Abstract
The emergence of multidrug-resistant (MDR) Klebsiella pneumoniae has resulted in a more frequent reliance on treatment using colistin. However, resistance to colistin (Colr) is increasingly reported from clinical settings. The genetic mechanisms that lead to Colr in K. pneumoniae are not fully characterized. Using a combination of genome sequencing and transcriptional profiling by RNA sequencing (RNA-Seq) analysis, distinct genetic mechanisms were found among nine Colr clinical isolates. Colr was related to mutations in three different genes in K. pneumoniae strains, with distinct impacts on gene expression. Upregulation of the pmrH operon encoding 4-amino-4-deoxy-l-arabinose (Ara4N) modification of lipid A was found in all Colr strains. Alteration of the mgrB gene was observed in six strains. One strain had a mutation in phoQ. Common among these seven strains was elevated expression of phoPQ and unaltered expression of pmrCAB, which is involved in phosphoethanolamine addition to lipopolysaccharide (LPS). In two strains, separate mutations were found in a previously uncharacterized histidine kinase gene that is part of a two-component regulatory system (TCRS) now designated crrAB. In these strains, expression of pmrCAB, crrAB, and an adjacent glycosyltransferase gene, but not that of phoPQ, was elevated. Complementation with the wild-type allele restored colistin susceptibility in both strains. The crrAB genes are present in most K. pneumoniae genomes, but not in Escherichia coli. Additional upregulated genes in all strains include those involved in cation transport and maintenance of membrane integrity. Because the crrAB genes are present in only some strains, Colr mechanisms may be dependent on the genetic background.
INTRODUCTION
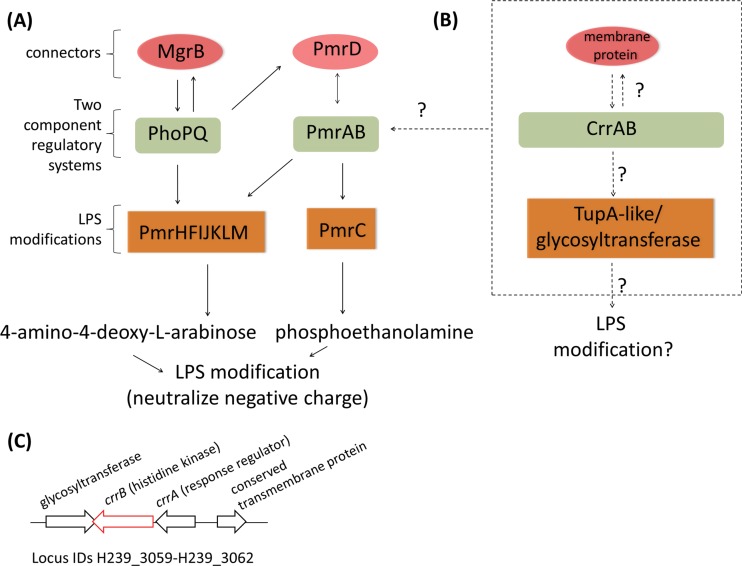
The increasing occurrence of multidrug-resistant (MDR) Klebsiella pneumoniae has expanded reliance on last-line therapies like colistin, a cationic antimicrobial peptide, for effective treatment (1). Of concern is the growing recovery of colistin-resistant (Colr) strains from clinical settings (2–4). Colistin disrupts membrane integrity through displacement of cations like Mg2+ and Ca2+ in the outer membrane, leading to cell lysis (5). Resistance mechanisms described to date involve lipopolysaccharide (LPS) modification, particularly through derivatization of lipid A phosphate moieties with a sugar or ethanolamine. These modifications reduce the electrostatic affinity between the cationic colistin and anionic LPS. Mutations in the transcriptional regulatory systems controlling these LPS modifications are a common genetic mechanism leading to colistin resistance. For example, the PhoPQ and PmrAB two-component regulatory systems (TCRS) regulate expression of the gene (pmrC) that codes for the addition of phosphoethanolamine (pETN) and genes encoding biosynthesis and lipid A transfer of 4-amino-4-deoxy-l-arabinose (Ara4N) (pmrHFIJKLM) (Fig. 1A). Other regulatory components in this pathway include PmrD and MgrB, two connector proteins that convey feedback between the PmrAB and PhoPQ TCRS (6–9) (Fig. 1A). Mutations in pmrAB, phoPQ, and mgrB have been identified as mechanisms conferring Colr in several Gram-negative pathogens, including K. pneumoniae (10–15).
FIG 1.
Model of Colr mechanisms. (A) Diagram of genes involved in colistin resistance based on published summaries from Gram-negative bacteria. (B) Model of newly identified proteins and potential interactions with previously described pathways of Colr. (C) Genome segment comprising differentially expressed genes, including the mutated histidine kinase gene in UHKPC26 and UHKPC28.
How colistin resistance mechanisms alter the global transcription profile and how transcriptome profiles vary in the genetic mechanism(s) that confers colistin resistance remain to be examined. By coupling complete genome sequence data and whole-genome transcriptional characterization of colistin-susceptible (Cols) and Colr K. pneumoniae, we were able to relate genetic mechanisms of colistin resistance to gene expression changes.
