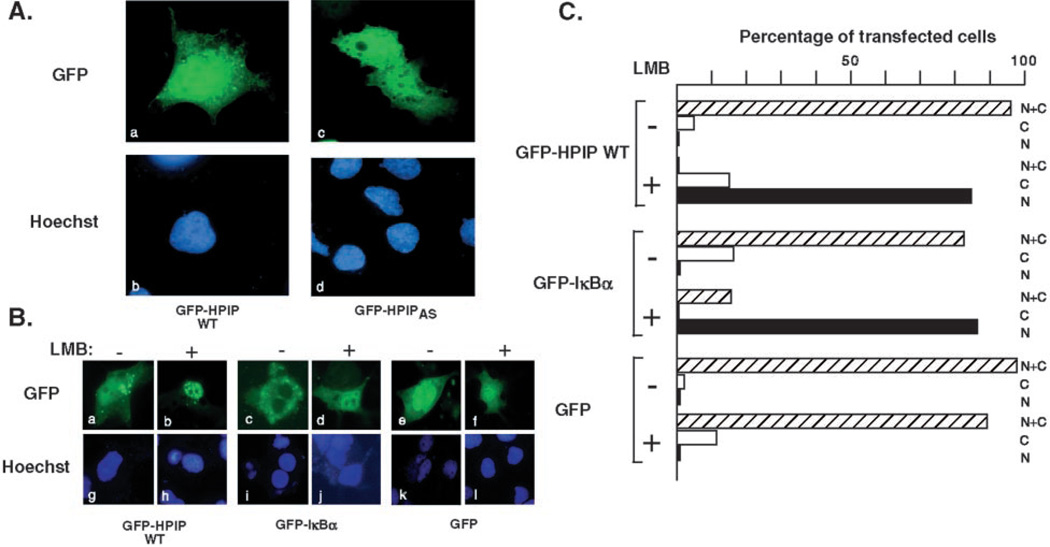
To determine the subcellular localization of HPIP, we used fluorescence microscopy to localize GFP-tagged versions of full-length HPIP in transiently transfected Cos-1 cells (Fig. 3A). Both wild type GFP-HPIP (panels a and c) and the naturally occurring splice variant GFP-HPIPAS (panels b and d) showed a similar distribution of fluorescence spread throughout the cell with a modest increase in signal within the nucleus compared with the cytoplasm. Fluorescence was excluded from the nucleoli and cytoplasmic vacuoles. GFP alone showed a similar widespread pattern, except that the nucleus was less clearly defined (data not shown).
Fig. 3. HPIP shuttles between the nucleus and the cytoplasm.
A, Cos-1 cells were transfected with expression plasmids (100 ng) encoding GFP-HPIP (panels a and c) and GFP-HPIPAS (panels b and d). The nuclei were counterstained with Hoechst dye 33258 (panels c and d). B, nuclear export of HPIP can be inhibited by LMB. Cos-1 cells expressing GFP-HPIP, GFP-IκBα, or GFP alone were seeded in duplicate, and 24 h post-transfection one set of samples was treated for 1 h with 20 ng/ml leptomycin B (+ LMB) or with vehicle alone (− LMB) and fixed for microscopy. The nuclei were counterstained with Hoechst 33258. C, quantitation of the experiment shown in B. At least 100 cells were scored for nuclear (N), cytoplasmic (C), and nuclear/cytoplasmic (N+C) distribution. The values are plotted as percentages of the total number of transfected cells examined.
To investigate the possibility that HPIP actively cycles between nuclear and cytoplasmic compartments, we treated GFP-HPIP expressing Cos-1 cells with leptomycin B (LMB), a specific inhibitor of the CRM1-mediated nuclear export pathway (34–37). This experiment is shown in Fig. 3B. In the absence of LMB, GFP-HPIP was distributed throughout the cell (Fig. 3B, panel a); however, in the presence of drug, GFPHPIP accumulated in the nucleus as discrete speckles with very little cytoplasmic fluorescence (panel b). As controls, cells were transfected with GFP-IκBα (panels c and d) and GFP alone (panels e and f). As previously reported, GFP-IκBα showed a dramatic redistribution from the cytoplasm to the nucleus following exposure to LMB (32). GFP alone was found in both the nucleus and the cytoplasm, and this distribution was unaltered by drug treatment. Fig. 3C shows quantitation of this data. At least 100 cells from each sample were scored for GFP signal in the nucleus (N), cytoplasm (C), or both compartments (N+C) and plotted as percentages of the total. The data clearly show that relocalization of GFP-HPIP in response to LMB treatment occurred in the majority of cells. Taken together, these results argue that GFP-HPIP is in a state of dynamic exchange between the cytoplasmic and nuclear compartments and that the CRM-1-mediated export pathway mediates its export from the nucleus into the cytoplasm.
