Abstract
In this case report of autosomal recessive pigmented hypomaturation amelogenesis imperfecta (AI), we identify a novel homozygous missense mutation (g.165151T>G; c.1317T>G; p.Leu436Arg) in SLC24A4, a gene encoding a potassium-dependent sodium-calcium exchanger that is critical for hardening dental enamel during tooth development.
Keywords: Inherited Diseases, Mutations, Enamel, Tooth, Calcium
“Amelogenesis Imperfecta” or AI has been used as a diagnosis for inherited enamel malformations that occur in the absence of defects outside of the dentition,1, 2 and to refer to the presence of enamel defects in syndromes (e.g. Platyspondyly with amelogenesis imperfecta; OMIM, 601216).3 AI is a diverse collection of inherited diseases featuring varied forms of enamel malformations (hypoplasia, hypocalcification, hypomaturation, pigmented hypomaturation, and local hypoplasia), and multiple patterns of inheritance (autosomal dominant; autosomal recessive, X-linked). Many AI diseases exhibit only enamel defects (isolated AI), while others have systemic problems (syndromic AI). In the United States isolated AI affects about 1/13,000 persons.4 A clinical diagnosis of isolated AI is based upon the appearance, thickness, and hardness of the patients’ enamel and analyses of oral radiographs and pattern of inheritance.2
The results of genetic studies have changed our perspective of AI. Inherited enamel defects can be caused by mutations in many genes and some distinctions between isolated and syndromic types have blurred. Junctional Epidermolysis Bullosa (JEB) is a recessive form of syndromic AI that features enamel malformations and skin fragility.5 Mutations in both alleles of LAMA3, LAMB3, LAMC2, and COL17A1 cause JEB, while defects in a single allele can cause isolated AI. Isolated enamel defects have been associated with defects in AMELX,6 ENAM,7 AMBN,8 MMP20,9 FAM83H,10 ITGB6,11, 12 C4orf26,13 KLK4,14 WDR72,15 and SLC24A4,16 COL17A1,17 and LAMB3.18, 19 As enamel malformations are often the only clinical manifestation evident at the time of diagnosis, clinicians are frequently uncertain as to whether the presenting case is isolated or syndromic. Syndromic AI and their genetic causes include Cone-Rod Dystrophy and AI (CNNM4),20 Enamel Renal Syndrome (FAM20A),21, 22 Tricho-Dento-Osseous syndrome (DLX3),23 Immunodeficiency 9 (ORAI),24 Immunodeficiency 10 (STIM1),25, 26 Oculodentodigital Dysplasia (GJA1),27, 28 Kohlschütter–Tönz Syndrome (ROGDI),29 and Nance Horan Syndrome (NHS).11, 30 Although some syndromic AI diseases have distinctive clinical features, identifying the gene/mutation that causes the disease provides a specific diagnosis, discerns between isolated and syndromic AI, and improves assessment of the patients’ prognosis.
CASE REPORT
A seven-year-old girl in the mixed dentition stage of dental development presented at the Pediatric Dentistry Clinic at Istanbul University. The chief complaint concerned the enamel layer of her dentition, which showed milky brown staining even as the teeth erupted (Fig. 1). The medical history was non-contributory, and no malformations outside of the enamel were evident clinically. Later, after finding that SLC24A4 defects caused the disease, we specifically inquired about organs that might express NCKX4 (the protein product of SLC24A4),31 such as lungs, but abnormalities outside of the dentition were not observed and problems with the lungs were specifically denied.
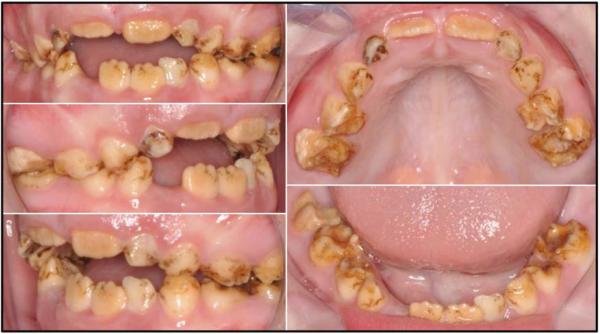
Figure 1.
Oral photographs of the female proband at age 6.