MATERIALS AND METHODS
Strains and genomic analyses.
Strains originated from a collection of KPC-producing K. pneumoniae isolates obtained from a consortium of tertiary-care hospitals previously described (16, 17). Colistin MICs were determined with a Sensititre system (17) and confirmed by Etest strips (bioMérieux). Genome sequencing was previously described (17) and consisted of Illumina HiSeq reads assembled with Newbler and annotated using the Comprehensive Microbial Resource annotation pipeline (18). Genome sequences were compared using Mauve (19), and gene content analysis used PanOCT (20). Single-nucleotide variants (SNVs) were determined using BWA for sequence read mapping (21) and Mpileup (22) for SNV detection in paired and closely related strains. A SNV-based phylogeny was constructed from kSNP software (23) output, as described in reference 17, with the inclusion of additional non-ST258 reference genomes. Two strains had paired isogenic Cols isolates (UHKPC57 with UHKPC179, and UHKPC27 with UHKPC52). SNVs were associated with Colr based on their presence in one of the Colr strains (Table 1) and the absence of that allele in all 48 Cols strains from reference 17 and additional genomes of the same MLST type in GenBank.
TABLE 1.
Klebsiella pneumoniae strains and predicted Colr-associated mutations
| Strain | GenBank accession no.a | SRA accession no.b | MLSTc | Locationd | Colistin MIC (μg/ml)e | KPC typec | Cols parent | Mutated geneg | Protein accession no. | Mutation |
|---|---|---|---|---|---|---|---|---|---|---|
| UHKPC45 | ARVO00000000 | SRR900179 | 258b | Cleveland OH, USA | 4 | 3 | mgrB | Deletion of 863 bp starting at mgrB38 | ||
| UHKPC52 | ARVN00000000 | SRR921382 | 258a | Cleveland OH, USA | 32 | Nonef | UHKPC27 | mgrB | IS1 family IS in mgrB with adjacent deletion, missing first 14 bp of mgrB | |
| UHKPC81 | APVQ00000000 | SRR900190 | 234 | Cleveland OH, USA | 4 | 2 | mgrB | IS4 family IS inserted at mgrB23 | ||
| VAKPC280 | APVZ00000000 | SRR900205 | 258a | Cleveland OH, USA | 16 | 2 | mgrB | IS5 family IS inserted at mgrB41 | ||
| 280_1220 | ARSQ00000000 | SRR900126 | 258a | Northeast Ohio, USA | 16 | 2 | mgrB | IS5 family IS inserted with adjacent deletion; full mgrB deletion | ||
| UHKPC179 | ARSM01000000 | SRR900157 | 15 | Cleveland OH, USA | 16 | Nonef | UHKPC57 | mgrB | EPO88337 | C28Y substitution |
| DMC1316 | ARSB00000000 | SRR900140 | 258b | Detroit, MI, USA | 1 | 3 | phoQ | EPO27247 | D434N substitution | |
| UHKPC26 | APVT00000000 | SRR900163 | 258a | Cleveland OH, USA | 16 | 2 | crrB | EOY97329 | L94M substitution | |
| UHKPC28 | ARRU00000000 | SRR900165 | 258a | Cleveland OH, USA | 16 | 2 | crrB | EPN94815 | Q10L substitution | |
| UHKPC27 | APVR00000000 | SRR900164 | 258a | Cleveland OH, USA | 0.25 | 2 | NA | Isogenic Cols strain | ||
| UHKPC57 | ARPR00000000 | SRR900182 | 15 | Cleveland OH, USA | 0.5 | 2 | NA | Isogenic Cols strain |
Accession number for the genomic sequence.
Accession number for the RNA-seq data set.
Multilocus sequence type (MLST) and blaKPC type determined as described in reference 15.
Geographic location of source hospital.
Determined by Etest.
Plasmid present in the paired Cols strain but not in the Colr strain.
NA, not applicable.
RNA-Seq experiments.
The 57 strains characterized in reference 17 were grown to mid-log phase at 37°C in LB broth. Cells were harvested and preserved with RNAprotect (Qiagen) until extraction with UltraClean RNA isolation kits (MoBio). cDNA libraries were constructed with ScriptSeq Complete Gold kits (Epicentre Biosciences) and were sequenced on an Illumina HiSeq instrument. Reads from each strain were mapped to the corresponding genome assembly and RPKM (number of mapped reads per kilobase of gene length per million total mapped reads) values were calculated in CLC (version 7.0.4). Genes with significantly different RPKM values were identified using the Significant Analysis for Microarray (SAM) (24) statistical analysis component of Multiexperiment Viewer (MeV version 4.9 [www.tm4.org]), where Colr and Cols strains represented the two unpaired classes.
Complementation assay.
