Abstract
Hematopoietic stem cells (HSCs) are rare, multipotent cells that generate via progenitor and precursor cells of all blood lineages. Similar to normal hematopoiesis, leukemia is also hierarchically organized and a subpopulation of leukemic cells, the leukemic stem cells (LSCs), is responsible for disease initiation and maintenance and gives rise to more differentiated malignant cells. Although genetically abnormal, LSCs share many characteristics with normal HSCs, including quiescence, multipotency and self-renewal. Normal HSCs reside in a specialized microenvironment in the bone marrow (BM), the so-called HSC niche that crucially regulates HSC survival and function. Many cell types including osteoblastic, perivascular, endothelial and mesenchymal cells contribute to the HSC niche. In addition, the BM functions as primary and secondary lymphoid organ and hosts various mature immune cell types, including T and B cells, dendritic cells and macrophages that contribute to the HSC niche. Signals derived from the HSC niche are necessary to regulate demand-adapted responses of HSCs and progenitor cells after BM stress or during infection. LSCs occupy similar niches and depend on signals from the BM microenvironment. However, in addition to the cell types that constitute the HSC niche during homeostasis, in leukemia the BM is infiltrated by activated leukemia-specific immune cells. Leukemic cells express different antigens that are able to activate CD4+ and CD8+ T cells. It is well documented that activated T cells can contribute to the control of leukemic cells and it was hoped that these cells may be able to target and eliminate the therapy-resistant LSCs. However, the actual interaction of leukemia-specific T cells with LSCs remains ill-defined. Paradoxically, many immune mechanisms that evolved to activate emergency hematopoiesis during infection may actually contribute to the expansion and differentiation of LSCs, promoting leukemia progression. In this review, we summarize mechanisms by which the immune system regulates HSCs and LSCs.
Facts
Hematopoiesis and leukemia are both hierarchically organized processes originating from HSCs and LSCs, respectively.
LSCs display many features of normal HSCs, including quiescence and self-renewal.
HSCs and LSCs crucially depend on signals from the BM microenvironment, the so-called niche.
The BM microenvironment contains innate and adaptive immune cells that regulate hematopoiesis during homeostasis, stress response and infections.
In leukemia, activated immune cells paradoxically contribute to disease progression.
Open Questions
What is the contribution of BM-infiltrating immune cells to the HSC and LSC niche?
What are the molecular mechanisms of the interaction between immune cells, LSCs and niche cells?
Do stress-induced alterations in hematopoiesis favor leukemia development and progression?
How can the knowledge about BM-resident immune cells be exploited to improve immunotherapy for leukemia?
The concept that cancer develops in a hierarchical tree from disease-originating cancer stem cells (CSCs) that self-renew and give rise to more differentiated, non-cancer-initiating cells by asymmetric division was first documented in leukemia two decades ago.1 The CSC hypothesis is now widely accepted and was extended and adapted to several solid tumors.2 Since the first description of leukemic stem cells (LSCs), our knowledge about their biology grew substantially and nowadays, LCSs are phenotypically well characterized in chronic myeloid leukemia (CML) and in some forms of acute myeloid leukemia (AML).3 From a clinical point of view, LSCs are of fundamental interest as they are resistant against most of our current cancer treatments such as irradiation and chemotherapy and probably also against more targeted therapies such as tyrosine kinase inhibitors and immunotherapy.4 Therefore, LSCs are the main reason for treatment failure and disease relapse. Different mechanisms may contribute to the resistance of LSCs to current therapies. LSCs express drug efflux proteins that lead to multidrug resistance.5 In addition, most cytotoxic drugs and irradiation depend on cell division in order