Oral examination of the mixed dentition revealed that the teeth were of normal size and contour, but mostly brown in color. There was significant attrition, particularly of the posterior teeth, suggesting that the enamel lacked hardness. The enamel surfaces were rough and pitted, largely due to chipping. The coarse areas were a deeper brown than surrounding parts of the crowns, suggesting a susceptibility to extrinsic staining. The enamel appeared most normal in the cervical regions, where attrition had not yet roughened the surface, and the enamel hadn’t stained. The hardening, or maturation, of enamel is accomplished by modulating (smooth-ended and ruffle-ended) ameloblasts and occurs more readily at the cervical margins where the enamel is thinner.32
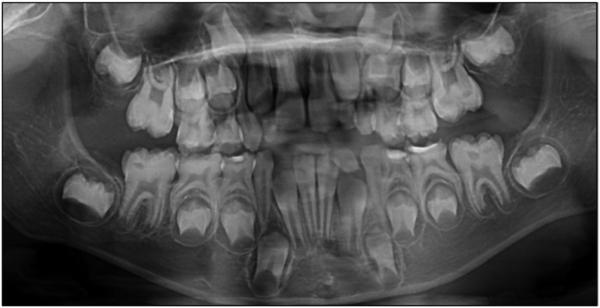
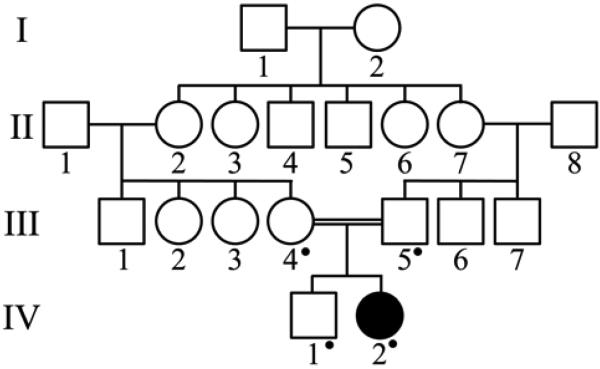
The enamel did not contrast well with dentin on the panoramic radiograph, although the images of most unerupted teeth showed a distinct white line at the dentino-enamel junction (DEJ) (Fig. 2). The thickness of enamel layer on unerupted teeth was within normal limits. A pedigree was drawn up based upon information provided by the mother of the proband (Fig. 3). The proband (IV-2) was the product of a consanguineous union of first cousins and was the only person affected. A clinical diagnosis was made of isolated autosomal recessive pigmented, hypomaturation amelogenesis imperfecta.
Figure 2.
Panorex radiograph of the female proband at age 6.
Figure 3.
Pedigree of the AI kindred.
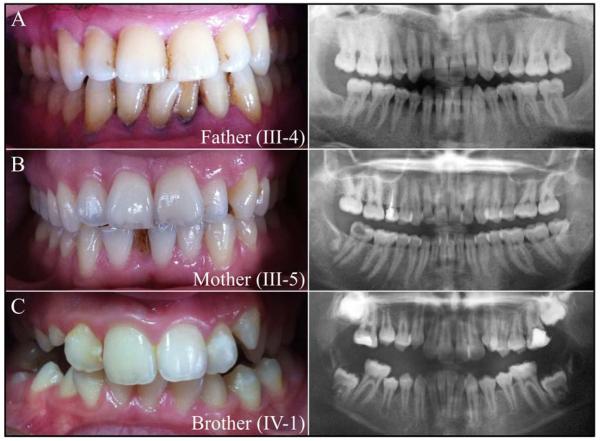
Oral and radiographic examinations of the parents (III-4, III-5) and sibling (IV-1) were within normal limits (Fig. 4). Poor oral hygiene and a high prevalence of dental decay is routinely observed in this population, so the finding of dental caries in other family members does not support a conclusion of increased caries susceptibility in the unaffected heterozygotes.
Figure 4.
Photos and panoramic radiographs of unaffected heterozygotes.