To determine whether the mutation in crrB was necessary to confer colistin resistance, a pUC-19-derived plasmid containing the wild-type crrB gene as well as the upstream and downstream flanking sequences was introduced into UHKPC26 and UHKPC28. To generate a plasmid vector with a zeocin marker that could be used for selection of transformants in the MDR background of these strains, the bla gene in the pUC19 vector was replaced with the open reading frame of the resistance gene for zeocin. Specifically, by using the primers Zeo_ORF_F and Zeo_ORF_R (primer sequences are shown in Table S1 in the supplemental material), a 412-bp PCR fragment representing the open reading frame (ORF) of the zeocin resistance gene was amplified from a pAF6-derived plasmid (25). A vector fragment ending with the bla promoter and terminator, but not extending into the bla ORF, was amplified as a 1,857-bp fragment from pUC19 using the primers Zeo-pUC19_F and Zeo-pUC19_R. These fragments were purified using a PCR clean-up kit (ZymoResearch) and combined using Gibson assembly (26). The assembly product was used for transformation of NEB5α Escherichia coli cells (New England BioLabs). Plasmids were prepared from the transformants using a miniprep kit (Qiagen) and confirmed using restriction analysis with SmaI (New England BioLabs).
To clone a fragment containing the crrB gene into the zeocin plasmid, a 2,125-bp fragment was amplified from the genomic DNA sample of the Cols strain UHKPC27. The crrB locus is wild type in this strain. This amplification was first performed using Q5 polymerase and the primers MW_F_5Phos and MW_R. The amplified fragment was purified using a PCR purification kit (ZymoResearch) and further amplified using PrimeSTAR MAX polymerase (Clontech) and the primers MW_F and MW_R. The amplified fragment included the crrAB ORFs and the 5′ and 3′ flanking sequences. The zeocin vector was amplified using PrimeSTAR Max polymerase and the primers pUC19_MW_GA_F and pUC19_MW_GA_R. The amplified vector and insert fragments were purified using a PCR purification kit and used in Gibson assembly. The assembly mixture was used to transform NEB5α cells. Plasmids were purified from the transformants and confirmed using restriction analysis with HinfI and Sanger sequencing.
Electroporation of UHKPC26 and UHKPC28 was conducted as described previously (27) except that >1 μg of DNA was introduced into the transformation mixture and cells were recovered in 500 μl low-salt LB broth. The transformed cells were selected on an agar plate containing low-salt LB broth and 1,250 μg/ml zeocin. The presence of this plasmid in the colonies was confirmed using PCR with the primers Jct_Up_F and Jct_Up_R, where the amplification of the product in this PCR required the junction between the crrA upstream sequence and the vector to be present.
A broth microdilution assay was used to determine the MIC of UHKPC26 and UHKPC28 strains carrying the empty vector or the vector with the crrAB genes. Overnight cultures of UHKPC26 and UHKPC28 grown in cation-adjusted Mueller-Hinton broth (MHB) plus 1,250 μg/ml zeocin were washed, resuspended in MHB broth, and inoculated into MHB with a series of colistin concentrations. The Cols strain UHKPC27 was used as a control.
Nucleotide sequence accession numbers.
Sequence reads are available from the Short Read Archive at NCBI under accession numbers SRR896011, SRR900124 to SRR900127, SRR900132 to SRR900135, SRR900138 to SRR900144, SRR900151 to SRR900158, SRR900160 to SRR900169, SRR900178 to SRR900184, SRR900187 to SRR900191, SRR900193, SRR900198, SRR900199, SRR900202 to SRR900208, and SRR921382.
RESULTS
Genomic characterization.
Nine independent clinical strains representing three MLST sequence types (15, 234, and 258) were previously sequenced as part of a survey of K. pneumoniae strains in Midwestern U.S. hospitals (17) (Table 1). The most common mechanism of Colr involved mgrB alteration, a previously identified Colr mechanism in K. pneumoniae (6, 28) (Table 1). MgrB is a negative regulator of PhoQ, and inactivation leads to overexpression of phoPQ (9). Six strains had independent mutations of mgrB. Four strains experienced mgrB disruption mediated by different classes of insertion sequence (IS) families inserted at different positions in the mgrB gene. The MgrB amino acid substitution in UHKPC179 (C28Y) occurred at a cysteine residue previously identified as involved in a key disulfide bond in MgrB (29); thus, the substitution of a tyrosine here likely interferes with its ability to repress PhoQ. Genome analysis indicated that an 863-bp deletion resulted in loss of the entire mgrB gene and an adjacent ORF from UHKPC45. Two of these strains (UHKPC52 and UHKPC179) had a matched Cols isolate (UHKPC27 and UHKPC57, respectively) from the same patient obtained before the initiation of colistin treatment. In each case, the mgrB gene was intact in the parental isolates and disrupted in the Colr isolate.
A second resistance mechanism was identified as a mutation in the phoQ gene in DMC1316. The PhoQ substitution (D434N) in the cytoplasmic GHKL domain (ATP-lid loop) in DMC1316 is likely critical for phosphate transfer to PhoP (30).