to induce cell death but LSCs are largely quiescent. Many stem cell characteristics including quiescence are determined by interactions with the niche. Growing evidence suggests that LSCs depend on similar niche signals as their normal counterpart, the hematopoietic stem cells (HSCs).6 Although HSCs are mobile and recirculate in the blood, most of them are found in the trabecular bone area of the bone marrow (BM),7, 8 where they reside in close proximity to sinusoids and other blood vessels.9 Endothelial and perivascular cells produce C-X-C motif chemokine 12 (CXCL12) and stem cell factor that are necessary for HSC and LSC maintenance.10, 11, 12 The role of other cell populations present in the BM in the regulation of HSC function is less clear. However, the sympathetic nervous system, adipocytes, macrophages and cells of the adaptive immune system have been shown to regulate hematopoietic stem and progenitor cells (HSPCs).13, 14
In a healthy individual, CD4+ and CD8+ T cells represent approximately 1.5% and 2.5% of the total BM cellularity, respectively. Up to 30% of all BM-resident CD4+ T cells are CD4+CD25+FOXP3+ regulatory T cells (Tregs).15 Interestingly, BM T cells including Tregs are also localized in the trabecular bone area in proximity to sinusoids. BM CD4+ and CD8+ T cells have a memory phenotype and secrete cytokines that are necessary for HSC maintenance, such as interleukin 3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF).16 Therefore, BM-resident T cells may contribute to the formation of the perivascular HSC niche. In response to an infection or BM stress, the cellular composition of the microenvironment as well as the cytokine milieu change fundamentally in order to meet the organism's requirement for demand-adapted hematopoiesis.17
Similarly, leukemia induces an adaptive and innate immune response and causes an inflammatory environment in the BM. Various leukemia antigens have been characterized and activated leukemia-specific CD4+ and CD8+ T cells have been documented.3, 18 Therefore, the BM microenvironment and the LSC niche changes dramatically with leukemia-specific effector T cells infiltrating the BM. These infiltrating effector T cells may potentially recognize and eliminate LSCs. However, mechanisms that evolved to protect normal HSCs from elimination and to regulate demand-adapted responses during inflammation most likely protect LSCs from immune-mediated elimination and may even contribute to the expansion of LSCs and leukemia progression. The understanding of these mechanisms may help to develop novel immunotherapies that allow targeting LSCs specifically.
HSCs and LSCs
HSCs are multipotent and self-renewing tissue-specific stem cells that initiate and maintain life-long hematopoiesis, the production of mature blood cells of all lineages.19 HSCs are rare cells that only comprise approximately 0.001–0.01% of total BM cells in mice20 and approximately 0.01–0.2% of total BM mononuclear cells in humans.21 Being at the top of the hematopoietic hierarchy, HSCs divide infrequently, giving rise to transient-amplifying multipotent (MPPs) and lineage-restricted progenitors that proliferate extensively and differentiate toward mature blood cells (Figure 1). Quiescence, also known as dormancy, warrants the genomic integrity of HSCs, as frequent chromosomal replications may introduce oncogenic DNA mutations. Dormancy also protects HSCs from uncontrolled proliferation, which would result in exhaustion.22 In addition, HSCs can undergo asymmetrical division, assuring that always one daughter cell remains an HSC. This mechanism, referred to as self-renewal, keeps the HSC pool constant. However, HSCs possess a non-exhaustive replication and proliferation capacity that can be initiated in stress situations, such as after cytotoxic chemotherapy, irradiation or during infections.22 Moreover, HSCs express receptors for cytokines, chemokines and danger-associated molecular patterns, allowing them to respond to signals from mature immune cells and to sense pathogens directly during inflammation or infection to adapt their cycling and differentiation behavior.17
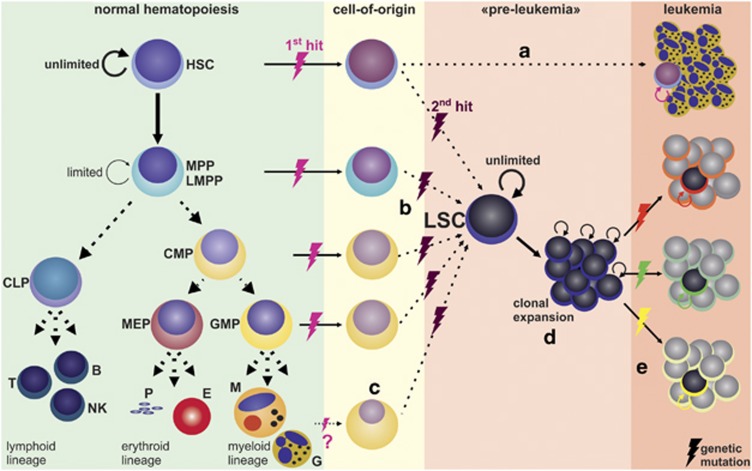
Figure 1.