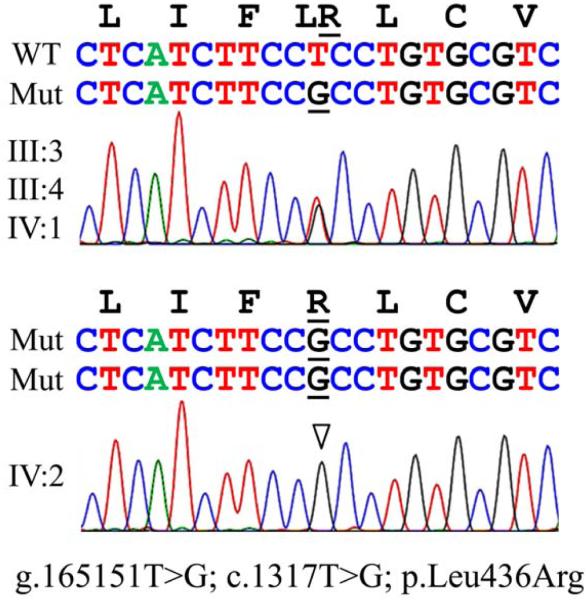
The nuclear family (III-4, III-5, IV-1, IV-2) was recruited into our study. The human study protocol and consents were reviewed and approved by the Ethics Committee at the University of Istanbul, Turkey and Institutional Review Board at the University of Michigan. Study participants signed appropriate written consents after an explanation of their contents and their questions about the study had been answered. Genomic DNA was extracted from blood samples obtained from the four recruited members and used for mutation analyses. The five Kallikrein 4 (KLK4) coding exons and adjacent intron borders were amplified and characterized by DNA sequencing as previously described,14 but no sequence variations were identified (data not shown). The 17 SLC24A4 coding exons and intron borders were sequenced as described previously33 and a sequence variation in the 13th coding exon was identified (g.165151T>G; c.1317T>G; p.Leu436Arg). The parents and sibling were heterozygous, while the proband was homozygous for this novel SLC24A4 mutation (Fig. 5), which is not listed in the 1000 Genomes database.34 PolyPhen-235 predicted the leucine to arginine substitution to be probably damaging and gave a score of 0.994.
Figure 5.
SLC24A4 chromatograms. The parents and sibling were heterozygous for SLC24A4 mutation g.165151T>G; c.1317T>G; p.Leu436Arg (top), while the proband showed the mutation on both SLC24A4 alleles. The gene and cDNA and mutation descriptions are based upon SLC24A4 reference sequences NG_023408.1 and NM_153646.3, respectively.
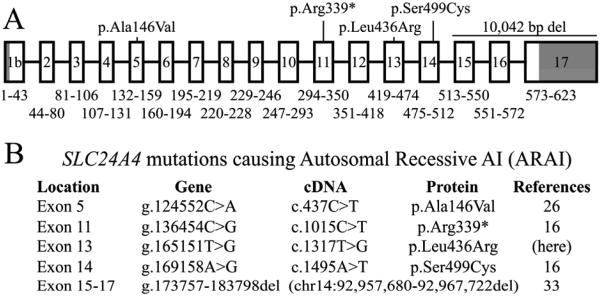
This is fifth SLC24A4 mutation reported to cause AI (Fig. 6).16, 26, 33 The enamel phenotype of the proband closely matches those of previously reported AI-causing SLC24A4 defects.
Figure 6.
SLC24A4 gene structure and disease-causing mutations. A: The SLC24A4 gene structure is for SLC24A4 transcript variant 1 (TV1; NM_153646.3). The 17 numbered boxes are exons. Introns are indicated by a black line. The intron and exon lengths are not drawn to scale. The numbers below each exon indicate the range of amino acids encoded by each exon. Darkened areas are noncoding. B: SLC24A4 mutations reported to cause AI. The gene and cDNA and mutation descriptions are based upon SLC24A4 reference sequences NG_023408.1 and NM_153646.3, respectively.
DISCUSSION
Much has been learned in the last decade about the genetic etiologies of inherited enamel defects, although we probably are only about half way to knowing the full spectrum of genes that cause AI.36, 37 More AI-causing genes have been discovered than anyone originally thought possible. Because so many known and unknown genes can cause AI, a genetic diagnosis is not easy to make and is currently accomplished only in the context of a genetics study. Familiarity with genotype-phenotype correlations (how the enamel looks when a specific gene is mutated) can help narrow down the possibilities.38 With a little luck, mutation analyses of a short list of prioritized candidate genes can discover the AI-causing mutation, as was done in this study. Increasingly, whole-exome sequencing (WES) is used to identify sequence variations in the most critical 2% of the genome (associated with protein coding regions). These variations are then analyzed to pinpoint which one causes the disease.39 This approach has greatly advanced our knowledge of the genes and mutations that cause rare inherited diseases.40 When most of the genes that cause AI are known, genetic testing will be used routinely to diagnose AI in patients.