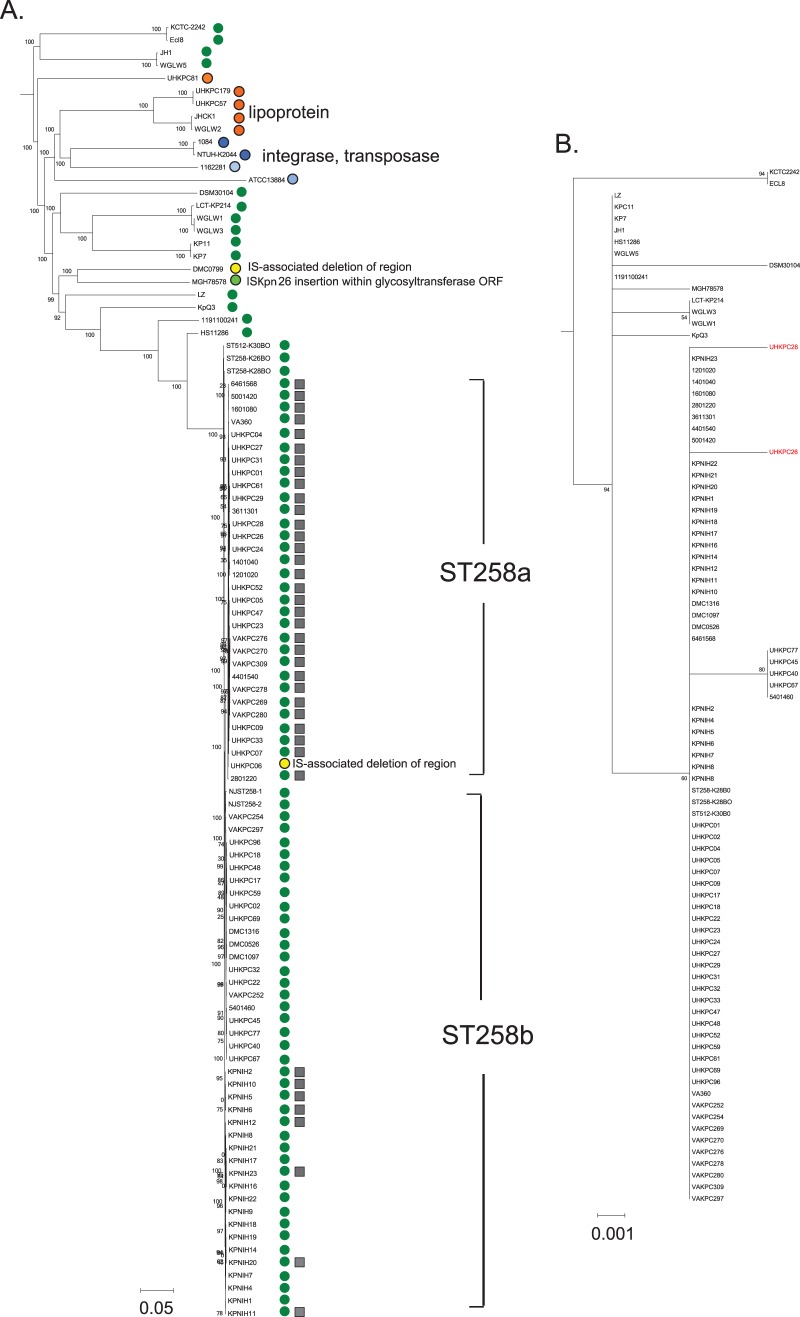
Strains UHKPC26 and UHKPC28 did not have any mutations in known colistin resistance pathways, but each strain possessed an independent mutation in the histidine kinase component of a previously uncharacterized two-component regulatory system, which we designate CrrAB to signify colistin resistance regulation. In each case, the “wild-type” allele is observed in Cols strains of ST258 (e.g., locus H239_3061 from strain UHKPC45) (Table 1). This histidine kinase gene (crrB) and an adjacent response regulator (crrA) are present in all K. pneumoniae ST258 strains with the exception of UHKPC06, where an insertion sequence (IS) event resulted in the deletion of the region, and in many other K. pneumoniae strain types (Fig. 2A). It appears that all K. pneumoniae strains have either the crr genes or an IS-mediated deletion or substitution of the region. The crrAB genes and the two flanking genes (Fig. 1C) do not have clear orthologs in E. coli or Salmonella, but orthologs of these genes are present several Enterobacter genomes but in a different genomic context than in K. pneumoniae.
FIG 2.
Comparative analyses of the crrAB region. (A) Presence of crrAB plus upstream (conserved hypothetical membrane protein ORF) and downstream (glycosyltransferase-like ORF) region among K. pneumoniae strains shown with a genome-wide SNV-based phylogeny constructed using kSNP output. Shared circle color indicates shared genome content and organization; gray squares indicate the presence of a shared ISKpn26 insertion upstream of the conserved hypothetical membrane protein H239_3059. (B) Maximum-likelihood tree of protein sequence alignment of CrrB (353 amino acids) in publicly available K. pneumoniae genomes.
Because no matched Cols parental strains were available for UHKPC26 or UHKPC28, we considered whether other variants in these strains might be involved in the Colr phenotype by identifying SNVs present in these strains relative to closely related Cols strains. The only shared nonsynonymous mutation in the two strains was in MenB (A244T), a naphthoate synthase protein that is part of the menaquinone biosynthesis pathway. Strain UHKPC28 had an additional mutation in RstA (F166Y), the response regulator component of RstAB, and a mutation in a transmembrane domain of WecA (G134A), which is involved in cell envelope synthesis. The complete list of sequence differences between UHKPC24, UHKPC26, and UHKPC28 genomes and the reference genome HS11286 are given in Table S2 in the supplemental material. None of these candidates were deemed to be stronger candidates as the Colr mutation than the crrB mutations.
