Abstract
A hierarchical hormonal cascade along the hypothalamic-pituitary-adrenal axis orchestrates bodily responses to stress. Although corticotropin-releasing hormone (CRH), produced by parvocellular neurons of the hypothalamic paraventricular nucleus (PVN) and released into the portal circulation at the median eminence, is known to prime downstream hormone release, the molecular mechanism regulating phasic CRH release remains poorly understood. Here, we find a cohort of parvocellular cells interspersed with magnocellular PVN neurons expressing secretagogin. Single-cell transcriptome analysis combined with protein interactome profiling identifies secretagogin neurons as a distinct CRH-releasing neuron population reliant on secretagogin’s Ca2+ sensor properties and protein interactions with the vesicular traffic and exocytosis release machineries to liberate this key hypothalamic releasing hormone. Pharmacological tools combined with RNA interference demonstrate that secretagogin’s loss of function occludes adrenocorticotropic hormone release from the pituitary and lowers peripheral corticosterone levels in response to acute stress. Cumulatively, these data define a novel secretagogin neuronal locus and molecular axis underpinning stress responsiveness.
Keywords: acute stress, Ca2+ sensor, HPA axis, vesicular release
Introduction
Stress is the body’s reaction to environmental conditions or noxious challenges. Fast adaptive responses to stress are reliant on a hierarchical hormonal cascade that adjusts and adapts to metabolic processes at the periphery (Selye & Fortier, 1949; Baxter & Forsham, 1972) to maximize energy expenditure in order to avoid adverse environments (Sapolsky et al, 1986; Swanson, 1991; Korte, 2001; de Kloet et al, 2005; McEwen, 2007). Stress hormone release is controlled by the hypothalamic-pituitary-adrenal (HPA) axis whose anatomical organization, allowing dynamic hormonal integration at the ligand and receptor levels and via extensive feedback and amplification loops, is well established (Kiss et al, 1984; Makino et al, 2002; Simmons & Swanson, 2009; Tasker & Herman, 2011). In contrast, the localization and molecular identity of many neurosecretory neurons in rodents (particularly mouse) and humans are less well characterized, partly due to the rapid, phase-locked nature of the synthesis and release of stress hormones, limiting their usefulness as cellular markers (Swanson & Simmons, 1989; Meister et al, 1990). In this context, the neuronal sites producing corticotropin-releasing hormone (CRH) (Spiess et al, 1981; Vale et al, 1981) in the paraventricular nucleus (PVN) of the hypothalamus (Swanson et al, 1986; Swanson & Simmons, 1989; Herman et al, 1992; Sawchenko et al, 1993), as well as rate-limiting steps of CRH release into the hypophyseal portal circulation at the median eminence, are of particular significance since CRH liberates adrenocorticotropic hormone (ACTH) as the initial step of the peripheral stress response (Rivier & Vale, 1983; Kovacs & Sawchenko, 1996). Nevertheless, detailed molecular underpinnings of CRH release remain to be defined.
The release of arginine–vasopressin (AVP) and oxytocin from magnocellular neurosecretory cells (Gainer et al, 2002), and of combinations of releasing and inhibitory hormones and other neuropeptides from parvocellular neurons (Swanson & Sawchenko, 1983), requires Ca2+-dependent quantal exocytosis of dense-core vesicles in response to afferent stimulation or direct metabolic challenges, entraining bursts of neuronal excitation in the PVN (Dutton & Dyball, 1979; Cazalis et al, 1985; Carafoli et al, 2001; Petersen et al, 2005; Dayanithi et al, 2012). The priming of vesicular release is regulated by a multi-protein complex sequentially recruiting Ca2+ sensors, SNARE proteins and the docking machinery embedded in the presynaptic terminal (Hata et al, 1993; Sollner et al, 1993; Sutton et al, 1998; Pang & Sudhof, 2010). Ca2+ sensors are proteins that undergo conformational changes upon Ca2+ binding, sculpting inter-molecular interactions with downstream effectors to propagate action potential-dependent release events (Brose et al, 1992; Sudhof & Rothman, 2009). As such, calbindin-D28k and calretinin were histochemically associated with magnocellular neurosecretory cells (Sanchez et al, 1992; Arai et al, 1993). In contrast, neuron-specific Ca2+ sensors were neither anatomically localized to nor functionally characterized in parvocellular hypothalamic neurons.
Recent neuroanatomical mapping places secretagogin, an EF-hand Ca2+-binding protein (Wagner et al, 2000), into the PVN and adjacent pre-autonomic areas (Mulder et al, 2009; Maj et al, 2012). Despite the unexpected abundance of neurons expressing secretagogin mRNA and protein (secretagogin+ neurons) in the mammalian hypothalamus, their phenotype, cumulatively defined by their fast neurotransmitter, neuropeptide and/or hormone contents, remains unknown. Moreover, and even if the proximity of secretagogin+ neurons to the ventricular system along the longitudinal axis of the forebrain might indirectly infer a conserved role in secretory processes (Mulder et al, 2009), secretagogin’s role in the nervous system remains elusive.
Here, we sought to address whether secretagogin marks a hitherto undescribed neurosecretory locus in the PVN. We have combined systems neuroanatomy, neurophysiology, single-cell transcriptomics and proteomics to unequivocally define secretagogin+ neurons as a cohort of CRH-expressing parvocellular cells, partly interspersed within magnocellular domains of the PVN, and to show that this protein is a Ca2+ sensor modulating CRH release. By exploiting a genetic approach, we show that secretagogin-mediated CRH secretion is indispensable to prime ACTH release into the general circulation upon acute noxious stress (Makara et al, 1981). Cumulatively, our results define a key molecular mechanism gating the HPA axis upon stress-induced activation of CRH release from a molecularly distinct subtype of neuroendocrine parvocellular neurons in the hypothalamus.
Results
Secretagogin+ neurons in the hypothalamic paraventricular nucleus
Recent neuroanatomical studies have identified abundant secretagogin expression in the hypothalamus (Mulder et al, 2009; Maj et al, 2012). Yet neither the detailed topology of secretagogin+ neurons nor their functional significance in neuroendocrine control is known. Herein, we localized secretagogin+ neurons, in descending order and along the longitudinal axis of the adult mouse brain, to paraventricular (PVN) = arcuate ≫ periventricular, dorsomedial and ventromedial hypothalamic nuclei (Fig 1A–A3 and B). A dense secretagogin+ cell group was found in the dorsolateral PVN, adjacent to and partly interspersed with magnocellular oxytocin+ and AVP+ magnocellular neurons, whose axons primarily descend into the posterior pituitary for the transient storage and “on-demand” release of these hormones (Bargmann, 1969; Gainer et al, 2002). Clearly, oxytocin, AVP and secretagogin segregated to non-overlapping cell cohorts (Fig 1B and B1). We extended this analysis to show that the supraoptic nucleus, an alternative site for oxytocin+ and AVP+ magnocellular neurons to reside (Swanson & Sawchenko, 1983), lacked appreciable secretagogin expression (Fig 1B2). These data suggest that secretagogin+ neurons belong to a discrete group of neurons interspersed with but not identical to oxytocin+ or AVP+ magnocellular neurons.
Figure 1. Secretagogin locus in the paraventricular nucleus of the hypothalamus.
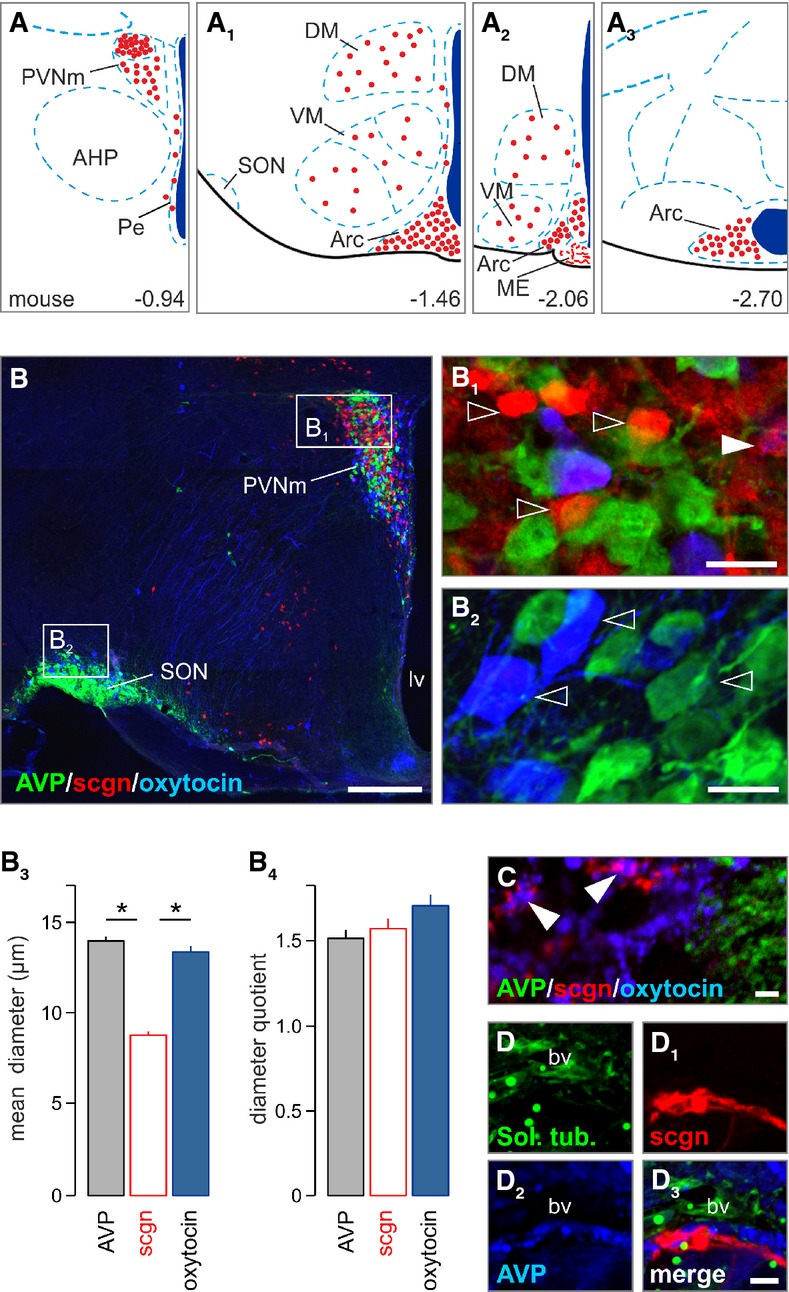
- A–A3 Secretagogin+ neurons populate the paraventricular, dorsolateral, ventromedial, periventricular and arcuate nuclei of the mouse hypothalamus (Paxinos & Franklin, 2001). Red circles denote the localization of neuronal perikarya and their relative densities. AHP, anterior hypothalamus; Arc, arcuate nucleus; DM, dorsomedial nucleus; ME, median eminence; Pe, periventricular nucleus; PVN, paraventricular nucleus, PVNm, magnocellular part; SON, supraoptic nucleus; VM, ventromedial nucleus.
- B–B2 Largely non-overlapping distribution of AVP+, oxytocin+ and secretagogin+ (sgcn+) neurons in the magnocellular PVN. The mouse supraoptic nucleus (SON) harbored vasopressin+ and oxytocin+ but not secretagogin+ neurons. Open arrowheads pinpoint single-labeled neurons. Solid arrowhead denotes AVP/secretagogin dual-labeling. lv, lateral ventricle; PVNm, magnocellular part of the paraventricular nucleus; scgn, secretagogin.
- B3, B4 Secretagogin+ neurons had smaller somatic diameters than AVP+ or oxytocin+ neurons yet without a difference in their diameter quotient, a measure of ovoid profiles (*P < 0.05, Student’s t-test).
- C–D3 Terminal-like profiles in the posterior pituitary (arrowheads in C) suggesting that secretagogin can co-exist, even if infrequently with oxytocin. Solanum tuberosum lectin (Sol. tub) was used to identify blood vessels.
Data information: Scale bars: 100 μm (B), 10 μm (B1, B2), 2 μm (C–D3)
Next, we employed quantitative morphometry of secretagogin+ neurons to cytoarchitecturally distinguish them from their oxytocin+ or AVP+ counterparts. Secretagogin+ soma were significantly (P < 0.05) smaller in diameter than (Fig 1B3), but equally round (Fig 1B4) as those of either oxytocin+ or AVP+ neurons, suggesting parvocellular identity (Swanson & Sawchenko, 1983). Nevertheless, secretagogin+ axons occasionally terminated around blood vessels in the posterior pituitary and formed Herring body-like specializations that at points contained moderate levels of oxytocin (Fig 1C–D3). Cumulatively, these data suggest that secretagogin is broadly expressed in the PVN in a molecularly distinct neuronal contingent. Here, we focused on deciphering the identity and function of small-diameter “parvocellular-like” PVN neurons.