HPIP Contains a Leucine-rich Nuclear Export Signal
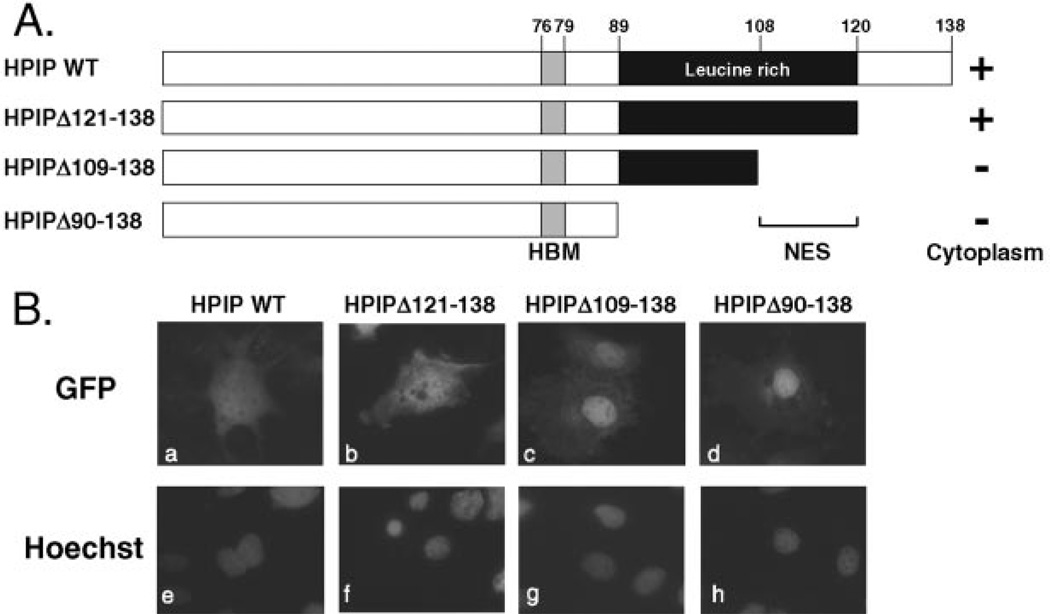
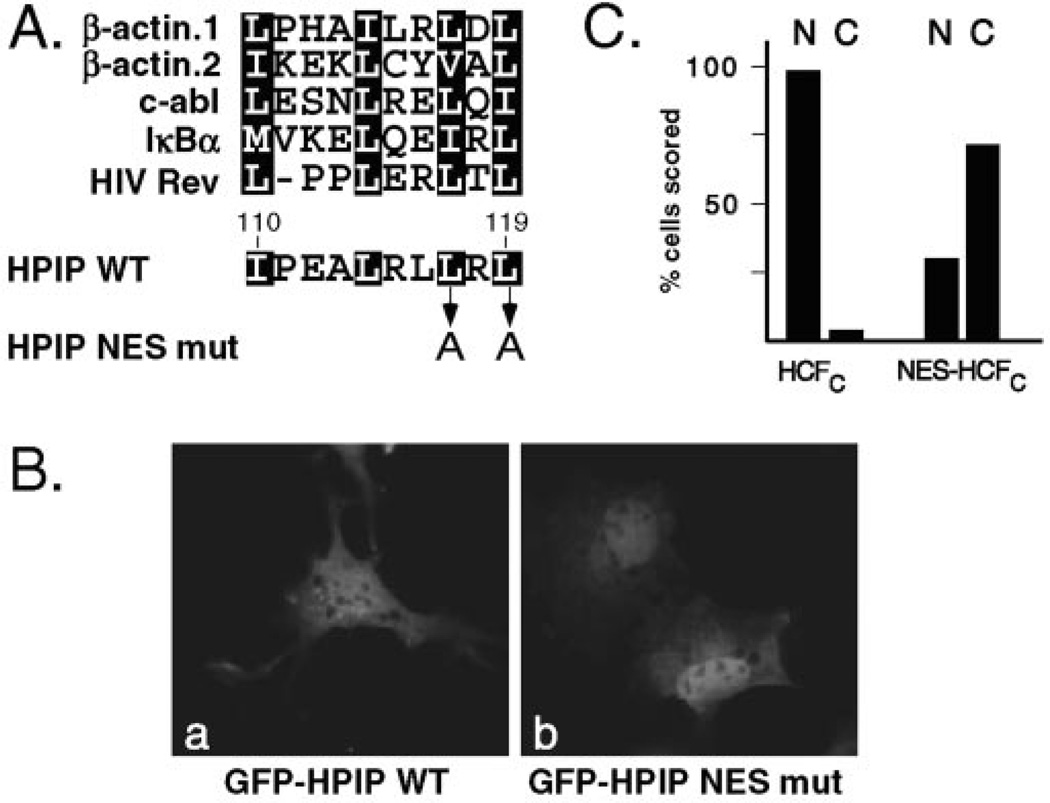
The CRM1/exportin 1 protein functions by recognizing one or more nuclear export signals (NESs) within the cargo protein (35, 38, 39). Typically, each NES corresponds to a short sequence defined by a series of appropriately spaced hydrophobic residues, most commonly leucines (40, 41). Examination of the HPIP sequence identified a leucine-rich region (residues 89–119) toward the C terminus of the polypeptide (Fig. 1A). Within this sequence, we identified an imperfect match (110IX3LXXLXL119) to the NES consensus: LX2–3LXXLXL. To determine whether this leucine-rich region contained an NES, we generated a series of C-terminal truncations (shown schematically in Fig. 4A) that were tested as GFP fusions in transiently transfected Cos-1 cells (Fig. 4B). Deletion of sequences beyond the leucine-rich region (GFP-HPIP Δ121–138, panel b) did not alter the distribution from the wild type fusion protein, whereas the next deletion (GFP-HPIP Δ109–138, panel c), which removed the putative NES, led to a striking accumulation in the nucleus. 94% of transfected cells showed predominantly nuclear fluorescence, whereas the remainder showed fluorescence in both the nucleus and the cytoplasm. Moreover, this nuclear localization was maintained (96% of transfected cells) when the entire leucine-rich region (GFP-HPIP Δ90–138, panel d) was removed. Immunoblotting of parallel transfections confirmed that the fusion proteins were of the expected size and expressed at similar levels (data not shown). These results indicate that HPIP contains a functional leucine-rich NES located between residues 108–120, the region that incorporates the near-consensus NES.
Fig. 4. The leucine-rich region is required for nuclear export.
A, schematic showing HPIP truncations. Cytoplasmic (+) versus nuclear (−) accumulation is indicated. B, representative Cos-1 cells transfected with plasmids (100 ng) encoding GFP-HPIP WT (panels a and e), GFP-HPIP Δ121–138 (panels b and f), GFP-HPIP Δ109–138 (panels c and g), or GFP-HPIP Δ90–138 (panels d and h). After incubation for 24 h, the cells were fixed and analyzed. The nuclei were counterstained with Hoechst 33258 (panels e–h).