Transcriptome characterization.
Two primary patterns of changes in gene expression were observed in Colr strains relative to Cols strains (Table 2). Common to both patterns was increased expression of the pmrHFIJKLM operon relative to Cols strains. This operon encodes genes responsible for the biosynthesis of Ara4N and modification of the lipid A component of LPS with this moiety. The six strains with mgrB modifications and the one phoQ mutant strain shared a pattern characterized by elevated phoPQ expression relative to Cols strains with no difference in pmrCAB transcript levels. The mutational spectrum in mgrB suggests loss-of-function mutations in this negative regulator of phoPQ, and each strain showed higher phoPQ expression levels than Cols strains from the collection. Of these strains, all but UHKPC52 also exhibited significantly elevated transcript levels for pmrD.
TABLE 2.
RPKM values for differentially expressed genes or those that have been previously associated with PhoPQ or PmrAB regulons in other organisms
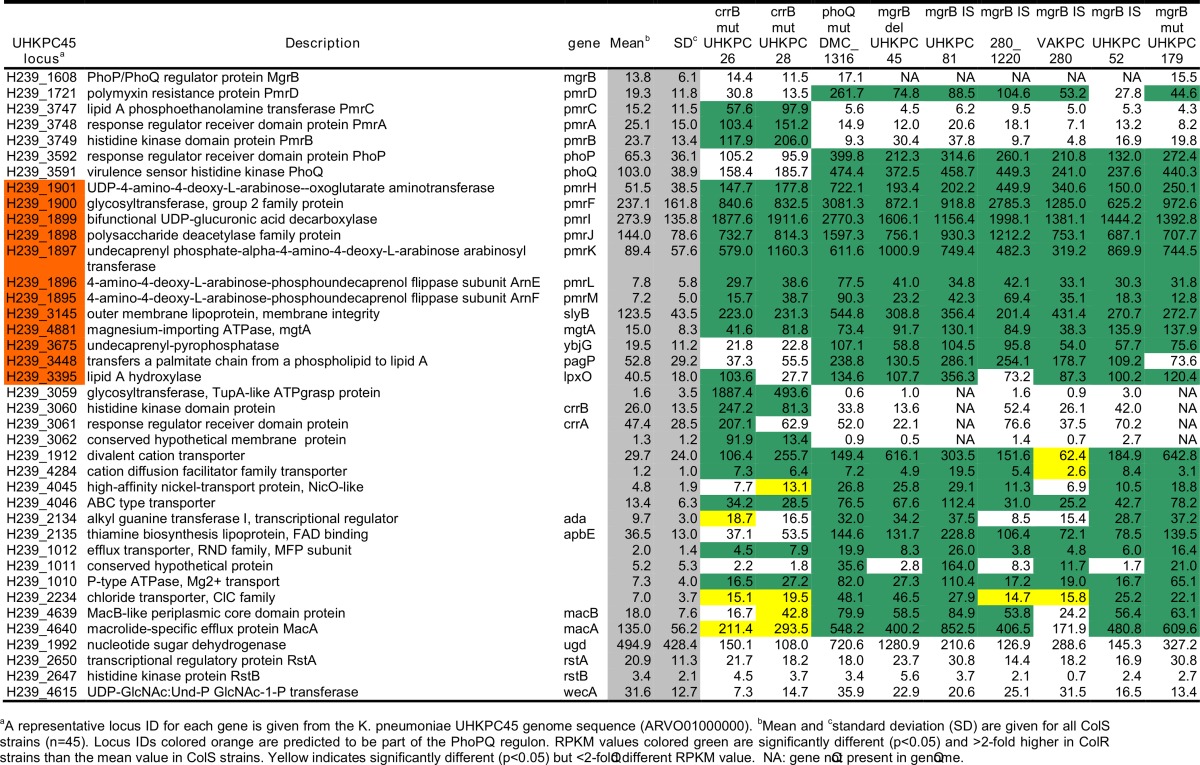
A representative locus ID for each gene from the K. pneumoniae UHKPC45 genome sequence (ARVO01000000) is given. Locus IDs in orange are predicted to be part of the PhoPQ regulon.
Means and standard deviations are given for all Cols strains (n = 45). RPKM values in green are significantly different (P < 0.05) and >2-fold higher in Colr strains than the mean value in Cols strains. Yellow indicates significantly different (P < 0.05) but <2-fold-different RPKM values. NA, gene not present in the genome.
SD, standard deviation.
In UHKPC26 and UHKPC28, the two strains with mutations in crrB, a distinct transcriptional pattern was observed, characterized by elevated transcription of pmrCAB, with no difference in phoPQ RPKM values compared to Cols strains. The sequences of the pmrCAB locus and flanking regions in these strains were identical to those in other ST258 strains. Expression levels were higher in UHKPC26 and UHKPC28 for crrB, a gene encoding an adjacent conserved hypothetical membrane protein, and a gene encoding an adjacent putative glycosyltransferase than in other Colr and Cols strains (Table 2 and Fig. 1C). In UHKPC26, the mutation is in the HAMP domain of the histidine kinase. In contrast, the histidine kinase mutation in UHKPC28 affects the predicted signal peptide domain, and transcript levels of the response regulator crrA were unaffected. Neither rstA nor wecA, two other genes with sequence variants in UHKPC28, had altered expression in UHKPC28 relative to Cols strains.