Secretagogin typifies a novel subclass of parvocellular neurons in the PVN
Protein localization at the subcellular level is a reliable predictor of protein function. Our high-resolution laser-scanning analysis (Fig 1B and B1) suggested that secretagogin, a predicted cytosolic protein (Birkenkamp-Demtroder et al, 2005), might be distributed ubiquitously, including dendrites and axons. We confirmed this notion at the ultrastructural level by pre-embedding immunoelectron microscopy (Fig 2A–C). In the PVN, silver-intensified immunogold particles, revealing secretagogin’s localization, were detected in the cytoplasm (Fig 2A), particularly in submembranous compartments or in likely contact with the plasmalemma (Fig 2A1), and along the endoplasmic reticulum (Fig 2A2). Likewise, secretagogin was localized to dendrite shafts, often along with their inner membrane surface (Fig 2B). Moreover, we demonstrated secretagogin immunoreactivity in close proximity to the plasmalemma in nerve endings and possibly to dense-core vesicles (Fig 2C). Quantitative analysis of gold-intensified silver particles in the PVN (soma, dendrites) and the median eminence (axonal nerve endings) demonstrated that the majority of secretagogin associates to (endo-)membranes in neurons (that is, a given particle was < 50 nm from the nearest membrane surface; Fig 2D). Nevertheless, we found an opposite configuration in secretagogin distribution when comparing particle numbers on the plasmalemma and in the cytosol in neuronal soma and axons: In neuronal soma, the majority of secretagogin labeling appeared intracellular (Fig 2D). In contrast, secretagogin localization along the plasma membrane of nerve endings outweighed cytosolic secretagogin content. These data suggest that secretagogin might play a role in controlling hormone or neuropeptide release.
Figure 2. Ultrastructural analysis suggests the prevalence of membrane-bound secretagogin.
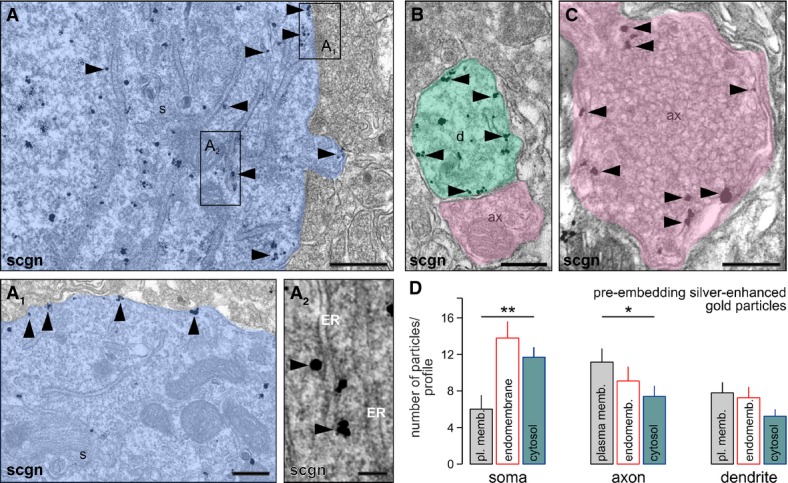
- Secretagogin (scgn) distribution at the ultrastructural level as revealed by pre-embedding silver-enhanced immunogold labeling. Secretagogin (arrowheads) was localized to membranous organelles in the perikarya (A), particularly the plasmalemma (A1) and endoplasmic reticulum (ER) in neuronal soma (s; A2). Open rectangles in (A) denote the location of insets. Semi-transparent shading is used to visually dissociate subcellular compartments in (A–C).
- Pre-embedding secretagogin labeling (arrowheads) was also seen in dendrite (d) segments.
- In axo-dendritic terminals (ax), secretagogin was closely associated with synaptic vesicles along the plasmalemma (arrowheads).
- Quantitative analysis of subcellular secretagogin distribution upon electron microscopy detection of silver-enhanced gold particles. Particles were considered as membrane bound when they were at < 50 nm of a membrane (plasma membrane or endomembrane; i.e., secretory vesicle, Golgi or endoplasmic reticulum). In the soma of PVN neurons, significantly more particles were found in the cytosol as along the plasma membrane (**P < 0.01). In contrast, membrane association predominated in axonal nerve endings in the median eminence (*P < 0.05). Note that a significant proportion of particles was found adjacent to endomembranes in all subcellular compartments studied.
Data information: Scale bars:, 1 μm (A), 500 nm (B, C), 200 nm (A1, A2).
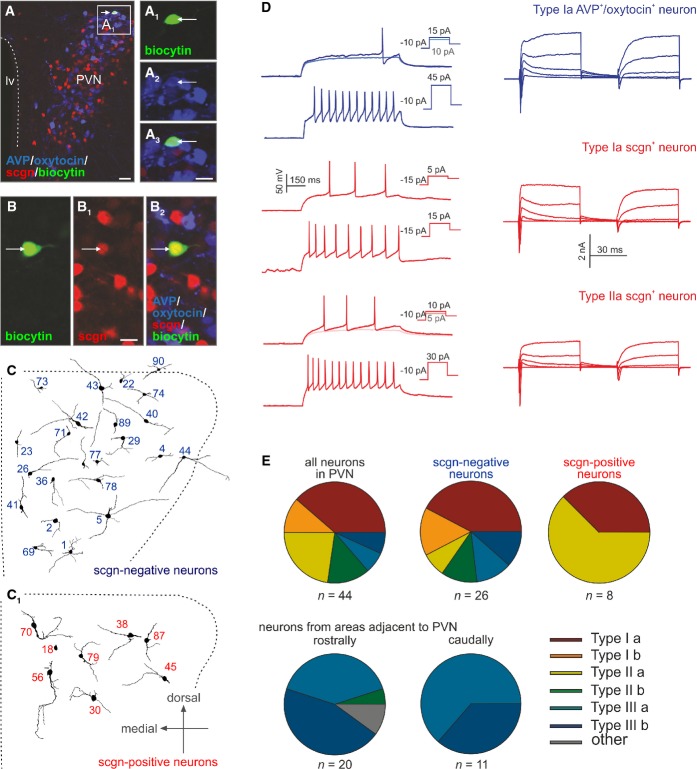
Next, we have taken a neurophysiological approach to gain insights into the biophysical properties of secretagogin+ neurons and distinguish these from magnocellular, parvocellular or pre-autonomic PVN neurons using their properties to generate action potentials (APs), as well as 16 additional parameters (Supplementary Table S1) as classification criteria (Luther & Tasker, 2000; Luther et al, 2002). In rat, magnocellular neurons are characterized by a delay in generating the first AP upon current stimulation after pre-hyperpolarization (type I) due to a relatively high amplitude of their A-type currents (Luther & Tasker, 2000; Lee et al, 2012). In contrast, AP delay is atypical for parvocellular neurons (type II) (Luther & Tasker, 2000; Lee et al, 2012). Pre-autonomic neurons generate low threshold spikes and bursts given the activity of voltage-gated T-type Ca2+ channels (Stern, 2001; Lee et al, 2008). In whole-cell patch-clamp experiments using ex vivo hypothalamus slice preparations, we have analyzed > 75 neurons in the PVN and pre-autonomic areas of juvenile mice (postnatal days 21–28). Using unified current- and voltage-clamp protocols, we designed novel classification criteria for mouse PVN neurons (Fig 3), distinguishing three primary neuron types, which could then be clustered into six subtypes (Fig 3E). In particular, type I neurons were reminiscent of magnocellular neurons from rat, including delayed AP generation after pre-hyperpolarization and high-amplitude A-type-like currents (Fig 3A–A3 and D). This neuron population could be divided into Ia and Ib subgroups, based on outward current properties in recording conditions inactivating A-type channels (Supplementary Fig S1). Ia neurons generated outward currents typical for slowly activated delayed-rectifying K+ channels (Fig 3D, Supplementary Fig S1). Meanwhile, Ib neurons exhibited transient, fast-activated currents upon depolarization (from −40 mV; Supplementary Fig S1). Post hoc immunohistochemistry defined biocytin-filled magnocellular neurons as exclusively belonging to the Ia group.
Figure 3. Electrophysiological classification of secretagogin+ parvocellular neurons.
- A–A3 Biocytin-filled (arrow) neuron immunonegative for secretagogin yet containing AVP/oxytocin (a mixture of magnocellular markers was used in triple-labeling experiments). Scale bars: 50 μm (A), 8 μm (A3). lv, lateral ventricle; PVN, paraventricular nucleus.
- B–B2 Secretagogin+ biocytin-filled neuron (arrow) lacking AVP/oxytocin immunosignal. Scale bars: 8 μm.
- C, C1 Dendritic reconstruction of secretagogin− and secretagogin+ neurons at their actual location in the paraventricular nucleus of the hypothalamus.
- D Electrophysiological characteristics of magnocellular and parvocellular neurons. The waveform of repetitive action potential firing is shown on the left, while differential channel characteristics are depicted on the right. Examples of secretagogin+ neurons are shown in red.
- E Pie diagrams showing the grouping of neurons in the PVN (upper row) and in adjacent pre-autonomic areas (lower row) cumulatively based on electrophysiological criteria listed in Supplementary Table S1. Secretagogin+ neurons typically belonged to Ia and IIa subtypes.
Neurons that had histochemically been identified as secretagogin positive (Fig 3B–B2) primarily belonged to type II mouse parvocellular neurons and were equivalent in biophysical properties to those described in rat (Lee et al, 2012) or mouse (Wamsteeker Cusulin et al, 2013). Using the classification criteria introduced for type I neurons above, the type II cohort was subdivided as IIa and IIb neurons (Supplementary Fig S1). Nevertheless, we also identified some secretagogin+ neurons with AP signatures similar but not identical to magnocellular cells (Fig 3D and E), reinforcing our hypothesis on immunohistochemically undetectable AVP and/or oxytocin levels and introducing a novel scale of molecular heterogeneity among magnocellular PVN neurons.
Lastly, type III neurons were secretagogin negative(−) low threshold and produced spike bursts upon somatic current injections (Supplementary Fig S1). These pre-autonomic cells, likewise sub-clustered as IIIa and IIIb (Fig 3E), therefore were excluded as being secretagogin+ neurons. Overall, these data suggest that the majority of secretagogin+ neurons in the cluster were parvocellular cells in the PVN.
Secretagogin+ parvocellular neurons express CRH
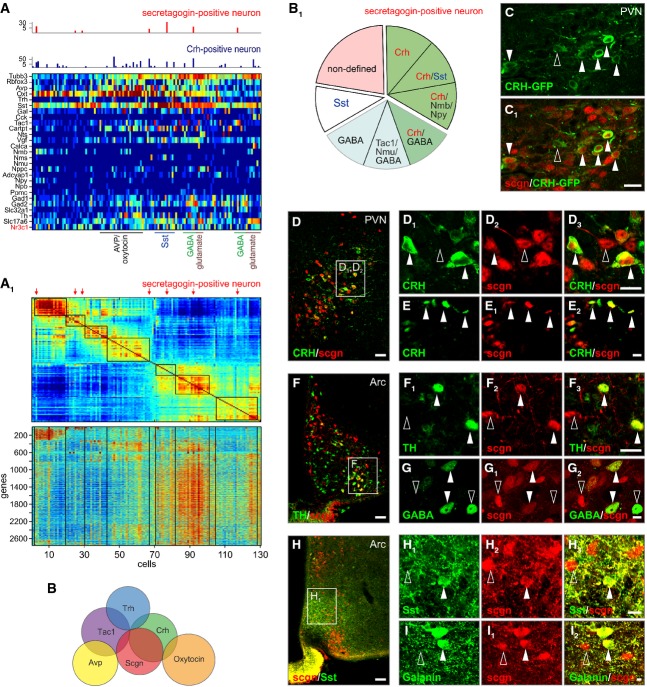
Parvocellular neurons in the PVN and other hypothalamic areas are diverse as to their neurochemical phenotypes (Swanson & Sawchenko, 1983; Everitt et al, 1986; Swanson et al, 1986), with their majority producing releasing or release-inhibiting hormones (Guillemin, 1978; Schally, 1978) that regulate trophormone secretion in the anterior pituitary via the rete mirabile of the hypothalamo-hypophyseal portal system, a concept originally described by Harris (1972). Considering that somatic neuropeptide and hormone detection is often difficult and relies on the permanent blockade of the anterograde axonal transport machinery (Cortes et al, 1990), we applied unbiased clustering analysis to single-cell transcriptome data (Tsafrir et al, 2005; Islam et al, 2014) generated from 130 cells dissociated from the mouse PVN. Overall, oxytocin, AVP, somatostatin, GABA and glutamate neurons exhibited discrete mRNA profiles in the PVN (“hot spots” in Fig 4A and A1; GEO accession number GSE63093, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63093). In contrast, secretagogin+ neurons, as well as CRH+ and thyrotropin-releasing hormone (TRH)+ neurons, did not form separate clusters.
Figure 4. Molecular identity of secretagogin+ paraventricular neurons.
Single-cell mRNA transcriptome profiling of dissociated cells from the mouse paraventricular nucleus.
- A Differential clustering based on secretagogin (Scgn), neuropeptide, hormone and hormone receptor mRNA expression. Secretagogin-expressing(+) neurons typically contained corticotropin-releasing hormone (Crh) and Nr3c1 mRNA transcripts.
- A1 Clusters of gene transcripts from 130 cells reveal the phenotypic segregation of PVN neurons. Increasing mRNA copy numbers were depicted by a color gradient from deep blue (not detected) to red (high numbers). Secretagogin+ neurons are indicated by red arrows.
- B Venn diagrams depicting the (non-)overlapping relationships of select mRNA transcripts used to molecularly define PVN neurons.
- B1 Cumulative ratio of phenotypic diversity among secretagogin+ neurons. Note that Crh and/or GABA (derived from Gad1, Gad2 and Slc32a1 mRNA expression) co-existed with secretagogin in parvocellular neurons.
- C, C1 Secretagogin co-localization with GFP in the PVN (solid arrowheads) of adult CRH-GFP (BAC) reporter mice (Alon et al, 2009). Open arrowhead pinpoints a single-labeled neuron.