Mutation of the NES Reduces Export
Having shown that deletion of the HPIP C terminus leads to a dramatic nuclear accumulation of the GFP fusion proteins, we focused on the putative NES located between residues 110 and 120. Fig. 5C shows an alignment of the putative NES with a selection of well characterized examples from other proteins. From this alignment it is clear that there is little sequence conservation of the variable residues interspersed between the hydrophobic position. To verify that the HPIP sequence is a bona fide NES, we changed two of the key leucine residues (Leu117 and Leu119) to alanine (HPIP NES mut). Analogous mutations have been shown to block recognition by the CRM1 protein and nuclear export (40–43). The mutations were generated in the context of full-length HPIP fused to GFP. As shown in Fig. 5B, mutation of the NES led to a partial accumulation in the nucleus. This was observed in 88% of transfected cells. In the remaining 12% of cells the pattern of fluorescence appeared indistinguishable from wild type GFP-HPIP. This relocalization was less dramatic than with the C-terminal deletions but still consistent with the presence of a leucine-rich NES. Conceivably there are additional sequences in the C terminus that can function as an export signal that are removed in HPIPΔ109–138, which shows a more overt nuclear accumulation (Fig. 4B).
Fig. 5. Identification of the HPIP nuclear export signal.
A, alignment of the HPIP sequence (residues 110–119) with known NES from β-actin (55), c-abl (56), IκBα (57, 58), and HIV Rev (40, 59). The two-leucine residues in HPIP (Leu117 and Leu119) targeted for alanine substitution mutagenesis (HPIP NES mut) are indicated. B, Cos-1 cells were transfected with plasmids (100 ng) encoding GFP-HPIP or GFP-HPIP NES mut. C, quantitation of nuclear versus cytoplasmic staining (expressed as a percentage) of Cos-1 cells transfected with plasmids encoding GFP-HCF-1C (n = 112) or GFP-NES-HCF-1C (n = 107).