One predictable consequence of genetic testing that identifies the genes/mutations that cause and define the AI disease will be expansion of the recognized clinical phenotype. Clinicians will start to notice and report subtle phenotypes that were not previously associated with AI. In mouse studies, Slc24a4 transcripts were detected in abundance in the brain, lungs, aorta, and thymus.31 In contrast only 26 SLC24A4 expressed sequence tags (ESTs) have been identified among over 3.3 million ESTs for normal tissues in the human EST database (Hs.385530), which did not sample developing teeth. There may be significant differences between mice and humans in the expression of SLC24A4. Slc24a4 null mice showed a reduced ability to locate an odorous source.41 As genetic testing to identify the genetic cause of AI in each patient becomes the standard of care, clinicians will likely notice additional phenotypes that go along with specific AI diseases and give us a better understanding of how these inherited conditions affect our patients.
Given the complexity of the AI diagnosis and the potential for hidden systemic conditions, it is recommended that clinicians request a genetic consultation or mediate enrollment of their AI patients into a genetics study.
ACKNOWLEDGMENTS
We thank the participants in this study. This work was supported by NIDCR/NIH Grant DE015846.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Witkop CJ., Jr. Heterogeneity in inherited dental traits, gingival fibromatosis and amelogenesis imperfecta. South Med J. 1971;64(Suppl1):16–25. doi: 10.1097/00007611-197102001-00005. [DOI] [PubMed] [Google Scholar]
- 2.Witkop CJ., Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol. 1989;17:547–53. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 3.Bertola DR, Antequera R, Rodovalho MJ, Honjo RS, Albano LM, Furquim IM, et al. Brachyolmia with amelogenesis imperfecta: further evidence of a distinct entity. Am J Med Genet A. 2009;149A:532–4. doi: 10.1002/ajmg.a.32661. [DOI] [PubMed] [Google Scholar]
- 4.Witkop CJ. Hereditary defects in enamel and dentin. Acta Genet Stat Med. 1957;7:236–9. doi: 10.1159/000150974. [DOI] [PubMed] [Google Scholar]
- 5.Kiritsi D, Has C, Bruckner-Tuderman L. Laminin 332 in junctional epidermolysis bullosa. Cell Adh Migr. 2013;7:135–41. doi: 10.4161/cam.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagerström M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, et al. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–5. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 7.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–7. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 8.Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, et al. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet in press. 2014:23. doi: 10.1093/hmg/ddu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, et al. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–5. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, et al. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82:489–94. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang SK, Choi M, Richardson AS, Reid BM, Lin BP, Wang SJ, et al. ITGB6 loss-of-function mutations cause autosomal recessive amelogenesis imperfecta. Hum Mol Genet. 2014;23:2157–63. doi: 10.1093/hmg/ddt611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulter JA, Brookes SJ, Shore RC, Smith CE, Abi Farraj L, Kirkham J, et al. A missense mutation in ITGB6 causes pitted hypomineralized amelogenesis imperfecta. Hum Mol Genet. 2013;18:18. doi: 10.1093/hmg/ddt616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry DA, Brookes SJ, Logan CV, Poulter JA, El-Sayed W, Al-Bahlani S, et al. Mutations in C4orf26, encoding a peptide with in vitro hydroxyapatite crystal nucleation and growth activity, cause amelogenesis imperfecta. Am J Hum Genet. 2012;91:565–71. doi: 10.1016/j.ajhg.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–9. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, et al. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 2009;85:699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parry DA, Poulter JA, Logan CV, Brookes SJ, Jafri H, Ferguson CH, et al. Identification of mutations in SLC24A4, encoding a potassium-dependent sodium/calcium exchanger, as a cause of amelogenesis imperfecta. Am J Hum Genet. 2013;92:307–12. doi: 10.1016/j.ajhg.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGrath JA, Gatalica B, Li K, Dunnill MG, McMillan JR, Christiano AM, et al. Compound heterozygosity for a dominant glycine substitution and a recessive internal duplication mutation in the type XVII collagen gene results in junctional epidermolysis bullosa and abnormal dentition. Am J Pathol. 