- D–I3 We found secretagogin (scgn) co-expressed with CRH in both neuronal soma (solid arrowheads; D1–D3) and axon terminal-like specializations (arrowheads; E–E2) in the PVN. Likewise, secretagogin+ neurons harbored, yet infrequently, tyrosine hydroxylase (TH; solid arrowheads; F–F3), GABA (G–G2), somatostatin (Sst; H–H3) and galanin (I–I2). Open rectangles denote the general location of insets. Open arrowheads indicate the lack of co-localization.
Data information: Scale bars: 40 μm (D, F, H), 25 μm (C1), 10 μm (D3, F3, G2, H3), 4 μm (E2, I2).
Next, we focused on which RNAs for hormones, neuropeptides and/or low-molecular-weight neurotransmitters were present in secretagogin+ neurons. We demonstrated the transcriptome identity of these neurons by analyzing the absolute number of mRNA transcripts from 151 cells dispersed from the mouse PVN (Fig 4A and A1). We first tested the abundance of AVP, oxytocin, CRH and TRH gene transcripts (Fig 4A and A1) and show their broad variations, with particular reference to AVP and oxytocin mRNAs (that is, variations at 3–4 orders of magnitude; Supplementary Fig S2A). We defined a GABAergic hypothalamic contingent of ˜30%, expressing Gad1 and/or Gad2 encoding respective glutamic acid decarboxylase 65 and 67 kDa (GAD65/GAD67) isoforms, as well as the vesicular GABA transporter (Slc32A1; Fig 4A). Tyrosine hydroxylase and vesicular glutamate transporter 2 mRNAs were present in ˜15 and ˜10% of the threshold-adjusted sample, respectively.
We then restricted our analysis of baseline mRNA expression values for each gene by ranking from minimum to maximum expression values (Supplementary Fig S2A) to increase homogeneity (Fig 4A1). As such, this cell population comprised 7.3% oxytocin+ neurons (n = 11), 6.0% AVP+ neurons (n = 9), 4.6% TRH+ neurons (n = 7) and 6.6% CRH+ neurons (n = 10). When dissecting the presence of gene transcripts of key pro-neuropeptides, our data from 151 cells revealed distinct somatostatin (n = 15 with high level), galanin (n = 11), cholecystokinin (n = 5), tachykinin 1 (n = 9), cocaine- and amphetamine-regulated transcript (CART; n = 24), neurotensin S (n = 2), VGF nerve growth factor-inducible protein (n = 21), calcitonin gene-related peptide (n = 4), neuromedin B (n = 4), neuromedin S (n = 2), natriuretic peptide C (n = 2) and adenylate cyclase activating polypeptide 1 (Adcyap1; n = 2). Of these, somatostatin, galanin, cholecystokinin, neurotensin S and CART were expressed at mRNA copy numbers exceeding 100 per cell, while other peptides were found expressed at low or moderate copy numbers. mRNA transcripts for neuropeptide Y, neuromedin U, neuropeptide B and pro-opiomelanocortin were infrequently present. The co-existence of fast neurotransmitters (GABA, dopamine, glutamate) with hormones and neuropeptides allowed for a refined molecular classification of the PVN and associated hypothalamic areas (Fig 4A and A1).
Lastly, we focused on secretagogin mRNA expression (Fig 4A and A1; GEO accession number GSE63093, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63093). From a total of 151 cells, we identified nine cells with secretagogin mRNA transcripts. Secretagogin+ cells in the sampling cohort lacked AVP, oxytocin, TRH, galanin, cholecystokinin, neurotensin S, calcitonin, neuromedin S, natriuretic peptide C and Adcyap1 mRNAs. In contrast, some secretagogin+ cells contained somatostatin, Tac1, CART, neuromedin B, neuromedin U, neuropeptide Y and/or neuropeptide B mRNA transcripts (Fig 4A and B). Secretagogin+ cells abundantly carried CRH mRNA transcripts and those of the promiscuous glucocorticoid receptor Nr3c1 (Fuxe et al, 1985) (Pearson’s correlation coefficient = 0.399, P < 0.001), the latter being involved in the stress-induced feedback regulation of CRH synthesis (Aguilera et al, 2007).
mRNA may not be translated into appreciable amounts of protein. Therefore, it was imperative to histochemically verify the neurochemical identity of secretagogin+ neurons. First, we used a reporter mouse line expressing GFP under the control of the CRH promoter (Alon et al, 2009) to reveal the abundant presence of CRH+/secretagogin+ neurons in the PVN (Fig 4C and C1). We then applied colchicine in vivo to provoke neuropeptide accumulation in somatic domains in parvocellular neurons (Fig 4D–I2) (Cortes et al, 1990). Indeed, secretagogin was found chiefly co-expressed with CRH in PVN in both neuronal soma and local axon collaterals (Fig 4D–E3) and, less frequently, with tyrosine hydroxylase (ventrolateral arcuate nucleus; Fig 4F–F3), GABA (dorsomedial arcuate nucleus; Fig 4G–G2), somatostatin (Fig 4H–H3) and galanin (Fig 4I–I2) with dual-labeled secretory terminals accumulating in the median eminence (Fig 5A–A2). These data define secretagogin+ neurons as a CRH-containing cell cohort.
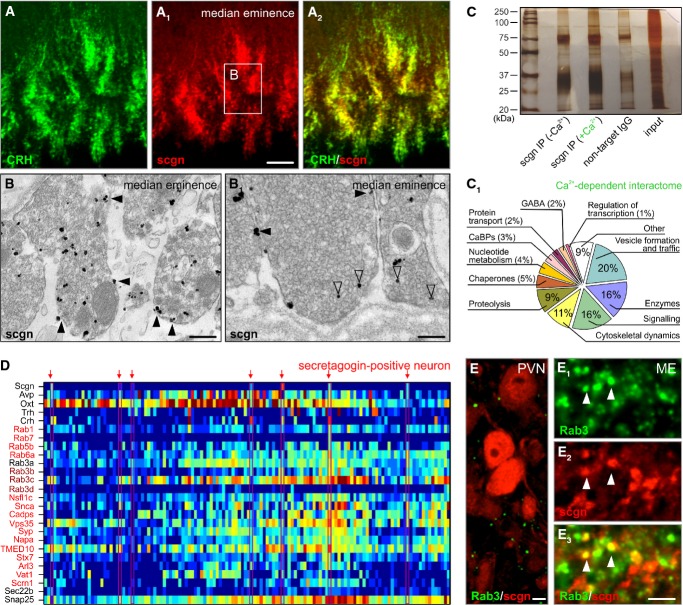
Figure 5. Secretagogin is a Ca2+ sensor protein.
- A–A2 Secretagogin co-existed in the majority of CRH+ nerve endings in the median eminence (ME). Open rectangle denotes the general location of (B, B1).
- B, B1 Large axon terminals in the median eminence were immunopositive for secretagogin with silver-intensified immunogold particles (open arrowheads) associated with axonal membrane and dense-core vesicles (see Fig 1D for quantitative data). Solid arrowheads denote silver deposit particles proximal to the plasmalemma.
- C Immunoprecipitation using an anti-secretagogin antibody in Ca2+-free and Ca2+-loaded conditions was subtractively used to decipher the Ca2+-dependent protein interactions. Silver-stained gel is shown.
- C1 Ontology classification of the 99 protein hits based on primary function assignments. Unbiased MALDI-TOF proteomics was used to identify interacting proteins. The most abundant hits (Supplementary Table S3 is referred to for details on individual proteins) are proteins implicated in vesicle fusion, trafficking, transport and formation and the regulation of vesicle exocytosis. “Other” refers to a group of proteins without known function.
- D Single-cell transcriptomics was used to validate the above proteome data by clustering mRNA transcripts encoding proteins that underpin vesicular release processes (red color labels highest mRNA abundance, whereas dark blue color indicates the lack of mRNA expression).
- E–E3 Rab3, a family of vesicular fusion and transport proteins (Schluter et al, 2006), was found co-localized with secretagogin (arrowheads) in the median eminence. Neuronal soma in the PVN lacked appreciable co-localization. Note that neither our MALDI-TOF analysis nor our histochemical probing allowed the precise identification of individual Rab3C-E family members.
Data information: Scale bars: 10 μm (A1, E3), 7 μm (E), 500 nm (B), 150 nm (B1).
Secretagogin+ neurons are tightly packed in the dorsolateral PVN. Dendritic reconstructions clearly distinguish these cells from magnocellular neurons (Fig 3C and C1). Nevertheless, we controlled our in vitro transcriptome profiling by plating dissociated PVN neurons from newborn mice and assessing their dendritic morphology. Secretagogin+ neurons had significantly smaller (P < 0.05; Supplementary Fig S2B and C) somatic diameter and more emanating (P < 0.05) and exceedingly arborizing (P < 0.05) processes than AVP+ neurons. In addition, the majority were secretagogin+/AVP− neurons. Yet a small secretagogin+ subpopulation co-expressed AVP, with their somatic diameters and dendrite complexities reminiscent of AVP+/secretagogin− neurons (Supplementary Fig S2B and C), lending further support to our in vivo findings (Fig 1). Ca2+-imaging experiments after KCl stimulation showed that parvocellular-like secretagogin+ neurons clearly segregated from their AVP+ or AVP+/secretagogin+ counterparts (Supplementary Fig S2D). Moreover, secretagogin+ neurons responded differentially to excitatory stimuli, including NMDA combined with glycine (P < 0.01), kainate (P < 0.001) and GABA (Haam et al, 2012) (P < 0.001; Supplementary Fig S3A–D1). These data not only demonstrate that secretagogin+ parvocellular neurons represent a molecularly and functionally distinct class of PVN neurons but also imply that secretagogin, a Ca2+-binding protein presumed associated with the SNARE machinery in vitro (Rogstam et al, 2007; Bauer et al, 2011), might sculpt their responsiveness and/or affect CRH release. However, direct in vivo evidence discerning Ca2+ “buffer” versus “sensor” roles for this protein remained elusive.
Secretagogin is a Ca2+ sensor expressed in the median eminence
Secretagogin+ neurons exhibited low-amplitude Ca2+ responses to depolarizing stimuli (Supplementary Fig S2D), as compared to AVP+/secretagogin− neurons in vitro. Therefore, we first tested whether secretagogin acts as a Ca2+ buffer. Upon transfecting human SH-SY5Y neuroblastoma cells with a plasmid encoding human secretagogin, we estimated their Ca2+ buffer capacity by population-wide (> 100 cells/experiment) Ca2+ imaging experiments in combination with post hoc assessment of neurochemical properties (Supplementary Fig S4A). The amplitude of Ca2+ responses in secretagogin+ neurons was not statistically different from control cells (Supplementary Fig S4B). Next, the relative level of secretagogin, scaled as immunofluorescence intensity (Supplementary Fig S4C), did not correlate with either the basal intracellular Ca2+ level or peak amplitudes evoked by carbachol or KCl treatments. Similarly, Ca2+ response kinetics failed to significantly correlate with secretagogin immunoreactivity at the level of individual cells (data not shown). Cumulatively, these results support that secretagogin is unlikely to function either as slow or as fast Ca2+ buffer, at least in vitro.
In view of the negative data on Ca2+ buffering, we hypothesized that secretagogin is a Ca2+ sensor in vivo. This notion is supported by secretagogin’s concentration and co-localization with CRH in the median eminence (Fig 5A–A2), the locus of the first capillary network of the hypothalamic artery that collects releasing hormones produced by neurosecretory cells, including CRH (Harris, 1972; Léránth et al, 1983; Swanson & Sawchenko, 1983). In particular, we employed pre-embedding silver-intensified immunogold labeling to show, at the ultrastructural level, that secretagogin is present in axon terminals with frequent association with the plasmalemma, and possibly also with the outer membrane of dense-core vesicles (Fig 5B and B1; for quantitative data in axons see Fig 2D). The presence of silver precipitates along the inner membrane of the nerve terminal reinforces a potential role for secretagogin as a Ca2+ sensor.
Next, we addressed whether secretagogin is a Ca2+ sensor to influence neurotransmitter transport and vesicular release by defining its Ca2+-dependent interactome using unbiased matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) proteomics (Fig 5C). We deployed a subtractive approach, whereby positive hits were defined as only recruited in the presence of 10 μM Ca2+ in the isolation buffers but not recovered under Ca2+-free conditions or by non-specific rabbit IgGs. Our hits (97 proteins in Ca2+-dependent interactions and 15 proteins without Ca2+ dependence (Supplementary Table S3); minimum number of peptide fragments ≥ 2, protein threshold: 99%, peptide threshold: 99%) therefore were interpreted as recruited upon Ca2+ influx (Fig 5C1, Supplementary Table S3), and chiefly included proteins implicated in: (i) vesicle formation, fusion and traffic; (ii) enzymatic reactions; (iii) signaling; and (iv) cytoskeletal dynamics and axonal transport (Fig 5C1) (Sudhof, 2004; Schluter et al, 2006; Wickner & Schekman, 2008). We also uncovered interactions with other Ca2+-binding proteins, glutamate decarboxylase and vesicular GABA transporter (Fig 5C1), reinforcing our transcriptome analysis. We validated the expression of mRNAs for many of these proteins in secretagogin+ neurons by targeted analysis of our single-cell transcriptome data (Fig 5D). This approach revealed the expression of multiple genes of interest involved in vesicular traffic and fusion in secretagogin+ parvocellular neurons (Fig 5D). Lastly, we performed immunohistochemistry in rat paraventricular neurons and median eminence with antibodies raised against Rab3 (all isoforms) to visualize secretagogin’s frequent co-existence with this family of proteins at the median eminence (Fig 5E–E3). In sum, these findings suggest that secretagogin is a Ca2+ sensor, which can affect CRH release from parvocellular neurons.