Other components of the PhoPQ regulon, such as slyB (encoding an outer membrane lipoprotein) and mgtA (encoding a magnesium-importing ATPase), were upregulated in all Colr strains (31–33), but pagP (encoding an outer membrane protein that adds a palmitate chain to lipid A), lpxO (encoding a dioxygenase that modifies lipid A) (34, 35), and ybjG (encoding a putative undecaprenyl pyrophosphate phosphatase) (36) were upregulated only in the mgrB and phoQ mutant strains. Other genes with altered transcription include those associated with cation transport, membrane integrity, and the macAB efflux transporters (Table 2). Expression of pmrD was elevated in some strains yet was not correlated with expression of pmrCAB under these experimental conditions for strains with mgrB or phoQ alterations. RPKM values for mgrB for strains with full-length sequences did not vary between Colr and Cols strains, indicating that the phoQ mutation in DMC1316 and related increased expression did not activate the feedback inhibition mediated by MgrB. No correlation was observed between the colistin MIC and the genetic mechanism of resistance: the MIC of strains with mgrB mutations ranged from 4 to 32 μg/ml. This could indicate that variation in additional genes, including genes in the phoPQ regulon, may modulate the resistance phenotype.
Confirmation of the role of crrB in colistin resistance.
To confirm that the crrB mutations in UHKPC26 and UHKP28 were necessary to confer colistin resistance, we cloned the wild-type crrAB genes from UHKPC27 into a pUC19-derived vector carrying a zeocin resistance marker and introduced this plasmid into the Colr strains. The UHKPC27 sequence is identical to that in UHKCP26 and UHKPC28 except for the hypothesized Colr-associated mutations. Colistin susceptibility was restored to UHKPC26 and UHKPC28 cells carrying the crrAB plasmid but not to cells with the vector alone (Table 3).
TABLE 3.
Complementation of crrB mutations
| Strain | MIC (μg/ml)a |
|---|---|
| UHKPC27 | <0.5 |
| UHKPC26 | >16 |
| UHKPC26 + vector | >16 |
| UHKPC26 crrAB | <0.5 |
| UHKPC28 | >16 |
| UHKPC28 + vector | >16 |
| UHKPC28 crrAB | <0.5 |
Determined by broth microdilution in cation-adjusted Mueller-Hinton broth.
DISCUSSION
Two distinct transcription profiles were observed; common to both was increased expression of the Ara4N pathway. Previous studies have shown linkage between Colr and pmrCAB expression (6, 12) and cross talk between pmrAB and phoPQ (37). The strains reported here demonstrate upregulation of either the pmrAB or phoPQ genes, but not both in the same Colr strain. The identification of the crr genes as additional regulators of colistin resistance expands the number of known genes that modulate this phenotype and highlights multifaceted ways that cells respond to antimicrobial peptide challenge.
The CrrAB proteins are not orthologs of other TCRS linked to PhoPQ or cell envelope stress response, such as RcsAB (38), RstAB (32), EvgAS (36), CpxAR (39), and BaeSR (40). Comparative analyses indicate that the crrAB region is variably present in K. pneumoniae, although it is present in nearly all ST258 strains (green circles in Fig. 2) and is found in other Klebsiella and Enterobacter species, albeit in different genomic contexts. In Enterobacter sp. strain SST3, a plant endophyte, the region is adjacent to the sap (sensitivity to antimicrobial peptides) operon (41). In K. pneumoniae genomes without these genes, either a lipoprotein of unknown function is present at this genomic location, or a phage-related integrase and transposase are present. The GC content of this region is approximately 40%, much lower than the average GC content of >50% for the K. pneumoniae chromosome, suggesting that it was laterally acquired. Genomic analysis also suggests that IS events are reshaping the region. ST258a strains described in reference 17 all have the same ISKpn26 insertion upstream of the conserved hypothetical protein (H239_3062), which also resulted in the deletion of 468 bp and truncation of an adjacent ABC transporter (H239_3063). Interestingly, eight ST258b strains described in reference 42 carried the same IS, ISKpn26, inserted at the same chromosomal position, suggesting a recombination event at this location. A phylogeny of CrrB is largely congruent with the SNV-based whole-genome phylogeny (Fig. 2B).
Both UHKPC26 and UHKPC28 had significantly elevated expression of an adjacent conserved hypothetical protein (H239_3059), a putative glycosyltransferase with a TupA-like ATP grasp domain (PF14305), which is predicted to be involved in surface polysaccharide biosynthesis, and an adjacent uncharacterized conserved membrane protein (H239_3062). These strains also had elevated expression of both the pmrH and pmrCAB operons, which encode the Ara4N and pETN pathways of LPS modification. Our working hypothesis is that CrrAB induces expression of the glycosyltransferase-like protein which transfers an as-yet-unidentified sugar to lipid A phosphate in a manner analogous to PmrC (Fig. 1B). The basis for the upregulation of pmrCAB has not been established. It is possible that CrrB, the novel TCRS histidine kinase, directly phosphorylates PmrA or that a yet-to-be-identified connector protein, perhaps the conserved hypothetical membrane protein (H239_3062), facilitates feedback between the two systems.