1996;148:1787–96. [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JW, Seymen F, Lee KE, Ko J, Yildirim M, Tuna EB, et al. LAMB3 mutations causing autosomal-dominant amelogenesis imperfecta. J Dent Res. 2013;92:899–904. doi: 10.1177/0022034513502054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulter JA, El-Sayed W, Shore RC, Kirkham J, Inglehearn CF, Mighell AJ. Whole-exome sequencing, without prior linkage, identifies a mutation in LAMB3 as a cause of dominant hypoplastic amelogenesis imperfecta. Eur J Hum Genet. 2014;22:132–5. doi: 10.1038/ejhg.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry DA, Mighell AJ, El-Sayed W, Shore RC, Jalili IK, Dollfus H, et al. Mutations in CNNM4 cause Jalili syndrome, consisting of autosomal-recessive cone-rod dystrophy and amelogenesis imperfecta. Am J Hum Genet. 2009;84:266–73. doi: 10.1016/j.ajhg.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, et al. Whole-exome sequencing identifies FAM20A mutations as a cause of Amelogenesis Imperfecta and Gingival Hyperplasia Syndrome. Am J Hum Genet. 2011;88:616–20. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SK, Aref P, Hu Y, Milkovich RN, Simmer JP, El-Khateeb M, et al. FAM20A Mutations Can Cause Enamel-Renal Syndrome (ERS) PLoS Genet. 2013;9:e1003302. doi: 10.1371/journal.pgen.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum Mol Genet. 1998;7:563–9. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- 24.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–85. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 25.Picard C, McCarl CA, Papolos A, Khalil S, Luthy K, Hivroz C, et al. STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity. N Engl J Med. 2009;360:1971–80. doi: 10.1056/NEJMoa0900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S-K, Choi M, Richardson A, Reid B, Seymen F, Yildirim M, et al. STIM1 and SLC24A4 are critical for enamel maturation. J Dent Res. 2014;93:94–100. doi: 10.1177/0022034514527971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Es RJ, Wittebol-Post D, Beemer FA. Oculodentodigital dysplasia with mandibular retrognathism and absence of syndactyly: a case report with a novel mutation in the connexin 43 gene. Int J Oral Maxillofac Surg. 2007;36:858–60. doi: 10.1016/j.ijom.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Himi M, Fujimaki T, Yokoyama T, Fujiki K, Takizawa T, Murakami A. A case of oculodentodigital dysplasia syndrome with novel GJA1 gene mutation. Jpn J Ophthalmol. 2009;53:541–5. doi: 10.1007/s10384-009-0711-6. [DOI] [PubMed] [Google Scholar]
- 29.Schossig A, Wolf NI, Fischer C, Fischer M, Stocker G, Pabinger S, et al. Mutations in ROGDI cause Kohlschutter-Tonz Syndrome. Am J Hum Genet. 2012;90:701–7. doi: 10.1016/j.ajhg.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdon KP, McKay JD, Sale MM, Russell-Eggitt IM, Mackey DA, Wirth MG, et al. Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance-Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am J Hum Genet. 2003;73:1120–30. doi: 10.1086/379381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XF, Kraev AS, Lytton J. Molecular cloning of a fourth member of the potassium-dependent sodium-calcium exchanger gene family, NCKX4. J Biol Chem. 2002;277:48410–7. doi: 10.1074/jbc.M210011200. [DOI] [PubMed] [Google Scholar]
- 32.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–61. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 33.Seymen F, Lee KE, Tran Le CG, Yildirim M, Gencay K, Lee ZH, et al. Exonal Deletion of SLC24A4 Causes Hypomaturation Amelogenesis Imperfecta. J Dent Res. 2014;14:14. doi: 10.1177/0022034514523786. [DOI] [PubMed] [Google Scholar]
- 34.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC. Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci. 2011;119:311–23. doi: 10.1111/j.1600-0722.2011.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, et al. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–83. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmer S, Estrella N, Milkovich R, Hu J. Autosomal dominant amelogenesis imperfecta associated with ENAM frameshift mutation p.Asn36Ilefs56. Clin Genet. 2012;29:1–3. doi: 10.1111/j.1399-0004.2012.01887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SK, Hu Y, Simmer JP, Seymen F, Estrella NM, Pal S, et al. Novel KLK4 and MMP20 Mutations Discovered by Whole-exome Sequencing. J Dent Res. 2013;92:266–71. doi: 10.1177/0022034513475626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaulieu CL, Majewski J, Schwartzentruber J, Samuels ME, Fernandez BA, Bernier FP, et al. FORGE Canada Consortium: Outcomes of a 2-Year National Rare-Disease Gene-Discovery Project. Am J Hum Genet. 2014;94:809–17. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan AB, Tobochnik S, Dibattista M, Wall CM, Reisert J, Zhao H. The Na(+)/Ca(2+) exchanger NCKX4 governs termination and adaptation of the mammalian olfactory response. Nat Neurosci. 2011;15:131–7. doi: 10.1038/nn.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]