The leukemic stem cell model. In normal hematopoiesis, rarely dividing hematopoietic stem cells (HSCs) with unlimited self-renewal capacity (indicated by circle arrow) give rise to transient-amplifying multipotent progenitors (MPPs) that have only limited self-renewal capacity.20 MPPs further differentiate toward oligopotent lineage-restricted progenitors (LRPs), such as common lymphoid and myeloid progenitors (CLPs, CMPs) and granulocyte-macrophage progenitors (GMPs) that have lost self-renewal capacity. LRPs proliferate intensely and produce all mature blood cell types required. The formation of a leukemic stem cell (LSC) in myeloid leukemia may result from mutations in cells in different stages of the hematopoietic hierarchy. (a) In chronic phase CML patients, the presence of BCR/ABL1 in all blood lineages suggests that the LSC is derived from an HSC or an early MPP with multilineage differentiation potential (HSC cell-of-origin).27, 28, 29 BCR/ABL1 is necessary and sufficient for the malignant phenotype, no further genetic lesions are required for chronic phase CML. (b) In contrast, in blast crisis CML and AML patients, LSCs exhibited immunophenotypes of LRPs, such as lymphoid-primed MPPs (LMPPs)31 or GMPs.30, 31 This supports the concept that other more differentiated cells can give rise to LSCs after re-acquisition of self-renewal (progenitor cell-of-origin). (c) In addition, the recent demonstration that some AML LSCs even express low amounts of lineage markers32 raised the question whether more differentiated hematopoietic cells may serve as cell-of-origin for LSCs as well. (d) In a ‘pre-leukemic' disease phase, genetically unstable, self-renewing LSCs clonally expand, facilitating the acquisition of further mutations and (e) the development of different leukemic clones. B, B cell; CML, chronic myeloid leukemia; E, erythrocyte; G, granulocyte; NK, natural killer cell; M, monocyte; MEP, megakaryocyte-erythrocyte progenitor; P, platelet; T, T cell
Leukemia is a paradigmatic disease of CSCs. According to the CSC hypothesis, tumors are composed of a bulk of cancer cells displaying marked morphological, genetic and functional heterogeneity. Within this bulk resides a small population of cells with stem cell characteristics that propagates the disease.2, 23 In leukemia, LSCs are thought to reside at the top of the leukemic hierarchy, like HSCs in hematopoiesis (Figure 1). LSCs produce more differentiated, heterogenic leukemic blasts that feature a high proliferative potential, a block in terminal differentiation and defective apoptosis or senescence mechanisms, leading to blast accumulation and clinical disease. Stem cell features, such as quiescence, the expression of high levels of ATP-binding cassette pumps and the localization in distinct niches, render CSCs resistant to all kinds of therapy.4, 6 Thus, cure of cancer implies the elimination of CSCs and persisting CSCs are a main cause of disease relapse.
In contrast to differentiated hematopoietic cells that are removed after fulfilling their functions, self-renewing HSCs persist for long periods of time, allowing accumulation of genetic damage and malignant transformation. Therefore, it has been suggested that HSCs serve as the cancer-initiating cells (cell-of-origin) for LSCs (Figure 1).24, 25 Experimental evidence supporting this theory came from seminal studies by John Dick and colleagues who first demonstrated in xenotransplants that all clonogenic capacity resided in lin−CD34+CD38− AML cells, whereas lin−CD34+CD38+ or lin+ AML cells failed to induce leukemias.1, 26 Furthermore, in CML, the break point cluster region/Abelson murine leukemia viral oncogene homolog 1 (BCR/ABL1) oncogene can be detected in several hematopoietic lineages, indicating that the cell-of-origin is an HSC with multilineage differentiation potential.27, 28, 29 In addition, DNA (cytosine-5)-methyltransferase 3A mutations have been found in HSCs, progenitor and mature cells of AML patients, but without coincident nucleophosmin 1 mutations present only in AML blasts.25 This indicates that AML evolves from pre-leukemic HSCs. However, the hypothesis that HSCs represent the cell-of-origin and undergo oncogenic transformation has been challenged by studies demonstrating that in some leukemias, more mature progenitor cell types or even cells expressing lineage markers can serve as leukemia-initiating cells giving rise to LSCs (Figure 1).30, 31, 32 This raised the question of how—besides the oncogenic events that are required for leukemia development—these leukemia-initiating cells re-acquire self-renewal capability to fully establish their LSC functions.23 In addition, these findings nurtured the alternative hypothesis of tumorigenesis, the stochastic model, claiming that every cancer cell has the ability to self-renew and to recapitulate the disease phenotype given that it enters a permissive environment, an event which has a very low probability.2
Thus, although the cell-of-origin may not be identified for all types of leukemia yet, these cells have to exhibit the essential stem cell characteristics of self-renewal and indefinite proliferative potential to give rise to LSCs that initiate and maintain the disease.