UHKPC26 and UHKPC28 do carry other single-nucleotide variants (SNVs) compared with Cols strains (see Table S1 in the supplemental material). RstA has been shown to be induced by PhoPQ in other Gram-negative pathogens (32, 43); however, the variant seen in UHKPC28 is unlikely to have contributed to the Colr phenotype as neither rstAB nor phoPQ expression was altered in UHKPC28. Furthermore, the UHKPC26 sequence was identical to Cols strains across this region, and the expression profiles are similar between the two strains. An additional variant was observed in a transmembrane region of WecA, which is involved in O-antigen biosynthesis, in strain UHKPC28 but is not predicted to be a key residue (44–46). However, the UHKPC26 wecA RPKM value was significantly lower than that in Cols strains even though there was no mutation in this gene. An examination of a potential role for WecA in LPS modification and a better characterization of what regulates its expression is needed to better understand any potential role in Colr.
Other transcription changes common to Colr strains included increased expression of cation transporters and other efflux pumps, which could be a response to altered membrane charge and difficulty in transporting cations through a LPS layer with a more neutral charge. Other genomic changes include the loss of blaKPC plasmids in UHKPC52 and UHKPC179 which were present in a closely related strain in the case of UHKPC52, or isogenic paired strain UHKPC57 in the case of UHKPC179. The loss of these plasmids suggests that there may be a fitness cost associated with acquiring colistin resistance that is partially mitigated by the plasmid loss, and this should be more rigorously explored with formalized fitness comparisons.
This study identified transcriptional changes of Colr strains during growth without colistin. Further experiments to examine how colistin exposure alters gene expression may provide additional insight into the resistance mechanism(s). Analysis of LPS and lipid A structures may identify novel modifications in UHKPC26 and UHKPC28. In addition, formal analysis of the fitness cost of resistance along with follow-up experiments to examine how the observed LPS modifications alter K. pneumoniae host interaction is important to understand the impact of colistin resistance on virulence.
Conclusions.
The combination of genomic and transcriptomic analysis revealed three genetic mechanisms conferring colistin resistance with distinct global gene expression profiles depending on the genetic mechanism. Interestingly, we found that each strain possessed unique mutations predicted to confer colistin resistance. Although the sample collection was not designed to be comprehensive or represent a formal epidemiological study, the absence of repeat observations of specific mutations suggests that independently derived Colr strains of K. pneumoniae may be more common than patient-to-patient spread of resistant strains. Moreover, the constellation of changes that can lead to a “final common phenotype” will challenge the future development of cationic antimicrobial peptides and molecular diagnostics, as targets may vary across strains. The fact that mutations were found in genes without E. coli orthologs highlights the importance of studying colistin resistance across the range of clinically significant pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded in part by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, under contract no. HHSN272200900007C. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers UM1AI104681 (R.A.B. and D.V.D.), R01AI072219 and R01AI063517 to R.A.B. and R01GM094403 to M.D.A. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program Award 1I01BX001974 and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B. In addition, K.K. is supported by the National Institute of Allergy and Infectious Diseases (NIAID); DMID Protocol Number: 10-0065; D.V.D. was awarded support from the Clinical and Translational Science Collaborative of Cleveland and award UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the NIH and NIH roadmap for Medical Research.
We thank the JCVI sequencing group for producing Illumina sequence data.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Veterans Administration.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04037-14.