Secretagogin controls CRH release in vitro and in vivo
We first determined whether secretagogin can affect CRH release by its RNAi-mediated knockdown in native PVN neurons (Fig 6A), followed by secretagogin histochemistry and the recovery of CRH in culture supernatants (Fig 6A1). We found significantly decreased CRH content in the culture media of hypothalamic neurons exposed to secretagogin-directed RNAi (Fig 6A1). Subsequently, we took the opposite approach and transiently overexpressed secretagogin in an immortalized CRH-containing embryonic mouse hypothalamic cell line (mHypoE-N44). Three days after transfection, mHypoE-N44 cells were stimulated with 15 μM ATP for 20 min, fixed and immunostained for secretagogin (Fig 6B and B1) with simultaneous intracellular CRH detection (Fig 6B2). CRH levels negatively correlated with that of secretagogin (Pearson’s correlation coefficient = −0.189, P = 0.016) upon secretagogin overexpression. In contrast, no significant correlation between cytosolic secretagogin and extracellular CRH content was determined in non-stimulated samples (Pearson’s correlation coefficient = 0.0379, P = 0.56). We concluded that in mHypoE-N44 cells, secretagogin can facilitate ATP-dependent CRH release without an effect on CRH synthesis per se.
Figure 6. Secretagogin regulates CRH release in vitro and in vivo.
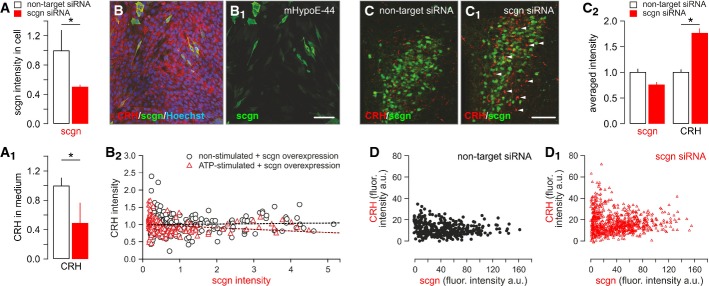
- A, A1 siRNA-mediated secretagogin (scgn) knockdown in cultured hypothalamic neurons, as indicated by reduced secretagogin immunoreactivity (A) and decreased CRH content in the culture medium (A1).
- B–B2 Transient overexpression of secretagogin in immortalized CRH-expressing mHypoE-N44 hypothalamic cells significantly reduced CRH immunofluorescence intensity in secretagogin+ cell bodies, indirectly supporting enhanced CRH release. All experiments were performed in triplicate. Scale bar: 50 nm.
- C, C1 siRNA-mediated in vivo silencing of secretagogin mRNA expression in the PVN provoked somatic CRH accumulation (arrowheads). Scale bar: 150 μm.
- C2 Quantitative analysis demonstrating significantly increased somatic CRH contents. Note that somatic secretagogin levels remained unchanged, which we interpret as data on a neuronal contingent not affected by siRNA silencing. The lack of secretagogin/CRH co-localization suggests that secretagogin expression fell below detection threshold in many CRH+ neurons.
- D, D1 Individual data points show maximal CRH fluorescence intensity (gray scale arbitrary unit (a.u.) expression) in PVN neurons that have low or no secretagogin expression after siRNA infusion.
Data information: Data in (C2) were normalized to those in non-targeting siRNA controls. *P < 0.05 versus control.
Next, we sought evidence that secretagogin knockdown affects CRH release in vivo by injecting siRNA (“Accell”) against secretagogin into the lateral ventricle and by performing quantitative double-label histochemistry in serial sections 3 days later. In the anterior PVN, which comprises CRH+ parvocellular neurons (Keegan et al, 1994), secretagogin knockdown occurred in 20–30% of CRH neurons (Fig 6C and C1). Since CRH, like other neuropeptides, undergoes axonal transport and Ca2+-dependent release after synthesis (Alonso et al, 1986), we observed CRH retention in parvocellular neuronal soma in PVN in RNAi-exposed hypothalami (P < 0.05; Fig 6C2, D and D1). We attributed the lack of change in secretagogin immunoreactivity, when measuring immunofluorescence intensity in individual cells, to the fact that our knockdown approach effectively arrested secretagogin expression in high CRH-expressing neurons, as supported by the relative scarcity of dual-labeled neurons. We considered our RNAi approach aimed to silencing secretagogin expression specific because neither magnocellular neurosecretory cells nor some neuropeptide systems localized to the PVN were affected upon experimental manipulations (Supplementary Fig S5A–D). Overall, our data suggest that secretagogin might modulate CRH release in the hypothalamus.
Secretagogin gates the CRH–ACTH axis in response to acute stress
Secretagogin’s involvement in modulating CRH release in vitro and in vivo suggests that this Ca2+-binding protein can be instrumental to modulate the stress-induced mobilization of the hypothalamic-pituitary-adrenal axis by priming the hierarchical CRH–ACTH cascade that underpins a peripheral corticosterone surge from the adrenal cortex (Swanson & Sawchenko, 1980; Makara et al, 1981; Swanson & Simmons, 1989). If so, secretagogin+ neurons are assumed to rapidly up-regulate their CRH content and function as “on-neurons” to activate hormonal responses to stress. We addressed this hypothesis by injecting formalin (4%) into one paw (Pacak et al, 1995; Naveilhan et al, 2001) and detecting activation of the immediate early gene c-fos (Ohtori et al, 2000) in the PVN 6h later (Fig 7A and B). Quantitative immunohistochemistry revealed that 62 ± 8% of c-fos+ neurons co-expressed secretagogin and CRH (Fig 7B1).
Figure 7. Secretagogin+ neurons gate the CRH–ACTH axis in response to acute stress.
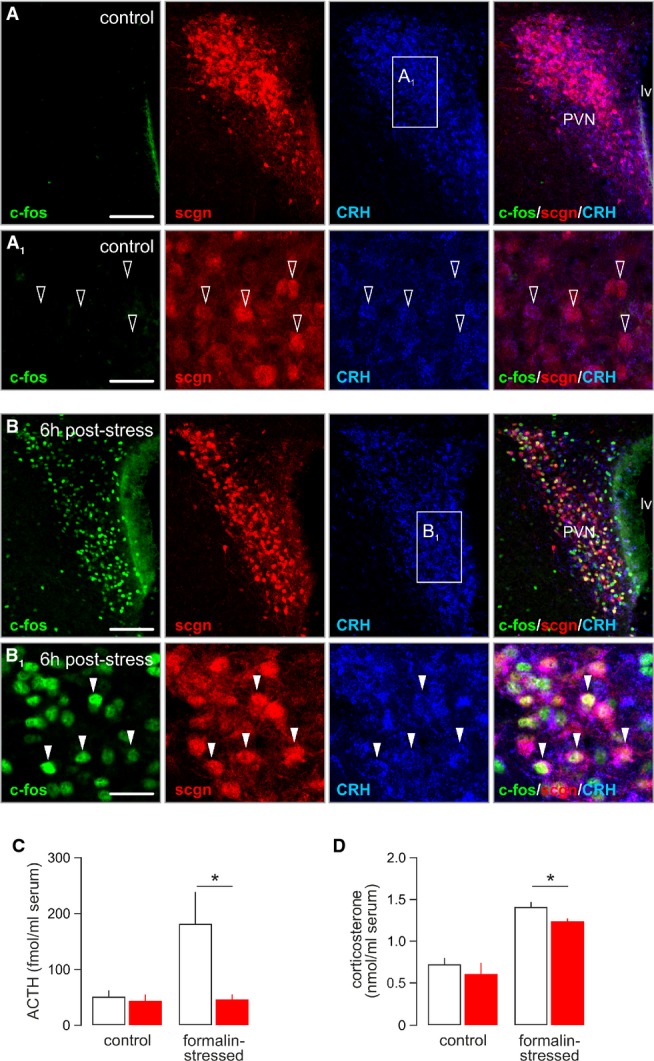
- A, A1 Paraventricular secretagogin-immunoreactive (scgn+) neurons weakly expressed CRH (open arrowheads) but not c-fos under control conditions. Scale bars: 300 μm (A), 25 μm (A1).
- B, B1 Stress induced by subcutaneous injection of formalin-triggered co-expression of c-fos and CRH in secretagogin+ paraventricular neurons (arrowheads). Scale bars: 300 μm (B), 25 μm (B1).
- C, D In vivo siRNA-mediated silencing of secretagogin expression in the PVN occluded the stress-induced surge of serum ACTH levels (C) and significantly reduced the increase in plasma corticosterone (D). *P < 0.05 versus control.
Since secretagogin is a Ca2+ sensor protein expressed in CRH+ cells recruited by acute stress, it is plausible to assume that its physiological role is the control of CRH release. To test this hypothesis, we performed secretagogin siRNA knockdown in vivo followed by determination of circulating ACTH and corticosterone levels 12 min after evoking formalin stress, coinciding with peak ACTH responses in blood (Cam & Bassett, 1983). Formalin injection in control animals (that is, injected with non-target siRNA 4 days prior) produced significant increase in ACTH content in plasma (Fig 7C). In contrast, infusion of secretagogin-specific siRNA into the lateral ventricle 4 days prior to formalin challenge blunted this ACTH response (P < 0.05; Fig 7C). Likewise, the ensuing corticosterone response was significantly reduced (Fig 7D) in mice treated with secretagogin siRNA. In sum, our data suggest that secretagogin is instrumental for CRH release and stress responsiveness.
Discussion
This study provides novel insights into the neurochemical organization of the mammalian hypothalamus. We report that secretagogin in many parvocellular neurons modulates CRH release. Unlike for small neurotransmitters, CRH, as many other neuropeptides/hormones, is not reused or recycled at the locus of its release. Moreover, CRH is transported over considerable distances from its somatic site of synthesis in the PVN to the median eminence (Supplementary Fig S6). Thus, hormonal secretion into the bloodstream is reliant on the coordination of regulated checkpoints, which might be either indirectly (vesicle formation and axonal transport; Supplementary Fig S6A) or directly (vesicular exocytosis) affected by secretagogin. From a technical standpoint, our study uses an interactive design of single-cell transcriptome and proteome discovery, verified at the level of systems neurobiology. Yet we did not apply real-time imaging of CRH release upon genetic manipulation of secretagogin expression. Therefore, we cannot entirely exclude that secretagogin, at least in part, primes the release machinery indirectly by recruitment of a multicomponent apparatus rather than directly interacting with its key components (Supplementary Fig S6B). Nevertheless, secretagogin was identified as a Ca2+ sensor protein whose proteome-wide interactions include α-SNAP, Rab family members, syntaxins, synaptophysin, TMED10 and CAPS1, many of which were implicated in vesicle formation, trafficking, exo- and endocytosis, and are integral components of synaptic vesicle membranes (Takamori et al, 2006; Tobin et al, 2012a,b). These interactions are compatible with the predominant association of secretagogin with the plasmalemma and endomembranes in hypothalamic neurons and nerve endings in the median eminence. We hypothesize that differential distribution of secretagogin in axons is due to the fusion of somatic endomembranes into the axonal plasma membrane, thus providing a structural conduit for secretagogin trafficking toward exocytosis release sites. Besides, secretagogin’s Ca2+-dependent interactions include molecular motors (kinesin-1, myosin-10), with kinesin-1 known to affect axonal transport (Hirokawa & Takemura, 2005), suggesting that secretagogin can intracellularly modulate the timing and velocity of cargoes moved along the axon and readied for subsequent exocytosis.
Although detailed neuroanatomy studies define at least seven neurochemically distinct sub-regions of the mammalian PVN (Simmons & Swanson, 2009), the classical cytoarchitectonic concept is that dorsolaterally located magnocellular neurons are spatially segregated from the parvocellular cell groups. While parvocellular neurons concentrate between the 3rd ventricle and magnocellular neuron clusters, CRF neurons neighbor magnocellular neurosecretory cells along the ventrolateral extremity of this hypothalamic area (Swanson & Sawchenko, 1980, 1983; Pelletier, 1991). This cytoarchitectonic organization is derived from a string of exquisite neuroanatomical studies spanning the past ˜40 years (Kiss et al, 1983; Palkovits, 1984; Kita et al, 1986; Swanson et al, 1986; Swanson & Simmons, 1989; Swanson, 1991; Horvath & Gao, 2005; Maejima et al, 2009; Simmons & Swanson, 2009; Vogt et al, 2014) and utilizing hormones/neuropeptides to mark neurosecretory cell identity, as well as somatic size measurements as classification criteria (Kiss et al, 1991). These classical methods often relied on colchicine-treated or stressed rodents to provide unequivocal phenotypic information on, for example, CRH neurons. Under these conditions, CRH cells co-express additional hormones and peptides, including vasopressin, neurotensin, angiotensin II and encephalin (Swanson et al, 1986; Ceccatelli et al, 1989). With regard to fast neurotransmitters, CRH neurons in rat are mainly glutamatergic (Hrabovszky et al, 2005) even though a subgroup was shown to be GABAergic (Meister et al, 1988). Functionally, the action of amino acid neurotransmitters to modulate pituitary hormone-producing endocrine cells may occur via local axon collaterals within the PVN. Local amino acid transmitter synthesis in, and release from, nerve endings in the external layer of the median eminence may yet modulate the release of CRH (and other releasing/inhibitory factors) into portal vessels by engaging presynaptic auto- and/or heteroreceptors.