The HSC Niche
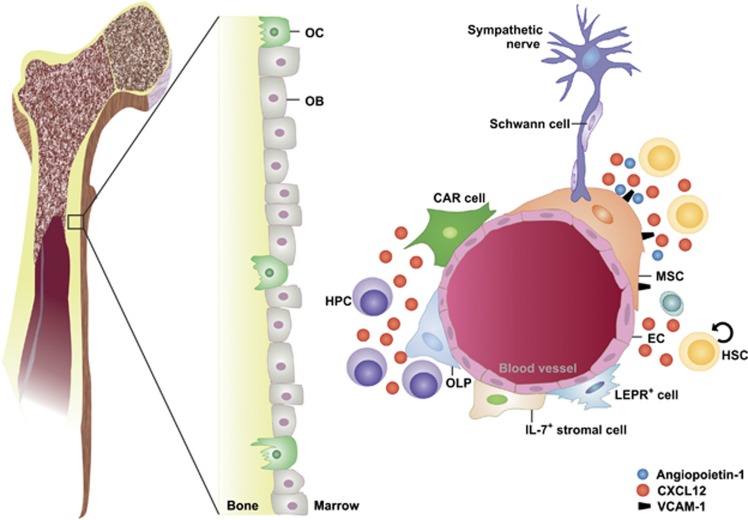
Many of the functional characteristics of HSCs and LSCs are driven by their surrounding microenvironment in the BM, the so-called HSC niche (Figure 2). The HSC niche has been initially defined as microenvironment that retains HSCs in their localization, avoids differentiation and ensures their stem cell phenotype.33 The functional and anatomical definition as well as the cellular composition of the HSC niche have been highly debated during the last decade. First, osteoblastic lineage cells have been described as critical components of the HSC niche (Figure 2). Bone-forming osteoblasts (OBs) that are located at the endosteal surface of the bone cavities and on trabeculae co-localized with HSCs and regulated the HSC pool size in vivo.34, 35 Furthermore, endosteal osteoclasts influence HSC maintenance and retention in the BM.36, 37 Later, Kiel and co-workers demonstrated that primitive HSCs defined by SLAM markers (CD150+, CD48−) preferentially localize in close proximity to sinusoidal endothelial cells (ECs) but not OBs, identifying sinusoidal blood vessels as HSC niche9 (Figure 2). Clinical observations confirm an important role for ECs in the formation of the HSC niche, as HSCs depend on an intact vasculature for recovery after myeloablation or BM transplantation.38, 39 More recently, perivascular nestin+ mesenchymal stem cells (MSCs) have been defined as central components of the HSC niche that regulate HSCs via the expression of stem cell factor, CXCL12, angiopoietin-1 and vascular cell adhesion molecule-1 (VCAM-1).12 In addition, it was demonstrated that β-adrenergic signals from the sympathetic nervous system mobilize HSCs13 and regulate circadian HSC egress.40
Figure 2.
The hematopoietic stem cell niche. Various cell types including osteoclasts (OCs), osteoblasts (OBs), osteolineage progenitor cells (OLPs), endothelial cells (ECs), mesenchymal stem/stromal cells (MSCs), specialized CXCL12-abundant reticular (CAR) cells and leptin receptor (LEPR)-positive cells contribute to the structure of the BM microenvironment. In addition, this microenvironment is innervated by sympathetic nerves fibers ensheated by nonmyelinating Schwann cells. Hematopoietic stem cells (HSCs) are located in the perivascular region of sinusoids and arterioles in close proximity to MSCs and ECs that regulate HSC maintenance and differentiation through soluble factors such as CXCL12 and angiopoietin-1 or cell contact-dependent signals such vascular cell adhesion molecule-1 (VCAM1). HPC, hematopoietic progenitor cell
These data suggested that at least two distinct HSC niches exist, an endosteal and a vascular niche (Figure 2). According to this hypothesis, the endosteal niche keeps HSCs quiescent and regulates their migration to the vascular niche, where differentiation occurs according to the organism's demand.22, 41 However, recent studies challenged the existence of an endosteal niche and a role of OBs in the maintenance of HSC quiescence. Transforming growth factor-β (TGF-β) is a niche factor that controls HSC dormancy.42 Nonmyelinating Schwann cells that are located around the blood vessels in the BM induce HSC dormancy by secreting TGF-β, suggesting that HSC quiescence is maintained in the vascular niche.43 Recently, these findings have been further elaborated by conditional deletion of CXCL12 in different cell types of the HSC niche.10, 44 HSC maintenance and self-renewal in the BM was primarily regulated by CXCL12 secreted from immature mesenchymal stem and progenitor cells and to a lesser extent from ECs. In contrast, CXCL12 secreted from OBs was dispensable for HSC function. In addition, retention of HSPCs in the BM was mediated by perivascular sinusoidal stromal cells, including CXCL12-abundant reticular cells45 and osteolineage progenitors that express leptin receptor and osterix.10, 44 Immunofluorescence imaging together with computational modeling revealed that quiescent HSCs associate with small arterioles in the endosteal BM. A rare subtype of pericytes (NG2+ pericytes) ensheated these arterioles and mediated HSC quiescence.46
In summary, there is strong evidence that HSCs reside in the perivascular region of the BM and that MSCs, ECs and pericytes regulate HSC maintenance and differentiation through soluble factors and cell contact-dependent signals8, 10, 44 (Figure 2).