REFERENCES
- 1.van Duin D, Kaye KS, Neuner EA, Bonomo RA. 2013. Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. Diagnostic Microbiol Infect Dis 75:115–120. doi: 10.1016/j.diagmicrobio.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis 53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capone A, Giannella M, Fortini D, Giordano A, Meledandri M, Ballardini M, Venditti M, Bordi E, Capozzi D, Balice MP, Tarasi A, Parisi G, Lappa A, Carattoli A, Petrosillo N, SEERBIO-GRAB Network . 2013. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect 19:E23–E30. doi: 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- 4.Marchaim D, Chopra T, Pogue JM, Perez F, Hujer AM, Rudin S, Endimiani A, Navon-Venezia S, Hothi J, Slim J, Blunden C, Shango M, Lephart PR, Salimnia H, Reid D, Moshos J, Hafeez W, Bheemreddy S, Chen TY, Dhar S, Bonomo RA, Kaye KS. 2011. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit, Michigan. Antimicrob Agents Chemother 55:593–599. doi: 10.1128/AAC.01020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 6.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Camacho E, Gomez-Gil R, Tobes R, Manrique M, Lorenzo M, Galvan B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoner FM, Alvarez-Tejado M, Garcillan-Barcia MP, De la Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 8.Luo SC, Lou YC, Rajasekaran M, Chang YW, Hsiao CD, Chen C. 2013. Structural basis of a physical blockage mechanism for the interaction of response regulator PmrA with connector protein PmrD from Klebsiella pneumoniae. J Biol Chem 288:25551–25561. doi: 10.1074/jbc.M113.481978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippa AM, Goulian M. 2009. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet 5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated to a single amino acid change in protein PmrB among Klebsiella pneumoniae of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Choi HJ, Ko KS. 2014. Differential expression of two-component systems, pmrAB and phoPQ, with different growth phases of Klebsiella pneumoniae in the presence or absence of colistin. Curr Microbiol 69:37–41. doi: 10.1007/s00284-014-0549-0. [DOI] [PubMed] [Google Scholar]
- 14.Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, St Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, Waterman PE. 2013. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J Infect Dis 208:1142–1151. doi: 10.1093/infdis/jit293. [DOI] [PubMed] [Google Scholar]
- 15.Cannatelli A, Pilato VD, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae associated with low-dosage colistin treatment. Antimicrob Agents Chemother 58:4399–4403. doi: 10.1128/AAC.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Duin D, Perez F, Rudin SD, Cober E, Hanrahan J, Ziegler J, Webber R, Fox J, Mason P, Richter SS, Cline M, Hall GS, Kaye KS, Jacobs MR, Kalayjian RC, Salata RA, Segre JA, Conlan S, Evans S, Fowler VG Jr, Bonomo RA. 2014. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 58:4035–4041. doi: 10.1128/AAC.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright MS, Perez F, Brinkac L, Jacobs MR, Kaye K, Cober E, van Duin D, Marshall SH, Hujer AM, Rudin SD, Hujer KM, Bonomo RA, Adams MD. 2014. Population structure of KPC-producing Klebsiella pneumoniae from Midwestern U.S. hospitals. Antimicrob Agents Chemother 58:4961–4965. doi: 10.1128/AAC.00125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, Sutton G. 2010. The comprehensive microbial resource. Nucleic Acids Res 38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts DE, Brinkac L, Beck E, Inman J, Sutton G. 2012. PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res 40:e172. doi: 10.1093/nar/gks757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. 1999. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol 1:239–251. doi: 10.1007/PL00011773. [DOI] [PubMed] [Google Scholar]
- 26.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 27.Fournet-Fayard S, Joly B, Forestier C. 1995. Transformation of wild type Klebsiella pneumoniae with plasmid DNA by electroporation. J Microbiol Methods 24:49–54. doi: 10.1016/0167-7012(95)00053-4. [DOI] [Google Scholar]
- 28.Gaibani P, Lombardo D, Lewis RE, Mercuri M, Bonora S, Landini MP, Ambretti S. 2014. In vitro activity and post-antibiotic effects of colistin in combination with other antimicrobials against colistin-resistant KPC-producing Klebsiella pneumoniae bloodstream isolates. J Antimicrob Chemother 69:1856–1865. doi: 10.1093/jac/dku065. [DOI] [PubMed] [Google Scholar]
- 29.Lippa AM, Goulian M. 2012. Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J Bacteriol 194:1457–1463. doi: 10.1128/JB.06055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marina A, Mott C, Auyzenberg A, Hendrickson WA, Waldburger CD. 2001. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J Biol Chem 276:41182–41190. [DOI] [PubMed] [Google Scholar]
- 31.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol 185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lejona S, Aguirre A, Cabeza ML, Garcia Vescovi E, Soncini FC. 2003. Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J Bacteriol 185:6287–6294. doi: 10.1128/JB.185.21.6287-6294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J Bacteriol 189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons HS, Reynolds CM, Guan Z, Raetz CR. 2008. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A Biochemistry 47:2814–2825. doi: 10.1021/bi702457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eguchi Y, Okada T, Minagawa S, Oshima T, Mori H, Yamamoto K, Ishihama A, Utsumi R. 2004. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J Bacteriol 186:3006–3014. doi: 10.1128/JB.186.10.3006-3014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng HY, Chen YF, Peng HL. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouslim C, Groisman EA. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol 47:335–344. doi: 10.1046/j.1365-2958.2003.03318.x. [DOI] [PubMed] [Google Scholar]
- 39.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1844:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Leblanc SK, Oates CW, Raivio TL. 2011. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol 193:3367–3375. doi: 10.1128/JB.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gan HM, McGroty SE, Chew TH, Chan KG, Buckley LJ, Savka MA, Hudson AO. 2012. Whole-genome sequence of Enterobacter sp. strain SST3, an endophyte isolated from Jamaican sugarcane (Saccharum sp.) stalk tissue. J Bacteriol 194:5981–5982. doi: 10.1128/JB.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam D, Choi E, Kweon DH, Shin D. 2010. The RstB sensor acts on the PhoQ sensor to control expression of PhoP-regulated genes. Mol Cells 30:363–368. doi: 10.1007/s10059-010-0126-8. [DOI] [PubMed] [Google Scholar]
- 44.Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A. 2008. Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J Bacteriol 190:7141–7146. doi: 10.1128/JB.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amer AO, Valvano MA. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571–582. [DOI] [PubMed] [Google Scholar]
- 46.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. 2007. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol 189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.