To determine whether the leucine-rich sequence was sufficient to function as an export signal, we fused the 10 residues of the putative NES sequence (110IPEALRLLRL119) to the C-terminal subunit of HCF-1 (residues 1436–2035). This fragment includes the HCF-1 NLS (residues 2014–2035) and is exclusively nuclear (12). As summarized in Fig. 5C, addition of the HPIP NES led to a substantial increase in cells with predominantly cytoplasmic fluorescence. This result confirms that the minimal peptide is sufficient to act as a NES when fused to a heterologous nuclear protein.
Elevated Expression of HPIP Leads to the Accumulation of HCF-1 in the Cytoplasm
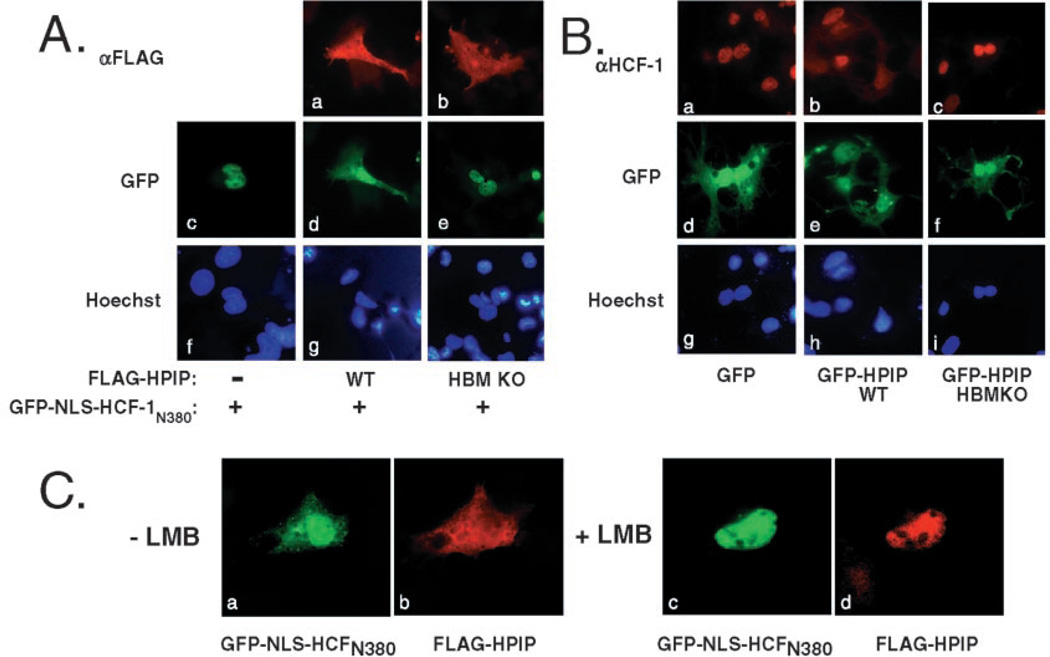
The identification of a functional NES suggests that HPIP might regulate HCF-1 function through association with the β-propeller domain and subsequent export into the cytoplasm. To address this, we cotransfected Cos-1 cells with a plasmid encoding the HCF-1 β-propeller domain fused to GFP (GFP-NLS-HCF-1N380) and FLAG-tagged versions of wild type and HBM mutant versions of HPIP (Fig. 6A). The GFP-HCF-1 fusion includes a single copy of the HCF-1 NLS to ensure that it is imported into the nucleus (panel c). Coexpression with FLAG-HPIP leads to a significant accumulation of GFP-NLS-HCF-1N380 protein in the cytoplasm (panel d), paralleling the diffuse fluorescence of FLAG-HPIP (panel a). This relocalization of GFP fluorescence was not seen using the mutant version of HPIP (panel e), even though FLAG-HPIP HBM KO shows a similar localization to HPIP wild type (compare panels a and b), indicating that direct association is required for relocalization of GFP-HCF-1.
Fig. 6. Expression of wild type HPIP promotes relocalization of HCF-1 to the cytoplasm.