The LSC Niche
Although LSCs harbor genetic abnormalities that result in increased proliferation and resistance to apoptosis, they still depend on similar interactions with niche cells as described for HSCs. Therefore, during leukemogenesis, LSCs ‘hijack' the niche and the signaling molecules from normal HSCs. Analogous to normal HSCs, transplanted leukemia cells preferentially migrate to CXCL12-expressing vascular niches.47 Moreover, the CXCL12 receptor C-X-C motif chemokine receptor 4 (CXCR4) regulates the migration of human AML cells in xenotransplant models.48 Signaling via CXCR4 leads to the upregulation of pro-survival signals and quiescence, both contributing to chemotherapy resistance. Importantly, CXCR4 is highly expressed on different types of human leukemia, including acute lymphoblastic leukemia and AML49, 50 and high CXCR4 expression on leukemia blasts correlates with poor outcome.51 Blocking CXCR4 in vitro using peptides resulted in reduced chemotaxis toward CXCL12-expressing cells and an inactivation of pro-survival signals. As a consequence, blocking CXCR4 increased the susceptibility of AML cells to chemotherapy.52 Similarly, preclinical studies demonstrate that the CXCL12-CXCR4 inhibitor plerixafor and novel monoclonal antibodies (mAbs) blocking CXCR4 increase the chemosensitivity of leukemia cells in vitro and in vivo.53, 54 In contrast to AML blasts, CML cells have a decreased expression of CXCR4 and therefore an attenuated chemotaxis toward CXCL12. Treatment with imatinib restores CXCR4 expression and CXCL12-mediated pro-survival signals. Thereby, imatinib may contribute to CXCL12/CXCR4 signaling-mediated resistance of the few remaining LSCs in CML.55
Similar to the role of quiescence induction in HSCs, TGF-β has been reported to induce quiescent G0 state of the cell cycle in AML cells. Blocking TGF-β by mAbs increased proliferation and susceptibility to cytarabine.56 In addition, TGF-β is a crucial regulator of protein kinase B (AKT/PKB) activation and controls forkhead-box protein O3a localization, thereby maintaining CML LSCs.57 Interestingly, it was shown that the effect of TGF-β on LSCs varies in different subtypes of leukemia. For instance, OB-specific activation of the parathyroid hormone receptor and subsequent secretion of TGF-β reduced LSCs in CML but increased LSC numbers in MLL/AF9-induced AML in mouse transplantation models.58
Niche cells not only interact with HSPCs via soluble factors but also via direct cell–cell interactions. For example, CD44, a transmembrane glycoprotein that exists in differently spliced isoforms mediates adhesion of LSCs through cell–cell and cell–extracellular matrix interactions by binding to hyaluronan that is concentrated at the endosteal region of the BM niche. In addition to its function as an adhesion molecule, CD44 also transduces intracellular signals that are involved in the regulation of cell proliferation and differentiation. Blocking CD44 in vivo prevented the migration of human AML and murine CML LSCs to the stem cell-supportive microenvironment in the BM and led to their eradication.59, 60