The advent of single-cell transcriptome analysis by RNA sequencing (Shapiro et al, 2013) revolutionized insights into cell diversity in many developing (Tang et al, 2009) and adult tissues (Islam et al, 2011; Kodama et al, 2012; Lovatt et al, 2014). In the nervous system, transcriptome analysis is predicted to reveal that neuronal diversity is far more complex than initially thought, with many non-classical neuronal sub-types emerging (that is, modalities or combinations of the mRNA landscape overarching several known cell subsets). These might be of particular relevance to specific behavioral contexts, environmental challenges or disease phenotypes. More specifically, in the mammalian hypothalamus, a significant expansion of the phenotypic divergence of functionally distinct neuroendocrine subtypes, carrying mixed phenotypic codes including sub-threshold levels of hormones or neuropeptides, is expected upon unbiased clustering of cellular transcriptomes. This will identify neuroendocrine cell cohorts differentially responding to not one but multiple stimuli. As such, secretagogin+ neurons in mouse were found to express mRNA transcripts for CRH, Gad1, Gad2, Slc32a1, Tac1 and somatostatin. This mRNA transcript repertoire confers, as said, a parvocellular origin (Dierickx & Vandesande, 1979; Alonso et al, 1986; Ceccatelli et al, 1989), yet with a broader mRNA spectrum than for a single classical subtype of parvocellular neuroendocrine cells. These considerations cumulatively have the following inferences: parvocellular neurons express a chief hormone, a neuropeptide. Nevertheless, they maintain the ability to co-express other (secondary) releasable products, including small molecule transmitters such as GABA and glutamate playing auxiliary roles. This allows dynamically switching mode of action upon a range of metabolic stimuli, underpinning fast “on/off” responses. Here, this hypothesis is reflected by the moderate presence of secretagogin in magnocellular neurons and their axon terminals in the posterior pituitary. Moreover, our single-cell transcriptome analysis revealed the coincidence of Scgn, CRH and Nr3c1 mRNA expression. Nr3c1 encodes a glucocorticoid receptor (nuclear receptor subfamily 3, group C member 1) (van Rossum et al, 2004). Nr3c1 can both function as a transcription factor that binds to glucocorticoid response elements in the promoters of glucocorticoid responsive genes to activate their transcription or act as a regulator of other transcription factors. Nr3c1 mutations manifest as generalized glucocorticoid resistance (Donner et al, 2013), characterized by the insensitivity of target tissues to glucocorticoids, resulting in the compensatory activation of the HPA axis and CRH hypersecretion into the systemic circulation. Thus, secretagogin may be the first molecular hinge linking Nr3c1-mediated feedback to CRH release from parvocellular neurons.
Since parvocellular neurons receive local-circuit, as well as long-range afferents (Kiss et al, 1984, 1996; Palkovits, 1987), the predominance of single gene products coincides with, and is driven by, the type of excitatory or inhibitory inputs they receive. Considering that hypothalamic neuronal circuitries are prone to synaptic reorganization upon metabolic challenges (Pinto et al, 2004; Horvath & Gao, 2005), parvocellular neurons might function as “fast integrators” in hypothalamic neuronal circuitries and can translate synaptic information into hormonal output with high fidelity. This arrangement is compatible with our electrophysiology classification showing that secretagogin+ neurons, although clearly parvocellular, belong to both Ia and IIa PVN subclasses. In addition, an evolutionary advantage of the broad functional diversity of neuronal contingents in the hypothalamus may be the ability of sustained neural control of secondary metabolism at the periphery, essential for the survival of the organism, even under pathological conditions associated with neuronal loss.
Action potentials represent the temporal code of electrical activity in neurons. Upon the arrival of a depolarization wave, a kaleidoscope of voltage-dependent ion channels regulates ion exchange along the plasma membrane. On the one hand, this influx of Ca2+ is to induce signaling events shaping the neurons’ transcriptional program to adapt to circuit requirements (Clapham, 2007). On the other hand, Ca2+ is the ubiquitous trigger for the quantal release of neurotransmitters and neuromodulators at axon terminals (Sudhof, 2004). In the hypothalamus, intracellular Ca2+ signaling regulates AVP and oxytocin release in magnocellular neurons (Dayanithi et al, 2012). Parvocellular neurons are equally dependent on Ca2+-dependent mechanisms (Hallbeck et al, 1996). In particular, a Ca2+/calmodulin-dependent signaling pathway was reported to modulate the transcription of CRH in neurons (Yamamori et al, 2004). Ca2+ waves are transduced into intelligible second messenger codes by Ca2+ sensor proteins with low affinity (> 1 μM) in protein–protein interactions. Alternatively, cytosolic Ca2+ buffers (Ca2+ affinity; < 1 μM) dissipate Ca2+ transients by reversibly binding 2–6 Ca2+ ions (Andressen et al, 1993). Although a clear separation of these two protein subfamilies is increasingly debated (Schwaller, 2009), a clear evidence for Ca2+ sensor function is the Ca2+-dependent recruitment of signaling proteins. Here, we used subtractive proteomics to define the Ca2+-dependent interactome of secretagogin in native hypothalamic preparations. Our data revealed that secretagogin interacts with protein subfamilies implicated in the formation, intracellular traffic, priming and docking of release vesicles. These findings are not entirely unexpected, since a pull-down analysis using purified secretagogin as bait reported similar results (Bauer et al, 2011). However, this is the first confirmed report of secretagogin’s interactome in situ in an identified neuronal cohort, which defines cell type-specific protein interactions for this Ca2+ sensor. Critically, our single-cell transcriptome data confirmed the presence of mRNAs for the proteins identified by mass spectrometry, providing unbiased support for our functional assays. The above data together with secretagogin’s localization in large axon terminals at the median eminence, and its association to the readily releasable pool of vesicles, as well as plasmalemmal compartments in the vicinity of release sites, show that secretagogin is ideally poised to gate neuropeptide secretion. Considering that secretagogin− nerve endings also exist in the median eminence, our results also suggest that as yet unknown and additional Ca2+-dependent or Ca2+-independent sensor proteins might exist to control the release of hormones distinct from CRH, galanin or somatostatin (Fig 5).
A primary drive of molecular neuroscience is the discovery of signaling events’ rate-limiting neural processes indispensable for the formation, maintenance and responsiveness of key neural axes. We demonstrated that acute noxious stress increases CRH expression in the PVN, particularly in secretagogin+ neurons. A causal relationship between secretagogin expression and CRH release is cumulatively highlighted by gain-of-function and loss-of-function analysis employing the coincident detection of the intracellular candidate (secretagogin) and the physiological output (CRH in media). Based on our protein interactome profiling, cell biology and neuroanatomy analyses, we propose that genetic impairment of secretagogin expression disrupts the traffic of CRH-laden neurosecretory vesicles toward release sites, as shown by the somatic accumulation of CRH immunoreactivity upon siRNA-mediated secretagogin silencing in vivo. Most notably, genetic manipulation of secretagogin availability led to the fallout of stress-induced ACTH elevation in peripheral blood, which is normally driven by the activation of CRH receptors on secretory cells of the anterior pituitary (Wynn et al, 1983). While ACTH expression is a multifactorial mechanism (Armario, 2006), we argue that the blunted ACTH response was due to the experimentally evoked attenuation of CRH release at the level of the median eminence. Nevertheless, corticosterone release in stressed animals was only partially suppressed. This may be due to our coincident ACTH and corticosterone sampling protocol focused on the primary ACTH response, and only capturing a relatively early up-shoot in the plasma corticosterone concentration.
Overall, we show that a spatially segregated parvocellular neuron population produces CRH and regulates its release via a novel Ca2+ sensor. Nevertheless, to what extent secretagogin is expressed in other neuron populations, e.g. the arcuate nucleus, and if secretagogin is also involved in the regulated release of many other neuropeptides/hormones remains to be determined. Therapeutic implications of these findings are vast, since they offer novel biomarkers for some of the most common metabolic disorders, and druggable targets for ceasing CRH release particularly in chronic stress, depression and genetic or sporadic forms of generalized glucocorticoid resistance.
Materials and Methods
Animals for histo- and biochemistry
Histochemical, biochemical and in vivo RNAi experiments were conducted in male C57BL/6N mice (n = 57, 12 weeks of age) and Sprague Dawley rats (n = 7; both from Scanbur). CRH-GFP reporter mice (all females, n = 3, 19 weeks of age) were generated using bacterial artificial chromosome technology (Alon et al, 2009). Hypothalamic cultures were prepared from mouse pups on postnatal day (P) 2. Animals were housed conventionally (12/12-h light cycle, 25% humidity). Adult mice (n = 4) received stereotaxic injections of colchicine (5 μl; Sigma) into the left lateral ventricle under deep anesthesia (5% isoflurane) and survived for an additional 24 h to facilitate somatic neuropeptide accumulation (Cortes et al, 1990). Experimental protocols were in accordance with the European Communities Council Directive (86/609/EEC) and approved by the regional ethical committee (Stockholms Norra Djurförsöksetiska Nämnd; N512/12). Particular effort was directed to minimize the number of animals and their suffering during the experiments.
Tissue preparation, immunohistochemistry and imaging
Wild-type animals were transcardially perfused with a fixative composed of 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.4) that was preceded by a short prerinse with physiological saline (anesthesia: 5% isoflurane). CRH-GFP mice were perfused with 0.1 M phosphate-buffered saline (PBS; 37°C, pH 7.4) followed by 4% PFA in 0.1 M PBS [37°C (20 ml) switched to ice-cold (50 ml)]. After post-fixation in the same fixative overnight and cryoprotection in 30% sucrose for at least 48 h, 30- or 50-μm-thick serial sections were cut on a cryostat microtome and processed for multiple immunofluorescence histochemistry according to the published protocols (Mulder et al, 2009). Alternatively, perfused brains were cryoprotected in 10% sucrose and sectioned onto SuperFrost+ glass slides at 20 μm thickness.
Free-floating sections were rinsed in PB (pH 7.4) and pre-treated with 0.3% Triton X-100 (in PB) for 1 h at 22–24°C to enhance the penetration of antibodies. Non-specific immunoreactivity was suppressed by incubating our specimens in a cocktail of 5% normal donkey serum (NDS; Jackson), 1% bovine serum albumin (BSA; Sigma) and 0.3% Triton X-100 (Sigma) in PB for 1 h at 22–24°C. Sections were then exposed to select combinations of primary antibodies diluted in PB to which 0.1% NDS and 0.3% Triton X-100 had been added for 16–72 h at 4°C (Supplementary Table S2). After extensive rinsing in PB, immunoreactivities were revealed by carbocyanine (Cy)2, 3 or 5-tagged secondary antibodies raised in donkey (1:200; Jackson; 2 h at 22–24°C). Sections were mounted onto fluorescence-free glass slides and coverslipped with Entellan (in toluene; Merck).
Double labeling of glass-mounted sections with the tyramide signal amplification method (TSA Plus; Perkin-Elmer) combined with the use of Cy2-conjugated secondary antibodies was performed as described (Shi et al, 2012). Glass-mounted sections were coverslipped with Aquamount (Dako). Images were acquired on Zeiss 700LSM, 710LSM or 780LSM confocal laser-scanning microscopes with maximal signal separation or spectral scanning. Composite figures were assembled in CorelDraw X5.
Electron microscopy
Male C57Bl6/J mice (n = 8) were anesthetized with pentobarbital (50 mg/kg) and transcardially perfused with physiological saline followed by 4% PFA and 0.25% glutaraldehyde in PB for 15 min. After removal of the brain, a diencephalic block was trimmed and cut in 100-μm frontal sections on a VT1200S vibratome (Leica). Sections containing the anterior hypothalamus were collected and immersed in 4% PFA in PB for an additional 2 h. Specimens were optionally stored in a mixture of 10% glycerol and 25% sucrose at −80°C. For ultrastructural analysis, sections were thawed, rinsed in PB (3×) and then incubated in 0.1 M Tris-NaCl buffer supplemented with 0.1% Tween-20 (TNT), 1% BSA and 1% glycine for 30 min. This was followed by exposure to anti-secretagogin antibodies [1:2,000 (L.W.), 1:800 (Sigma) in PB-TNT-BSA; Supplementary Table S2] at 4°C overnight. Sections were then rinsed in the above buffer and incubated with goat anti-rabbit F(ab’)2 coupled to 0.8 nm gold particles (1:100; Aurion) in PB-TNT-BSA for 2 h at 22–24°C. After fixation with 1% glutaraldehyde (10 min) and rinsing in water, gold particles were silver-enhanced using the HQ Silver Enhancement Kit (Nanoprobes; 5 min at 22–24°C) prior to post-fixation in 1% OsO4 for 1 h. Next, sections were flat-embedded (Epoxy resin) between two plastic coverslips. After polymerization, regions of interest were identified under a stereomicroscope, isolated with a razor blade and re-embedded in Epon. Ultra-thin sections (60 nm) perpendicular to the thickness of the vibratome section were obtained and optionally contrasted by uranyl acetate and lead citrate. Specimens were finally observed by using an H7650 Hitachi electron microscope.