A, Cos-1 cells were transfected with plasmids (100 ng) expressing GFP-NLS-HCF-1N380 alone (panels c and f) or together with plasmids encoding FLAG-tagged HPIP WT (panels a, d, and g) or HPIP HBM KO (panels b, e, and h). After 24 h, the cells were fixed and probed using αFLAG primary antibody followed by a mouse IgG secondary antibody conjugated to Texas Red. The cells were stained with Hoechst dye to visualize the nuclei (panels f–h). B, as in A except that cells were transfected with plasmids encoding GFP alone (panels a, d, and g), GFP-HPIP WT (panels b, e, and h), and GFP-HPIP HBM KO (panels c, f, and i) and probed with an antibody against HCF-1 (panels a–c) to detect the endogenous HCF-1 protein. GFP (panels d–f) and DNA (panels g–i) were visualized by fluorescence. C, cytoplasmic accumulation of HCF-1 is inhibited by leptomycin B. Cos-1 cells were transfected with plasmids (100 ng) encoding GFP-NLS-HCF-1N380 and FLAG-tagged HPIP WT, reseeded in duplicate, and 24 h post-transfection treated for 1 h with vehicle alone (− LMB, panels a and b) or with 20 ng/ml leptomycin B (+ LMB, panels c and d).
We then asked whether the localization of endogenous HCF-1 could be regulated by HPIP expression. As above, Cos-1 cells were transfected with plasmids encoding GFP, GFP-HPIP, and the GFP-HPIP HBM KO mutant, and the localization of endogenous HCF-1 protein was visualized by indirect immunofluoresence using an HCF-1-specific antibody (rabbit polyclonal αrHCF) (21). Transfected cells were identified by green fluorescence. In cells expressing GFP alone, HCF-1 staining was restricted to the nucleus (Fig. 6B, panel a) in 100% of transfected cells. In marked contrast, expression of GFP-HPIP led to a significant accumulation of HCF-1 protein in the cytoplasm, seen in at least 70% of transfected cells and paralleling the diffuse fluorescence of GFP-HPIP (panels b and e). This redistribution was not observed with the GFP-HPIP HBM KO mutant, indicating that direct physical association is required (panel c). This result shows that ectopic expression of HPIP is capable of redirecting HCF-1 into the cytoplasm.
Cytoplasmic Accumulation of HCF-1 Can Be Inhibited by LMB
To determine whether HCF-1 was being actively exported from the nucleus or simply trapped in the cytoplasm following synthesis, we treated transfected cells with LMB to disable CRM1-mediated export (Fig. 6C). As we had observed previously (Fig. 6A), a significant fraction of GFP-NLS-HCF-1N380 was found in the cytoplasm when cotransfected with FLAG-HPIP. This was observed in 85% of transfected cells (panel a and b), whereas the remaining 15% showed a more obvious localization to the nucleus (not shown). Treatment with LMB resulted in all cells showing an exclusively nuclear pattern for both GFP-NLS-HCF-1N380 and FLAG-HPIP (panels c and d). This result implies that GFP-NLS-HCF-1N380 is able to enter the nucleus following synthesis but is then exported back to the cytoplasm through association with HPIP.
DISCUSSION
We describe the identification of HPIP, a previously unknown cellular polypeptide that interacts specifically with the β-propeller domain of HCF-1. When expressed by transient transfection in cultured cells, GFP-tagged HPIP is found in both the cytoplasmic and nuclear compartments of the cell, and treatment with leptomycin B, an inhibitor of the CRM1 export receptor, leads to a rapid accumulation in the nucleus. These observations argue that the localization of HPIP is dynamic and that HPIP shuttles in and out of the nucleus. To this end, we identified a leucine-rich NES between residues 110 and 119 and show that it is a key determinant for nuclear export. The HPIP NES resembles canonical NESs with respect to the distribution of hydrophobic residues and is therefore likely to function through direct interaction with the CRM1 protein. Although leucine is most common at these positions, several known CRM-binding sites utilize isoleucine and/or valine in place of one or more leucines (40, 44). Deletion and point mutagenesis of the HPIP NES leads to nuclear accumulation of GFP-HPIP, reminiscent of LMB treatment, and we also show that this sequence is sufficient for cytoplasmic localization of a nuclear protein.
The sequence of HPIP provides few clues to its function as an HCF-1-binding protein. The only identifiable motifs are the HBM used for recognition of the HCF-1 β-propeller and the NES, which mediates nuclear export. This leads to the hypothesis that HPIP functions as a shuttle factor that regulates HCF-1 localization, and this scenario is shown schematically in Fig. 7. Two observations support the idea that the accumulation of cytoplasmic HCF-1 reflects nuclear export rather than some form of trapping or sequestration. Firstly, ectopically expressed GFP-HPIP was able to relocalize endogenous HCF-1 (Fig. 6B) as well as coexpressed FLAG-HCF-1 (Fig. 6A). Because endogenous HCF-1 is presumably already nuclear when HPIP is first expressed, HPIP must therefore first enter the nucleus before it can redirect HCF-1. In a similar vein, we show that LMB treatment inhibits the cytoplasmic accumulation of GFP-NLS-HCF-1N380, again arguing that HPIP must first enter the nucleus before it can export HCF-1 (Fig. 6C).