Taken together, there is strong evidence that LSCs at least partially depend on similar signals from the microenvironment as HSCs do. These requirements may differ in the distinct subtypes of myeloid leukemia. In addition, accumulating evidence suggests that molecular changes in the BM niche actually contribute to leukemia development. For example, Walkley et al. reported that a dysfunction of the tumor suppressor retinoblastoma protein or of the retinoic acid receptor in the BM microenvironment results in myeloproliferative disease.61, 62 Similarly, conditional knockdown of DICER1, a gene that regulates microRNAs, in osteoblastic precursors results in leukemia predisposition.50 More recently, Kode and colleagues showed that a single activating β-catenin mutation in OBs is sufficient to alter the differentiation of myeloid progenitors, leading to AML with common chromosomal aberrations and cell-autonomous progression.63
Immune Cells Contributing to the Niche
Although recent evidence documents that MSCs and ECs are fundamental regulators of HSC maintenance and quiescence, many other cell types that are present in the BM contribute to the microenvironment. These include adipocytes, fibroblasts and immune cells (Figure 3). Besides its main function as a hematopoietic organ, the BM serves as a primary and secondary lymphoid organ, hosting various mature immune cells including T and B cells, plasma cells, dendritic cells (DCs), neutrophils and macrophages (reviewed in Mercier et al.,64 Figure 4). These immune cells provide an ‘immune niche' that is involved in the regulation of HSC homeostasis and emergency hematopoiesis.17, 64 Lymphocytes represent a major fraction of total BM mononuclear cells, are widely distributed throughout the BM parenchyma and are occasionally organized as small lymphoid aggregates, typically consisting of mature CD3+ T cells64 (Figure 3). Clinical and experimental approaches investigating engraftment after BM transplantation suggested a fundamental role of CD4+ T cells in hematopoiesis.65, 66 T-cell-depleted allogeneic BM failed to engraft in a majority of the patients. In addition, HSC maintenance and successful long-term reconstitution depends on the expression of major histocompatibility complex class II, implying a role for CD4+ T cells in maintaining HSC function.67, 68 The mouse BM contains ∼1.5% of CD4+ T cells.15 Most of these cells have an activated memory phenotype and a diverse Vβ T cell receptor repertoire.69, 70 Observations in mice lacking the common gamma chain (γc−/−) indicated that hematopoiesis-promoting cytokines such as IL-3 and GM-CSF secreted by activated T cells in the BM modulate normal hematopoiesis.71 In addition, adoptive transfer of CD4+ T cells but not CD8+ T cells restored defective myeloid differentiation in T-cell-deficient mice, suggesting that especially antigen-activated CD4+ T cells maintain basal hematopoiesis in the BM.16
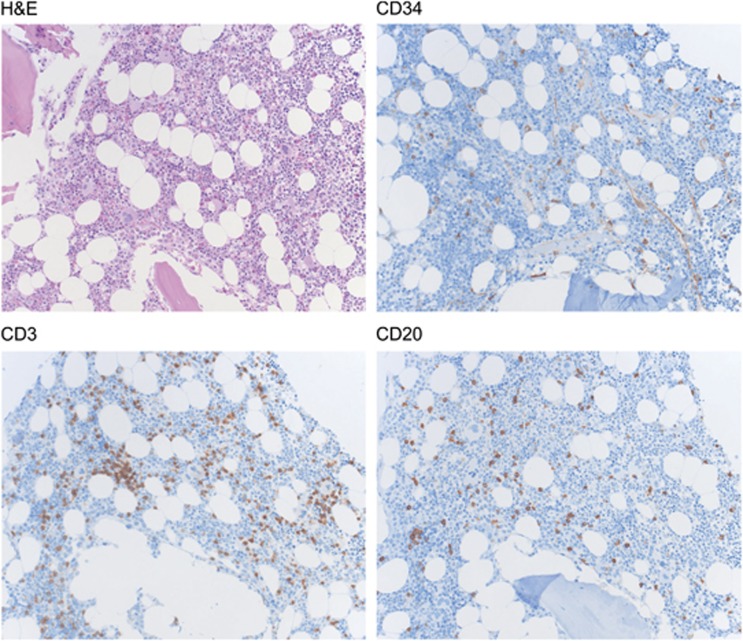
Figure 3.