The number of silver-enhanced gold particles in neuronal profiles of three compartments (that is, soma, dendrites and axonal nerve endings) in the PVN and external median eminence was quantified. In every compartment, we determined the distribution of these particles in a membrane-related domain (either the plasma- or an endomembrane) and in a cytosolic domain where no obvious organelle could be detected underneath. For this purpose, we measured the distance between particles and membranes on photomicrographs from n = 5 mice and considered a particle membrane bound if its distance was < 50 nm from the closest membrane profile. The number of counted profiles was as follows: soma: plasma membrane = 25, endomembrane = 29 and cytosol = 27; dendrite: plasma membrane = 24, endomembrane = 21 and cytosol = 15; and axon: plasma membrane = 28, endomembrane = 25 and cytosol = 25. Data were expressed as the number of particles per neuronal profile ± s.e.m.
Patch-clamp electrophysiology
Male C57Bl6/N mice (n = 22; 21–28 days) were deeply anesthetized (5% isoflurane) and decapitated. Brains were rapidly removed and immersed in ice-cold pre-oxygenated (95% O2/5% CO2) cutting solution containing (in mM) 90 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 10 HEPES–NaOH, 5 sodium ascorbate, 5 sodium pyruvate, 0.5 CaCl2, 8 MgSO4 and 20 glucose. Subsequently, 300-μm-thick coronal slices were cut on a vibratome (VT1200S, Leica). Slices encompassing the PVN region were selected and equilibrated in artificial cerebrospinal fluid (ACSF) containing (in mM) 124 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 2 CaCl2 and 2 MgSO4 at 22–24°C for 1–5 h before recording. Whole-cell recordings in current-clamp or voltage-clamp mode were made by using a HEKA ECP-10USB amplifier and PatchMaster Software (HEKA, Germany). During measurements, slices were continuously perfused with ASCF. The internal pipette solution contained (in mM) 114 potassium gluconate, 6 KCl, 10 HEPES, 5 EGTA, 4 ATP-Mg, 0.3 GTP (pH was adjusted to 7.3 with KOH) and 0.5% biocytin (Sigma) for post hoc cell identification. After recordings, brain slices were immersion-fixed with 4% PFA at 4°C overnight. Electrophysiological data were analyzed using Clampfit 10.0 (Molecular Devices) and SigmaPlot (Systat Software Inc.) for the parameters listed in Supplementary Table S1.
Single-cell transcriptome analysis
The PVN region of juvenile mice was isolated from 300-μm-thick coronal slices under microscopy guidance and dissociated using the Papain Dissociation System (Worthington). Isolated single cells were concentrated by centrifugation to a density of 300–500 cells/μl and loaded into a C1-AutoPrep system (Fluidigm) for single-cell capture, lysis, cDNA synthesis and amplification (Islam et al, 2014). Each capture site was imaged with 192 cells selected for RNA sequencing on an Illumina HiSeq2000 sequencer. Sequencing data at a depth of 500,000–2,000,000 reads/cell were analyzed (Islam et al, 2014). Out of 151 confirmed single cells, 130 cells provided data above a threshold of 10,000 transcript molecules/cell (excluding repeats and mitochondrial transcripts). Data on molecule counts were next subjected to clustering. In brief, ˜2,700 genes were selected based on their expression (average more than one molecule/cell and correlation > 0.5 with other 10 genes). Based on these criteria, the 1D order of cells and genes was performed using the SPIN algorithm (Tsafrir et al, 2005).
Immunoprecipitation and unbiased proteomics
Hypothalamic regions containing the PVN were isolated from P7 rats (three animals), divided into left and right parts with one hemisphere incubated in ACSF (“control stimulation”), while another placed in ACSF in which 52.5 mM NaCl was replaced by KCl to depolarize neurons by inducing Ca2+ influx (“treatment condition”). After 10 min of incubation, samples were collected in lysis buffer containing 50 mM NaCl, 20 mM HEPES, 10 μM CaCl2, 1 mM EDTA, 0.2% Triton X-100 and a cocktail of protease inhibitors (Roche; pH was adjusted to 7.4). Meanwhile, samples treated by depolarizing solution were transferred into the above lysis buffer supplemented with 10 μM CaCl2 without EDTA. Tissues were homogenized and centrifuged at 18,000 g for 30 min, and supernatants were used in subsequent experiments. After pre-clearance, samples were incubated with rabbit anti-secretagogin primary antibody (1:2,000; provided by L.W.) overnight at 4°C. An aliquot of each sample was exposed to rabbit IgG (Santa Cruz Biotechnology) to control non-specific binding. Subsequently, samples were incubated with GammaBind Plus Sepharose beads (GE Healthcare) for 90 min. After repeated rinses, proteins were eluted with Laemmli buffer and separated on 10% SDS–PAGE under denaturing conditions. After silver in-gel staining, proteins in each lane were identified by matrix-associated laser desorption ionization–time-of-flight mass spectrometry (Proteomics Science Facility at Karolinska Institutet) (Linden et al, 2012). The MS/MS data file generated was analyzed using the Mascot algorithm (version 2.5, Matrix Science) against the NCBInr database selected for rat (Rattus norvegicus) taxonomy, trypsin as the cleavage enzyme, carbamidomethyl as a fixed modification of cysteines and methionine oxidation and then deamidation of glutamines and asparagines as variable modifications (Tortoriello et al, 2014). All of the three “pool” mgf files within each treatment were merged before searching. Peptides were selected with a fragment tolerance of 0.1 Da and parent tolerance of 0.05 Da, and with a confidence threshold of > 99%. Proteins were unambiguously identified with 2 or more peptides and a protein threshold of > 99%. Additional data analysis was performed using Scaffold (version 4.3.4).
Cell culture and secretagogin manipulation
Human SH-SY5Y neuroblastoma cells and embryonic mouse hypothalamic cell line N44 (mHypoE-N44; Cedarline ®) were seeded at a density of 50,000 cells/well on poly-D-lysine (PDL)-coated 24-well plates and cultured in DMEM:Glutamax (Invitrogen) containing 10% fetal bovine serum overnight (without antibiotics). Lipofectamine 2000 (2 μl/well; Invitrogen) was used to transfect both cell lines with a pcDNA3.1 plasmid containing the human scgn gene (0.5 μg/well). Forty-eight hours after transfection, the cells were used for Ca2+ imaging and post hoc immunocytochemistry. mHypoE-N44 cells were routinely serum-starved for 1 day in DMEM:Glutamax also containing 25 μM forskolin. Subsequently, cells were stimulated by ATP (15 μM) for 20 min before immersion fixation in 4% PFA in PB.
Hypothalami were isolated on ice after decapitation of mice on P2. After enzymatic dissociation, cells were plated at a density of 300,000 cells/well in PDL-coated 24-well plates and grown for 7–8 days in vitro (DIV). For Ca2+ imaging, 50,000 cells were plated in each well. Cultures were maintained in DMEM/F12 containing B27 supplement (2%), L-glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 mg/ml; all from Invitrogen). For post hoc immunocytochemistry, cells on coverslips were fixed in 4% PFA for 30 min on ice and subsequently immunostained using select combinations of primary antibodies (Supplementary Table S2). Parameters processed for quantitative morphometry of single and dual-labeled cells included: (i) the somatic diameter, (ii) number of neurites, (iii) the maximum length of processes and (iv) the number of neurite branching (“bifurcations”; Supplementary Fig S2C; for analysis procedures see Keimpema et al, 2010). For secretagogin knockdown, primary hypothalamic neurons were grown for 4 days in vitro followed by exposure to 1 μM Accell secretagogin-specific siRNA (Thermo Fisher) for an additional 3 days. CRH release was determined after adding fresh medium containing 25 μM forskolin (Sigma) and incubation for 18 h by ELISA detection (CusaBio).
Ca2+ imaging and quantitative immunocytochemistry
Ca2+ responses were measured using Fura-2AM (Invitrogen) as intracellular Ca2+ indicator. Ratiometric imaging was performed on a CoolSnap HQ2 camera (Photometrics) and analyzed by MetaFluor software (Mollecular Devices). After recording, the cells on coverslips were immersed in ice-cold 4% PFA in PB and processed for immunocytochemistry as above. For quantitative immunocytochemistry (fluorescence densitometry), orthogonal z-stacks were captured on an LSM700 laser-scanning microscopy (Zeiss). Fluorescence intensity of single cells was determined by using the ImarisPro software (Bitplane) and correlated with corresponding Ca2+ responses (Supplementary Figs S3 and S4).
In vivo stress and siRNA-mediated secretagogin knockdown
First, we tested whether formalin stress induces c-fos accumulation in secretagogin+ PVN neurons by subcutaneous injection of 4% PFA (50 μl). After 6 h, mice (n = 2/time point) were transcardially perfused as above, and their PVN was processed for multiple immunofluorescence detection of c-fos, secretagogin and CRH (Supplementary Table S2). The number of c-fos+, secretagogin+ and c-fos+/secretagogin+ neurons was counted in 8,000–18,000 μm2 frames superimposed over the PVN (ZEN2010 software, Zeiss). C-fos+ nuclei were only counted if their fluorescence intensity exceeded tissue fluorescence by > 4-fold.
Second, the contribution of secretagogin in stress-induced hormone release in vivo was assessed in male mice (n = 24, 12 weeks of age). Animals (n = 12) received stereotaxic injections of either 250 μM SMARTpool Accell Scgn siRNA (1 nmol; Thermo Fisher Scientific) or Accell non-targeting siRNA (n = 12) in the left lateral cerebral ventricle (4 μl). A recovery period of 96 h was used before stress induction. Formalin stress was induced by injection of 4% PFA into the left paw. Blood (n = 24) was collected with a lag time of 12 min at the expected peak of circulating ACTH.
Third, the quantitative analysis of secretagogin knockdown efficacy was performed by analyzing the distribution and fluorescence intensity of CRH+ neurons in the PVN after in vivo knockdown as above with 2 μl of either 250 μM SMARTpool Accell Scgn siRNA (0.5 nmol; n = 3) or Accell non-targeting siRNA (n = 3). After transcardial perfusion and immunohistochemistry, projection images of orthogonal z-stacks (interval depth: 2 μm, optical thickness: 2 μm) were analyzed using the maximal intensity projection function of the ZEN2010 software (Zeiss; Fig 6C and C1).
ACTH concentrations were determined in unextracted plasma samples using a radioimmunoassay (RIA). Synthetic human ACTH1-39 was used as standard and for labeling with 125I (I-RB-4, Institute for Isotopes, Budapest, Hungary). An ACTH antibody (no. 8514), directed against the mid-portion of the human ACTH1-39 molecule, was raised in rabbit at the Institute of Experimental Medicine, Hungarian Academy of Sciences (Budapest, Hungary) (Barna & Koenig, 1992). This antibody is highly specific, showing 0.2% cross-reaction with α-MSH and no significant cross-reaction with γ-MSH, CLIP, ACTH11-24, ACTH25-39, ACTH1-14, and ACTH1-19. The intra- and interassay coefficients of variation were 4.7 and 7%, respectively.
Plasma corticosterone was measured from 10 μl of unextracted plasma with an RIA by use of a specific antiserum (Institute of Experimental Medicine, Hungarian Academy of Sciences) against corticosterone-3-carboxymethyloxim-BSA. An 125I-labeled carboxymethyloxim-tyrosine methyl ester derivative was used as tracer (I-RBO-36, Institute for Isotopes, Budapest, Hungary). The corticosterone antibody only crossreacts with other naturally occurring adrenal steroids at < 0.05%, except desoxycorticosterone (1.5%) and progesterone (2.3%). Final dilution of the antibody was 1:40,000. Incubation time was 24 h at 4°C, and a second antibody (anti-rabbit from goat) and 6% polyethylene glycol were used for separation. A calibration curve was prepared from corticosterone (Calbiochem), ranging from 0.27 to 40 pmol/tube. The intra- and interassay coefficients were 12.3 and 15.33%, respectively. All hormone measurements were performed in duplicate.
Statistical analysis
Data were analyzed using Statistical Package for the Social Sciences (version 21.0, SPSS Inc.) and SigmaPlot (Systat Software Inc.). Data were expressed as means ± s.e.m. A P-value of < 0.05 was considered statistically significant and calculated by Student’s t-test (on independent samples), Mann–Whitney rank-sum test or one-way analysis of variance (ANOVA with Dunn’s post hoc test), as appropriate.
Acknowledgments
The authors thank T.L. Horvath (Yale School of Medicine) and L. Swanson (University of Southern California) for their critical comments on this manuscript. Lisa E. Pomeranz and Kaamashri N. Latcha (The Rockefeller University) are acknowledged for providing brain tissues from CRH-GFP mice. The expert technical assistance of Mrs. Eva Dobozi Kovacs is greatly appreciated. The authors also thank the Bordeaux Bioimaging Center (CNRS UMS 3420) for technical support. This work was supported by the Swedish Research Council (T.Ha., T.Hö.), Hjärnfonden (T.Ha.), the Petrus and Augusta Hedlunds Foundation (T.Ha., T.Hö.), the Novo Nordisk Foundation (T.Ha., T.Hö.), Karolinska Institutet (T.Hö.), the Medical University of Vienna (T.Ha.), ANR (France, ANR-10-INBS-04-01, M.L.), the European Commission (PAINCAGE grant, T.Ha.) and the Wellcome Trust (094476/Z/10/Z, equipment grant, C.H.B. and postdoctoral research fellowship, M.F.).