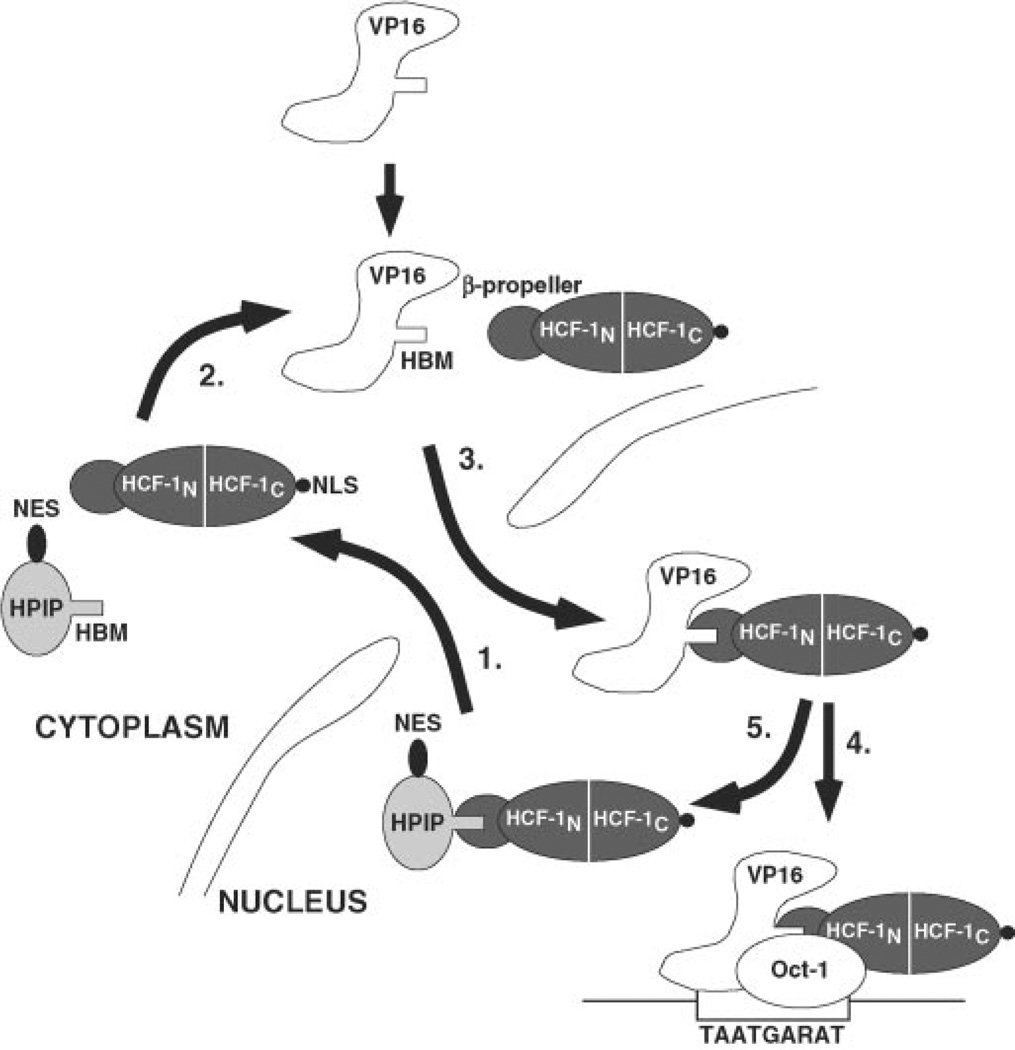
Fig. 7. Model for HPIP-mediated nucleocytoplasmic shuttling of HCF-1.
Step 1, nuclear HCF-1 is exported to the cytoplasm through association with HPIP, which interacts with the N-terminal β-propeller domain (HCF-1N), which recognizes the HPIP HBM. The CRM-1 export protein (not illustrated) recognizes the NES in HPIP. Step 2, HCF-1 may dissociate from HPIP and return to the nucleus or return as complex with HPIP. Step 3, in herpes simplex virus-infected cells, VP16 is released into the cytosol together with other tegument components. The HCF-1 β-propeller domain interacts with the high affinity HBM of VP16, and the resulting complex is transported through the nuclear pore. Nuclear import of HCF-1 is mediated by its C-terminal NLS, which is recognized by the importin complex (11, 12). Once in the nucleoplasm, VP16, still complexed to HCF-1, can further associate with Oct-1 and the TAATGARAT sequence found in the viral immediate-early gene promoters. The resulting VP16-induced complex activates transcription by way of the potent C-terminal activation of VP16.