Differentiated immune cells in the bone marrow (BM). Representative example of a healthy human BM stained for hematoxylin/eosin (HE), for CD34+ HSPCs, CD3+ T cells and CD20+ B cells
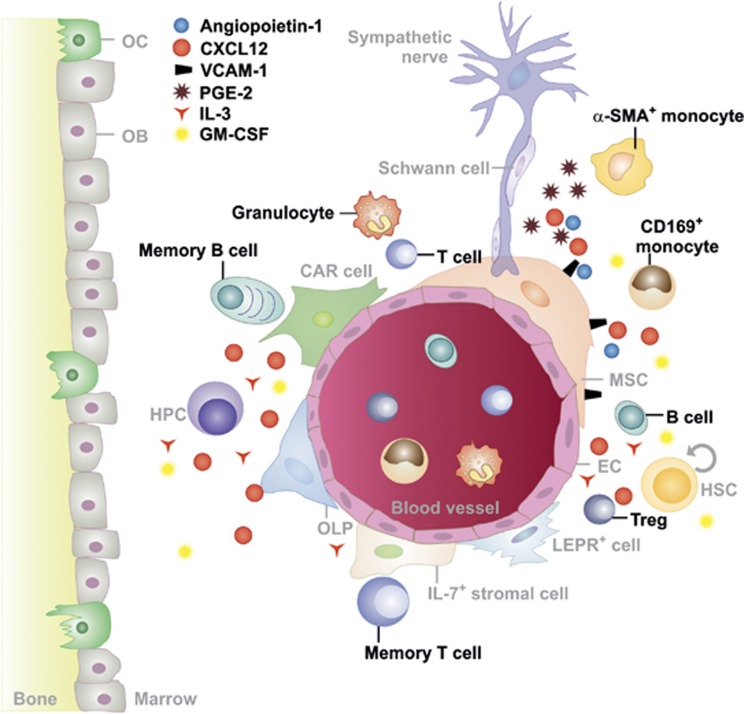
Figure 4.
The ‘immune niche'. The BM microenvironment hosts various mature immune cell types including T and B cells, dendritic cells, neutrophils and macrophages. These immune cells contribute to the BM microenvironment (‘immune niche') and regulate hematopoietic stem cells (HSCs) during steady-state and emergency hematopoiesis directly by secretion of hematopoiesis-promoting cytokines such as IL-3 and GM-CSF.16 In addition, immune cells can indirectly regulate HSPCs through signaling via mesenchymal stem/stromal cells (MSCs), for example, by prostaglandin E2 that increases the expression of CXCL12, angiopoietin-1 and vascular cell adhesion molecule-1 (VCAM-1) in MSCs.77 α-SMA, monocytes/macrophages expressing high levels of α-smooth muscle actin; CAR cell, CXCL12-abundant reticular cell; EC, endothelial cell; HPC, hematopoietic progenitor cell; OB; osteoblast; OC, osteoclast; OLP, osteolineage progenitor cell; Treg, CD4+CD25+FOXP3+ regulatory T cell
Tregs represent one-third of all CD4+ T cells in the BM.69, 70 This proportion is substantially higher than in lymph nodes and spleen, where the frequency of Tregs is approximately 5–10%. Depletion experiments and co-transfer of BM with or without Tregs indicated that Tregs suppress colony formation and myeloid differentiation of HSPCs.72 High-resolution in vivo imaging demonstrated that Tregs colocalize with HSPCs in the endosteum. Furthermore, Tregs provide an immune-privileged niche in the BM, protecting HSPCs from immune destruction.73
In addition to cells of the adaptive immune system, mononuclear phagocytes contribute to the regulation of HSCs in the BM. Depletion of mononuclear phagocytes using clodronate liposomes increased the number of circulating HSCs.74, 75, 76 CD169+ macrophages in the BM secrete soluble factors that stimulate nestin+ MSCs to express HSC retention factors, such as CXCL12, angiopoietin-1 and VCAM-1. In addition, the same macrophages are involved in steady-state and stress-induced erythropoiesis.77 Moreover, a rare population of monocytes and macrophages expressing high levels of α-smooth muscle actin and cyclooxygenase 2 induce prostaglandin E2 production and upregulation of CXCL12 on nestin+ MSCs.78 Furthermore, macrophages have been identified as central regulators of HSC egress from the BM after phagocytosis of aged neutrophils79 (Figure 4).
The BM Microenvironment During Immune Activation
The BM not only assures the continuous supply of different blood lineages during homeostasis, it also responds to the organism's increased demands during stress situations, such as infections or chemotherapy. Many of the mechanisms that regulate HSPCs during demand-adapted hematopoiesis may also regulate LSCs and leukemic progenitor cell function (Table 1, Figure 5).
Table 1. Shared molecular pathways in the regulation of HPSCs during infection and LSCs in leukemia.