Author contributions
THa conceived the general idea of this study. RAR, AA, M-DZ, HHS, SL, THö and THa designed the experiments. RAR, AA, M-DZ, AZ, ML, MF, SLS, RS, ÁD, MO, LS, MP, AC and CHB performed the experiments and analyzed the data, JM, HM, MU and LW contributed unique reagents. RAR, AA, THö and THa wrote the manuscript. All authors commented on the manuscript and approved its submission.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplemenatry Figure S1
Supplemenatry Figure S2
Supplemenatry Figure S3
Supplemenatry Figure S4
Supplemenatry Figure S5
Supplemenatry Figure S6
Supplemenatry Table S1
Supplemenatry Table S2
Supplemenatry Table S3
Supplementary Information
Review Process File
References
- Aguilera G, Kiss A, Liu Y, Kamitakahara A. Negative regulation of corticotropin releasing factor expression and limitation of stress response. Stress. 2007;10:153–161. doi: 10.1080/10253890701391192. [DOI] [PubMed] [Google Scholar]
- Alon T, Zhou L, Perez CA, Garfield AS, Friedman JM, Heisler LK. Transgenic mice expressing green fluorescent protein under the control of the corticotropin-releasing hormone promoter. Endocrinology. 2009;150:5626–5632. doi: 10.1210/en.2009-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso G, Szafarczyk A, Assenmacher I. Immunoreactivity of hypothalamo-neurohypophysial neurons which secrete corticotropin-releasing hormone (CRH) and vasopressin (Vp): immunocytochemical evidence for a correlation with their functional state in colchicine-treated rats. Exp Brain Res. 1986;61:497–505. doi: 10.1007/BF00237574. [DOI] [PubMed] [Google Scholar]
- Andressen C, Blumcke I, Celio MR. Calcium-binding proteins: selective markers of nerve cells. Cell Tissue Res. 1993;271:181–208. doi: 10.1007/BF00318606. [DOI] [PubMed] [Google Scholar]
- Arai R, Jacobowitz DM, Deura S. Immunohistochemical localization of calretinin-, calbindin-D28k- and parvalbumin-containing cells in the hypothalamic paraventricular and supraoptic nuclei of the rat. Brain Res. 1993;618:323–327. doi: 10.1016/0006-8993(93)91284-y. [DOI] [PubMed] [Google Scholar]
- Armario A. The hypothalamic-pituitary-adrenal axis: what can it tell us about stressors? CNS Neurol Disord Drug Targets. 2006;5:485–501. doi: 10.2174/187152706778559336. [DOI] [PubMed] [Google Scholar]
- Bargmann W. The neurosecretory diencephalon-hypophyseal system and its synaptic connections. J Neurovisc Relat. 1969;31(Suppl 9):64–77. [PubMed] [Google Scholar]
- Barna I, Koenig JI. Effects of mediobasal hypothalamic lesion on immunoreactive ACTH/beta-endorphin levels in cerebrospinal fluid, in discrete brain regions, in plasma, and in pituitary of the rat. Brain Res. 1992;593:69–76. doi: 10.1016/0006-8993(92)91265-g. [DOI] [PubMed] [Google Scholar]
- Bauer MC, O’Connell DJ, Maj M, Wagner L, Cahill DJ, Linse S. Identification of a high-affinity network of secretagogin-binding proteins involved in vesicle secretion. Mol BioSyst. 2011;7:2196–2204. doi: 10.1039/c0mb00349b. [DOI] [PubMed] [Google Scholar]
- Baxter JD, Forsham PH. Tissue effects of glucocorticoids. Am J Med. 1972;53:573–589. doi: 10.1016/0002-9343(72)90154-4. [DOI] [PubMed] [Google Scholar]
- Birkenkamp-Demtroder K, Wagner L, Brandt SF, Bording AL, Gartner W, Scherubl H, Heine B, Christiansen P, Orntoft TF. Secretagogin is a novel marker for neuroendocrine differentiation. Neuroendocrinology. 2005;82:121–138. doi: 10.1159/000091207. [DOI] [PubMed] [Google Scholar]
- Brose N, Petrenko AG, Sudhof TC, Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- Cam GR, Bassett JR. The plasma levels of ACTH following exposure to stress or nicotine. Arch Int Pharmacodyn Ther. 1983;264:154–167. [PubMed] [Google Scholar]
- Carafoli E, Santella L, Branca D, Brini M. Generation, control, and processing of cellular calcium signals. Crit Rev Biochem Mol Biol. 2001;36:107–260. doi: 10.1080/20014091074183. [DOI] [PubMed] [Google Scholar]
- Cazalis M, Dayanithi G, Nordmann JJ. The role of patterned burst and interburst interval on the excitation-coupling mechanism in the isolated rat neural lobe. J Physiol. 1985;369:45–60. doi: 10.1113/jphysiol.1985.sp015887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccatelli S, Cintra A, Hokfelt T, Fuxe K, Wikstrom AC, Gustafsson JA. Coexistence of glucocorticoid receptor-like immunoreactivity with neuropeptides in the hypothalamic paraventricular nucleus. Exp Brain Res. 1989;78:33–42. doi: 10.1007/BF00230684. [DOI] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Cortes R, Ceccatelli S, Schalling M, Hokfelt T. Differential effects of intracerebroventricular colchicine administration on the expression of mRNAs for neuropeptides and neurotransmitter enzymes, with special emphasis on galanin: an in situ hybridization study. Synapse. 1990;6:369–391. doi: 10.1002/syn.890060410. [DOI] [PubMed] [Google Scholar]
- Dayanithi G, Forostyak O, Ueta Y, Verkhratsky A, Toescu EC. Segregation of calcium signalling mechanisms in magnocellular neurones and terminals. Cell Calcium. 2012;51:293–299. doi: 10.1016/j.ceca.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Dierickx K, Vandesande F. Immunocytochemical localization of somatostatin-containing neurons in the rat hypothalamus. Cell Tissue Res. 1979;201:349–359. doi: 10.1007/BF00236995. [DOI] [PubMed] [Google Scholar]
- Donner KM, Hiltunen TP, Janne OA, Sane T, Kontula K. Generalized glucocorticoid resistance caused by a novel two-nucleotide deletion in the hormone-binding domain of the glucocorticoid receptor gene NR3C1. Eur J Endocrinol. 2013;168:K9–K18. doi: 10.1530/EJE-12-0532. [DOI] [PubMed] [Google Scholar]
- Dutton A, Dyball RE. Phasic firing enhances vasopressin release from the rat neurohypophysis. J Physiol. 1979;290:433–440. doi: 10.1113/jphysiol.1979.sp012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BJ, Meister B, Hokfelt T, Melander T, Terenius L, Rokaeus A, Theodorsson-Norheim E, Dockray G, Edwardson J, Cuello C. The hypothalamic arcuate nucleus-median eminence complex: immunohistochemistry of transmitters, peptides and DARPP-32 with special reference to coexistence in dopamine neurons. Brain Res. 1986;396:97–155. doi: 10.1016/s0006-8993(86)80192-5. [DOI] [PubMed] [Google Scholar]
- Fuxe K, Wikstrom AC, Okret S, Agnati LF, Harfstrand A, Yu ZY, Granholm L, Zoli M, Vale W, Gustafsson JA. Mapping of glucocorticoid receptor immunoreactive neurons in the rat tel- and diencephalon using a monoclonal antibody against rat liver glucocorticoid receptor. Endocrinology. 1985;117:1803–1812. doi: 10.1210/endo-117-5-1803. [DOI] [PubMed] [Google Scholar]
- Gainer H, Yamashita M, Fields RL, House SB, Rusnak M. The magnocellular neuronal phenotype: cell-specific gene expression in the hypothalamo-neurohypophysial system. Prog Brain Res. 2002;139:1–14. doi: 10.1016/s0079-6123(02)39003-4. [DOI] [PubMed] [Google Scholar]
- Guillemin R. Peptides in the brain: the new endocrinology of the neuron. Science. 1978;202:390–402. doi: 10.1126/science.212832. [DOI] [PubMed] [Google Scholar]
- Haam J, Popescu IR, Morton LA, Halmos KC, Teruyama R, Ueta Y, Tasker JG. GABA is excitatory in adult vasopressinergic neuroendocrine cells. J Neurosci. 2012;32:572–582. doi: 10.1523/JNEUROSCI.3826-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallbeck M, Blomqvist A, Hermanson O. Ca2+/calmodulin-dependent kinase II immunoreactivity in the rat hypothalamus. NeuroReport. 1996;7:1957–1960. doi: 10.1097/00001756-199608120-00019. [DOI] [PubMed] [Google Scholar]
- Harris GW. Humours and hormones. J Endocrinol. 1972;53:2–23. [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Herman JP, Schafer MK, Thompson RC, Watson SJ. Rapid regulation of corticotropin-releasing hormone gene transcription in vivo. Mol Endocrinol. 1992;6:1061–1069. doi: 10.1210/mend.6.7.1324419. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Horvath TL, Gao XB. Input organization and plasticity of hypocretin neurons: possible clues to obesity’s association with insomnia. Cell Metab. 2005;1:279–286. doi: 10.1016/j.cmet.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Hrabovszky E, Wittmann G, Turi GF, Liposits Z, Fekete C. Hypophysiotropic thyrotropin-releasing hormone and corticotropin-releasing hormone neurons of the rat contain vesicular glutamate transporter-2. Endocrinology. 2005;146:341–347. doi: 10.1210/en.2004-0856. [DOI] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La MG, Zajac P, Kasper M, Lonnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Keegan CE, Herman JP, Karolyi IJ, O’Shea KS, Camper SA, Seasholtz AF. Differential expression of corticotropin-releasing hormone in developing mouse embryos and adult brain. Endocrinology. 1994;134:2547–2555. doi: 10.1210/endo.134.6.8194481. [DOI] [PubMed] [Google Scholar]
- Keimpema E, Barabas K, Morozov YM, Tortoriello G, Torii M, Cameron G, Yanagawa Y, Watanabe M, Mackie K, Harkany T. Differential subcellular recruitment of monoacylglycerol lipase generates spatial specificity of 2-arachidonoyl glycerol signaling during axonal pathfinding. J Neurosci. 2010;30:13992–14007. doi: 10.1523/JNEUROSCI.2126-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss JZ, Palkovits M, Zaborszky L, Tribollet E, Szabo D, Makara GB. Quantitative histological studies on the hypothalamic paraventricular nucleus in rats: I. Number of cells and synaptic boutons. Brain Res. 1983;262:217–224. doi: 10.1016/0006-8993(83)91011-9. [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Cassell MD, Palkovits M. Analysis of the ACTH/beta-End/alpha-MSH-immunoreactive afferent input to the hypothalamic paraventricular nucleus of rat. Brain Res. 1984;324:91–99. doi: 10.1016/0006-8993(84)90625-5. [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Martos J, Palkovits M. Hypothalamic paraventricular nucleus: a quantitative analysis of cytoarchitectonic subdivisions in the rat. J Comp Neurol. 1991;313:563–573. doi: 10.1002/cne.903130403. [DOI] [PubMed] [Google Scholar]
- Kiss A, Palkovits M, Aguilera G. Neural regulation of corticotropin releasing hormone (CRH) and CRH receptor mRNA in the hypothalamic paraventricular nucleus in the rat. J Neuroendocrinol. 1996;8:103–112. doi: 10.1111/j.1365-2826.1996.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Kita T, Chihara K, Abe H, Minamitani N, Kaji H, Kodama H, Chiba T, Fujita T, Yanaihara N. Regional distribution of gastrin-releasing peptide- and somatostatin-like immunoreactivity in the rabbit hypothalamus. Brain Res. 1986;398:18–22. doi: 10.1016/0006-8993(86)91245-x. [DOI] [PubMed] [Google Scholar]
- de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Kodama T, Guerrero S, Shin M, Moghadam S, Faulstich M, du Lac S. Neuronal classification and marker gene identification via single-cell expression profiling of brainstem vestibular neurons subserving cerebellar learning. J Neurosci. 2012;32:7819–7831. doi: 10.1523/JNEUROSCI.0543-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte SM. Corticosteroids in relation to fear, anxiety and psychopathology. Neurosci Biobehav Rev. 2001;25:117–142. doi: 10.1016/s0149-7634(01)00002-1. [DOI] [PubMed] [Google Scholar]
- Kovacs KJ, Sawchenko PE. Sequence of stress-induced alterations in indices of synaptic and transcriptional activation in parvocellular neurosecretory neurons. J Neurosci. 1996;16:262–273. doi: 10.1523/JNEUROSCI.16-01-00262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Han TH, Sonner PM, Stern JE, Ryu PD, Lee SY. Molecular characterization of T-type Ca(2+) channels responsible for low threshold spikes in hypothalamic paraventricular nucleus neurons. Neuroscience. 2008;155:1195–1203. doi: 10.1016/j.neuroscience.2008.06.055. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee S, Shin SY, Ryu PD, Lee SY. Single cell analysis of voltage-gated potassium channels that determines neuronal types of rat hypothalamic paraventricular nucleus neurons. Neuroscience. 2012;205:49–62. doi: 10.1016/j.neuroscience.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Léránth C, Antoni FA, Palkovits M. Ultrastructural demonstration of ovine CRF-like immunoreactivity (oCRF-LI) in the rat hypothalamus: processes of magnocellular neurons establish membrane specializations with parvocellular neurons containing oCRF-LI. Regul Pept. 1983;6:179–188. doi: 10.1016/0167-0115(83)90011-3. [DOI] [PubMed] [Google Scholar]
- Linden M, Lind SB, Mayrhofer C, Segersten U, Wester K, Lyutvinskiy Y, Zubarev R, Malmstrom PU, Pettersson U. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 2012;12:135–144. doi: 10.1002/pmic.201000810. [DOI] [PubMed] [Google Scholar]
- Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, Ulyanova AV, Yeldell SB, Griepenburg JC, Buckley PT, Kim J, Sul JY, Dmochowski IJ, Eberwine J. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther JA, Tasker JG. Voltage-gated currents distinguish parvocellular from magnocellular neurones in the rat hypothalamic paraventricular nucleus. J Physiol. 2000;523(Pt 1):193–209. doi: 10.1111/j.1469-7793.2000.t01-1-00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther JA, Daftary SS, Boudaba C, Gould GC, Halmos KC, Tasker JG. Neurosecretory and non-neurosecretory parvocellular neurones of the hypothalamic paraventricular nucleus express distinct electrophysiological properties. J Neuroendocrinol. 2002;14:929–932. doi: 10.1046/j.1365-2826.2002.00867.x. [DOI] [PubMed] [Google Scholar]
- Maejima Y, Sedbazar U, Suyama S, Kohno D, Onaka T, Takano E, Yoshida N, Koike M, Uchiyama Y, Fujiwara K, Yashiro T, Horvath TL, Dietrich MO, Tanaka S, Dezaki K, Oh I, Hashimoto K, Shimizu H, Nakata M, Mori M. Nesfatin-1-regulated oxytocinergic signaling in the paraventricular nucleus causes anorexia through a leptin-independent melanocortin pathway. Cell Metab. 2009;10:355–365. doi: 10.1016/j.cmet.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Maj M, Milenkovic I, Bauer J, Berggard T, Veit M, Ilhan-Mutlu A, Wagner L, Tretter V. Novel insights into the distribution and functional aspects of the calcium binding protein secretagogin from studies on rat brain and primary neuronal cell culture. Front Mol Neurosci. 2012;5:84. doi: 10.3389/fnmol.2012.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makara GB, Stark E, Karteszi M, Palkovits M, Rappay G. Effects of paraventricular lesions on stimulated ACTH release and CRF in stalk-median eminence of the rat. Am J Physiol. 1981;240:E441–E446. doi: 10.1152/ajpendo.1981.240.4.E441. [DOI] [PubMed] [Google Scholar]
- Makino S, Hashimoto K, Gold PW. Multiple feedback mechanisms activating corticotropin-releasing hormone system in the brain during stress. Pharmacol Biochem Behav. 2002;73:147–158. doi: 10.1016/s0091-3057(02)00791-8. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Meister B, Hokfelt T, Geffard M, Oertel W. Glutamic acid decarboxylase- and gamma-aminobutyric acid-like immunoreactivities in corticotropin-releasing factor-containing parvocellular neurons of the hypothalamic paraventricular nucleus. Neuroendocrinology. 1988;48:516–526. doi: 10.1159/000125058. [DOI] [PubMed] [Google Scholar]
- Meister B, Villar MJ, Ceccatelli S, Hokfelt T. Localization of chemical messengers in magnocellular neurons of the hypothalamic supraoptic and paraventricular nuclei: an immunohistochemical study using experimental manipulations. Neuroscience. 1990;37:603–633. doi: 10.1016/0306-4522(90)90094-k. [DOI] [PubMed] [Google Scholar]
- Mulder J, Zilberter M, Spence L, Tortoriello G, Uhlen M, Yanagawa Y, Aujard F, Hokfelt T, Harkany T. Secretagogin is a Ca2+-binding protein specifying subpopulations of telencephalic neurons. Proc Natl Acad Sci U S A. 2009;106:22492–22497. doi: 10.1073/pnas.0912484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveilhan P, Hassani H, Lucas G, Blakeman KH, Hao JX, Xu XJ, Wiesenfeld-Hallin Z, Thoren P, Ernfors P. Reduced antinociception and plasma extravasation in mice lacking a neuropeptide Y receptor. Nature. 2001;409:513–517. doi: 10.1038/35054063. [DOI] [PubMed] [Google Scholar]
- Ohtori S, Takahashi K, Chiba T, Takahashi Y, Yamagata M, Sameda H, Moriya H. Fos expression in the rat brain and spinal cord evoked by noxious stimulation to low back muscle and skin. Spine (Phila Pa 1976) 2000;25:2425–2430. doi: 10.1097/00007632-200010010-00002. [DOI] [PubMed] [Google Scholar]
- Pacak K, Palkovits M, Kvetnansky R, Yadid G, Kopin IJ, Goldstein DS. Effects of various stressors on in vivo norepinephrine release in the hypothalamic paraventricular nucleus and on the pituitary-adrenocortical axis. Ann N Y Acad Sci. 1995;771:115–130. doi: 10.1111/j.1749-6632.1995.tb44675.x. [DOI] [PubMed] [Google Scholar]
- Palkovits M. Neuropeptides in the hypothalamo-hypophyseal system: lateral retrochiasmatic area as a common gate for neuronal fibers towards the median eminence. Peptides. 1984;5(Suppl 1):35–39. doi: 10.1016/0196-9781(84)90262-6. [DOI] [PubMed] [Google Scholar]
- Palkovits M. Anatomy of neural pathways affecting CRH secretion. Ann N Y Acad Sci. 1987;512:139–148. doi: 10.1111/j.1749-6632.1987.tb24956.x. [DOI] [PubMed] [Google Scholar]
- Pang ZP, Sudhof TC. Cell biology of Ca2+-triggered exocytosis. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic Press; 2001. [Google Scholar]
- Pelletier G. Anatomy of the hypothalamic-pituitary axis. Methods Achiev Exp Pathol. 1991;14:1–22. [PubMed] [Google Scholar]
- Petersen OH, Michalak M, Verkhratsky A. Calcium signalling: past, present and future. Cell Calcium. 2005;38:161–169. doi: 10.1016/j.ceca.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Pinto S, Roseberry AG, Liu H, Diano S, Shanabrough M, Cai X, Friedman JM, Horvath TL. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- Rivier C, Vale W. Influence of the frequency of ovine corticotropin-releasing factor administration on adrenocorticotropin and corticosterone secretion in the rat. Endocrinology. 1983;113:1422–1426. doi: 10.1210/endo-113-4-1422. [DOI] [PubMed] [Google Scholar]
- Rogstam A, Linse S, Lindqvist A, James P, Wagner L, Berggard T. Binding of calcium ions and SNAP-25 to the hexa EF-hand protein secretagogin. Biochem J. 2007;401:353–363. doi: 10.1042/BJ20060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum EF, Voorhoeve PG, te Velde SJ, Koper JW, Delemarre-van de Waal HA, Kemper HC, Lamberts SW. The ER22/23EK polymorphism in the glucocorticoid receptor gene is associated with a beneficial body composition and muscle strength in young adults. J Clin Endocrinol Metab. 2004;89:4004–4009. doi: 10.1210/jc.2003-031422. [DOI] [PubMed] [Google Scholar]
- Sanchez F, Alonso JR, Arevalo R, Carretero J, Vazquez R, Aijon J. Calbindin D-28K- and parvalbumin-reacting neurons in the hypothalamic magnocellular neurosecretory nuclei of the rat. Brain Res Bull. 1992;28:39–46. doi: 10.1016/0361-9230(92)90228-p. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Krey LC, McEwen BS. The neuroendocrinology of stress and aging: the glucocorticoid cascade hypothesis. Endocr Rev. 1986;7:284–301. doi: 10.1210/edrv-7-3-284. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Imaki T, Potter E, Kovacs K, Imaki J, Vale W. The functional neuroanatomy of corticotropin-releasing factor. Ciba Found Symp. 1993;172:5–21. doi: 10.1002/9780470514368.ch2. [DOI] [PubMed] [Google Scholar]
- Schally AV. Aspects of hypothalamic regulation of the pituitary gland. Science. 1978;202:18–28. doi: 10.1126/science.99816. [DOI] [PubMed] [Google Scholar]
- Schluter OM, Basu J, Sudhof TC, Rosenmund C. Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J Neurosci. 2006;26:1239–1246. doi: 10.1523/JNEUROSCI.3553-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller B. The continuing disappearance of “pure” Ca2+ buffers. Cell Mol Life Sci. 2009;66:275–300. doi: 10.1007/s00018-008-8564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selye H, Fortier C. Adaptive reactions to stress. Res Publ Assoc Res Nerv Ment Dis. 1949;29:3–18. [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Shi TJ, Xiang Q, Zhang MD, Tortoriello G, Hammarberg H, Mulder J, Fried K, Wagner L, Josephson A, Uhlen M, Harkany T, Hokfelt T. Secretagogin is expressed in sensory CGRP neurons and in spinal cord of mouse and complements other calcium-binding proteins, with a note on rat and human. Mol Pain. 2012;8:80. doi: 10.1186/1744-8069-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DM, Swanson LW. Comparison of the spatial distribution of seven types of neuroendocrine neurons in the rat paraventricular nucleus: toward a global 3D model. J Comp Neurol. 2009;516:423–441. doi: 10.1002/cne.22126. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Spiess J, Rivier J, Rivier C, Vale W. Primary structure of corticotropin-releasing factor from ovine hypothalamus. Proc Natl Acad Sci U S A. 1981;78:6517–6521. doi: 10.1073/pnas.78.10.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JE. Electrophysiological and morphological properties of pre-autonomic neurones in the rat hypothalamic paraventricular nucleus. J Physiol. 2001;537:161–177. doi: 10.1111/j.1469-7793.2001.0161k.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE. Paraventricular nucleus: a site for the integration of neuroendocrine and autonomic mechanisms. Neuroendocrinology. 1980;31:410–417. doi: 10.1159/000123111. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE. Hypothalamic integration: organization of the paraventricular and supraoptic nuclei. Annu Rev Neurosci. 1983;6:269–324. doi: 10.1146/annurev.ne.06.030183.001413. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE, Lind RW. Regulation of multiple peptides in CRF parvocellular neurosecretory neurons: implications for the stress response. Prog Brain Res. 1986;68:169–190. doi: 10.1016/s0079-6123(08)60238-1. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Simmons DM. Differential steroid hormone and neural influences on peptide mRNA levels in CRH cells of the paraventricular nucleus: a hybridization histochemical study in the rat. J Comp Neurol. 1989;285:413–435. doi: 10.1002/cne.902850402. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Biochemical switching in hypothalamic circuits mediating responses to stress. Prog Brain Res. 1991;87:181–200. doi: 10.1016/s0079-6123(08)63052-6. [DOI] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Tasker JG, Herman JP. Mechanisms of rapid glucocorticoid feedback inhibition of the hypothalamic-pituitary-adrenal axis. Stress. 2011;14:398–406. doi: 10.3109/10253890.2011.586446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin V, Leng G, Ludwig M. The involvement of actin, calcium channels and exocytosis proteins in somato-dendritic oxytocin and vasopressin release. Front Physiol. 2012a;3:261. doi: 10.3389/fphys.2012.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin V, Schwab Y, Lelos N, Onaka T, Pittman QJ, Ludwig M. Expression of exocytosis proteins in rat supraoptic nucleus neurones. J Neuroendocrinol. 2012b;24:629–641. doi: 10.1111/j.1365-2826.2011.02237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoriello G, Morris CV, Alpar A, Fuzik J, Shirran SL, Calvigioni D, Keimpema E, Botting CH, Reinecke K, Herdegen T, Courtney M, Hurd YL, Harkany T. Miswiring the brain: Delta9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J. 2014;33:668–685. doi: 10.1002/embj.201386035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21:2301–2308. doi: 10.1093/bioinformatics/bti329. [DOI] [PubMed] [Google Scholar]
- Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- Vogt MC, Paeger L, Hess S, Steculorum SM, Awazawa M, Hampel B, Neupert S, Nicholls HT, Mauer J, Hausen AC, Predel R, Kloppenburg P, Horvath TL, Bruning JC. Neonatal insulin action impairs hypothalamic neurocircuit formation in response to maternal high-fat feeding. Cell. 2014;156:495–509. doi: 10.1016/j.cell.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L, Oliyarnyk O, Gartner W, Nowotny P, Groeger M, Kaserer K, Waldhausl W, Pasternack MS. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J Biol Chem. 2000;275:24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- Wamsteeker Cusulin JI, Fuzesi T, Watts AG, Bains JS. Characterization of corticotropin-releasing hormone neurons in the paraventricular nucleus of the hypothalamus of Crh-IRES-Cre mutant mice. PLoS ONE. 2013;8:e64943. doi: 10.1371/journal.pone.0064943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn PC, Aguilera G, Morell J, Catt KJ. Properties and regulation of high-affinity pituitary receptors for corticotropin-releasing factor. Biochem Biophys Res Commun. 1983;110:602–608. doi: 10.1016/0006-291x(83)91192-0. [DOI] [PubMed] [Google Scholar]
- Yamamori E, Asai M, Yoshida M, Takano K, Itoi K, Oiso Y, Iwasaki Y. Calcium/calmodulin kinase IV pathway is involved in the transcriptional regulation of the corticotropin-releasing hormone gene promoter in neuronal cells. J Mol Endocrinol. 2004;33:639–649. doi: 10.1677/jme.1.01415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemenatry Figure S1
Supplemenatry Figure S2
Supplemenatry Figure S3
Supplemenatry Figure S4
Supplemenatry Figure S5
Supplemenatry Figure S6
Supplemenatry Table S1
Supplemenatry Table S2
Supplemenatry Table S3
Supplementary Information
Review Process File