There is circumstantial evidence that the presence of HCF-1 in the cytoplasm is likely to be important, if not essential, for lytic replication by herpes simplex virus. During infection of permissive cells, virions are disassembled at the plasma membrane, releasing the viral capsid and tegument into the cytoplasm (45). The capsid is thought to associate with microtubules and migrate to the nuclear pore complex (46). Several tegument components, including VP16, function in the nucleus and must therefore also be transported across the cytoplasm and into the nucleus (47–49). O’Hare and co-workers (11) have shown that VP16 does not possess its own nuclear localization signal and instead relies on association of HCF-1 for translocation into the nucleus. This seems paradoxical given the predominantly nuclear location of HCF-1 in most cells and raises the intriguing question of how VP16 interacts with HCF-1 after it is released from the virion. The results presented here suggest that at least some molecules of HCF-1 actively shuttle between the nuclear and cytoplasmic compartments and presumably it is the latter population that first encounters VP16 that has been released from the virion tegument.
HCF-1 is expressed in almost all cells, and VP16 may have evolved to exploit an active and ubiquitous transport mechanism. Although the relative affinities of the HBMs from VP16 and HPIP have not been compared, we anticipate the VP16 HBM to have the highest affinity for the HCF-1 β-propeller. Certainly, in our hands VP16 can be coimmunoprecipitated from transfected extracts more readily than HPIP, consistent with a more stable association (data not shown). A difference in affinity would allow the small number of VP16 molecules released by a virion (~1000 molecules/virion) to efficiently commandeer the available pool of cytoplasmic HCF-1, thereby ensuring rapid translocation to the nucleus and activation of the viral IE genes. In contrast to VP16, LZIP is probably not dependent on HCF-1 for nuclear localization. LZIP contains a short membrane-spanning sequence and is tethered to the endoplasmic reticulum (50). In response to appropriate stimuli, LZIP is cleaved between the bZIP domain and membrane tether, allowing the N terminus containing the activation and basic zipper domains to translocate to the nucleus. As is the case with other bZIP proteins, the basic region is probably sufficient for nuclear localization (51–53). Interestingly, mutation of the HBM does not prevent nuclear localization by HPIP (Fig. 6, A and B). The sequence is devoid of clustered basic amino acids that might serve as a NLS, and it is possible that HPIP associates with additional nuclear proteins; this could be addressed by further mutagenesis in the context of the disrupted HBM.
Antibodies against HPIP are not yet available, and there is currently no information on the relative levels or subcellular localization of HPIP in different cell types. Our Northern analysis and searches of the expressed sequence tag data bases suggest a relatively broad distribution of HPIP mRNA but does not address protein levels. In principal, cells expressing highest levels of HPIP might show the greatest proportion of cytoplasmic HCF-1. A case in point could be sensory neurons in which HCF-1 is known to be largely cytoplasmic (16), and it is tempting to speculate that HPIP contributes to the unique distribution of HCF-1 in these cells. This attractive hypothesis can be tested once appropriate reagents have been generated. Another interesting issue is whether HCF-1 and HPIP remain in a complex in the cytoplasm. Conceivably, HCF-1 is released from HPIP following export, allowing it to return to the nucleus, possibly in conjunction with a cargo protein such as VP16. Alternatively, the intact HCF-1-HPIP complex may simply be reimported, and overall distribution would reflect a balance between rates of export and import. If this is the case, VP16 will need to disrupt the complex and gain access to the HBM.
Acknowledgments
We thank Minoru Yoshida for the generous gift of leptomycin B. Ranjan Sen, Michael Garabedian, Stavros Giannakopoulos, and Ian Mohr provided helpful discussions, reagents or advice. The manuscript was greatly improved by constructive comments from Naoko Tanese.
Footnotes
This work was supported by National Science Foundation Grant MCB-98-16856 and National Institutes of Health Grant GM61139.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY116892.
Supported by the New York University School of Medicine Office of Minority and Multicultural Affairs Summer Research Program.
The abbreviations used are: IE, immediate-early; NES, nuclear export sequence; NLS, nuclear localization sequence; LMB, leptomycin B; GFP, green fluorescence protein; HA, hemagglutinin; HBM, HCF-binding motif; PBS, phosphate-buffered saline.
REFERENCES
- 1.Roizman B, Sears AE. In: Fields Virology. 3rd Ed. Fields BN, Knipe DM, Howley PM, editors. Vol. 2. Philadelphia, PA: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 2.O’Hare P. Semin. Virol. 1993;4:145–155. [Google Scholar]
- 3.Wilson AC, Cleary MA, Lai J-S, LaMarco K, Peterson MG, Herr W. Cold Spring Harbor Symp. Quant. Biol. 1993;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Thompson CC, McKnight SL. Trends Genet. 1992;8:232–236. [Google Scholar]
- 5.Stern S, Herr W. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 6.Gerster T, Roeder RG. Proc. Natl. Acad. Sci. U. S. A. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Hare P, Goding CR. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 8.Lai J-S, Herr W. Mol. Cell. Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Triezenberg SJ, Kingsbury RC, McKnight SL. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 10.Greaves R, O’Hare P. J. Virol. 1989;63:1641–1650. doi: 10.1128/jvi.63.4.1641-1650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaBoissière S, Hughes T, O’Hare P. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson AC, Boutros M, Johnson KM, Herr W. Mol. Cell. Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frattini A, Faranda S, Redolfi E, Zucchi I, Villa A, Patrosso MC, Strina D, Susani L, Vezzoni P. Genomics. 1994;23:30–35. doi: 10.1006/geno.1994.1455. [DOI] [PubMed] [Google Scholar]
- 14.Wilson AC, Parrish JE, Massa HF, Nelson DL, Trask BJ, Herr W. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Yang P, O’Hare P, Misra V. Mol. Cell. Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristie TM, Vogel JL, Sears AE. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wysocka J, Reilly PT, Herr W. Mol. Cell. Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. J. Biol. Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 19.Vogel JL, Kristie TM. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9425–9430. doi: 10.1073/pnas.160266697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson AC, Peterson MG, Herr W. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 21.Wilson AC, LaMarco K, Peterson MG, Herr W. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 22.Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. Mol. Cell. Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmen KA, Newell A, Robinson M, Mills JS, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. J. Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes TA, La Boissiere S, O’Hare P. J. Biol. Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- 25.LaBoissière S, Walker S, O’Hare P. Mol. Cell. Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freiman RN, Herr W. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, Yang P, Padmakumar S, Misra V. J. Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luciano RL, Wilson AC. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10757–10762. doi: 10.1073/pnas.190062797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaves RF, O’Hare P. J. Virol. 1990;64:2716–2724. doi: 10.1128/jvi.64.6.2716-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan MA, Samuels HH. Mol. Cell. Biol. 2000;20:5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tam WF, Lee LH, Davis L, Sen R. Mol. Cell. Biol. 2000;20:2269–2284. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriuchi H, Moriuchi M, Cohen JI. J. Virol. 1995;69:4693–4701. doi: 10.1128/jvi.69.8.4693-4701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 35.Fornerod M, Ohno M, Yoshida M, Mattaj IW. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 36.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff B, Sanglier JJ, Wang Y. Chem. Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 38.Stade K, Ford CS, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 39.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. Curr. Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 40.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Mol. Cell. Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim FJ, Beeche AA, Hunter JJ, Chin DJ, Hope TJ. Mol. Cell. Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forgues M, Marrogi AJ, Spillare EA, Wu CG, Yang Q, Yoshida M, Wang XW. J. Biol. Chem. 2001;276:22797–22803. doi: 10.1074/jbc.M101259200. [DOI] [PubMed] [Google Scholar]
- 44.Gorlich D, Kutay U. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 45.Knopf CW. Acta Virol. 2000;44:289–307. [PubMed] [Google Scholar]
- 46.Ye GJ, Vaughan KT, Vallee RB, Roizman B. J. Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly M, Elliott G. J. Virol. 2001;75:2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elliott G, O’Hare P. J. Virol. 2000;74:2131–2141. doi: 10.1128/jvi.74.5.2131-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrison EE, Stevenson AJ, Wang YF, Meredith DM. J. Gen. Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 50.Lu R, Misra V. J. Virol. 2000;74:934–943. doi: 10.1128/jvi.74.2.934-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams SC, Angerer ND, Johnson PF. Gene Expr. 1997;6:371–385. [PMC free article] [PubMed] [Google Scholar]
- 52.Liu JL, Lee LF, Ye Y, Qian Z, Kung HJ. J. Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varagona MJ, Raikhel NV. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- 54.Lu R, Misra V. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wada A, Fukuda M, Mishima M, Nishida E. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee SH, Hannink M. J. Biol. Chem. 2001;276:23599–23606. doi: 10.1074/jbc.M011197200. [DOI] [PubMed] [Google Scholar]
- 58.Johnson C, Van Antwerp D, Hope TJ. EMBO J. 1999;18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]