| Effector molecule | Expression of receptor | Effect in demand-adapted myelopoiesis | Effect in leukemia |
|---|---|---|---|
| IFNα | HSCs, MPPs LSCs | Dormant HSCs enter the cell cycle88, 89 Permanent signaling leads to exhaustion of HSCs88, 89, 90 | Modulation of gene expression (CML)143 Pro-apoptotic and anti-proliferative signals (CML)144 Adhesion to microenvironment (CML)145 Downregulation of BCR/ABL (CML)146 Immunomodulation (CML)147 |
| IFNγ | HSCs, MPPs, CMPs, MEPs, GMPs LSCs | Apoptosis of HSCs in vitro94 Proliferation of HSCs and MPPs in vivo96, 102 Increased myeloid differentiation in vivo Permanent signaling leads to suppression of HSCs95 | Pro-apoptotic effects (CML) Proliferation of CD34+ cells (CML) Proliferation of LSCs and leukemia progression in vivo (CML) |
| TNFα | HSCs LSCs | Reduced colony formation in vitro and reconstitution in vivo104, 146 Increased colony formation capacity in vitro105 Suppression of HSC proliferation in vivo106 | promotes NF-κB activity, LSC survival and expansion (CML, AML)110, 111 |
| IL-1β | HSCs LSCs | Increased granulocyte numbers in BM148 | Inhibits self-renewal capacity of LSCs (AML)149 |
| IL-6 | MPPs Leukemia MPPs | Reduced erythropoiesis115 Increased myelopoiesis102, 115 | Directs myeloid differentiation and sustains leukemia development (CML)116 |
| CD70-CD27 | HSCs, MPPs, CMPs, GMPs LSCs | Negative feedback signal to leukocyte differentiation122 | Increased proliferation of LSCs and leukemia progression in CML123 and AML (unpublished results) |
Abbreviations: AML, acute myeloid leukemia; BM, bone marrow; CML, chronic myeloid leukemia; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; HSCs, hematopoietic stem cells; HPSCs, hematopoietic stem- and progenitor cells; IFN, interferon; IL, interleukin; LSCs, leukemic stem cells; MPPs, multipotent progenitors; MEPs, megakaryocyte-erythrocyte progenitors; NF-κB, nuclear factor κB; TNF, tumor necrosis factor
Figure 5.
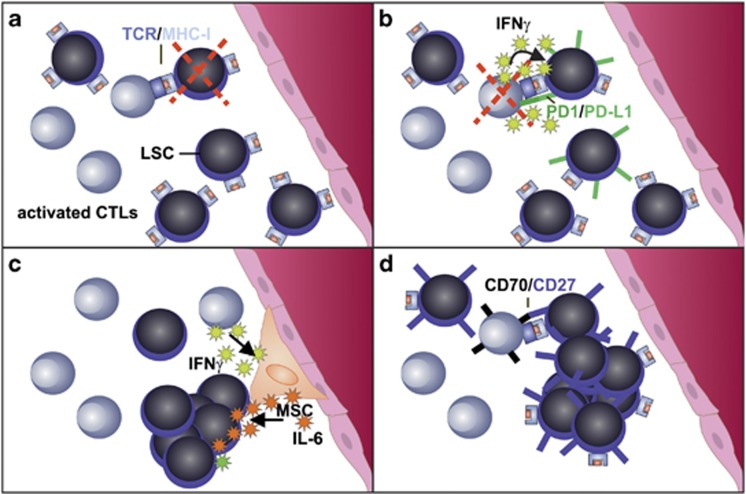
The interaction of activated cytotoxic effector CD8+ T cells (CTLs) with CML leukemic stem cells (LSCs). In CML, the BM is infiltrated by activated leukemia antigen-specific CTLs. (a) LSCs express MHC class I and present peptides derived from leukemia antigens to specific CTLs. Specific CTLs can eliminate LSCs in vitro and donor-derived CTLs can eliminate LSCs after allogeneic HSC transplantation.103, 141 Whether autologous CTLs can eliminate LSCs in vivo is currently unclear. (b) LSCs express programmed death ligand 1 (PD-L1) that interacts with programmed death 1 (PD-1) on activated CTLs and leads to CTL inhibition and ultimately deletion. CTL-secreted IFNγ further leads to upregulation of PD-L1 on LSCs and therefore protection of LSCs from CTL attack.103 (c) LSCs express the IFNγ receptor and can directly respond to IFNγ resulting in proliferation of LSCs and leukemia progression.103 In addition, IFNγ activates mesenchymal stem/stromal cells (MSCs) to produce IL-6.102 IL-6 secretion induces myeloid differentiation at the level of MPPs. In CML, IL-6 is secreted by BCR/ABL1-expressing leukemic cells, leading to a paracrine feedback loop.116 (d) LSCs and leukemic progenitors express the TNFR molecule CD27. CD27 is ligated by CD70 expressed on activated CTLs.123 CD27 signaling leads to activation of the Wnt pathway, proliferation of LSCs and leukemia progression
During an infection, antigenic stimulation drives clonal expansion of naïve and memory lymphocytes to meet the increased demand for T and B cells.80 In contrast, granulocytes are short-lived and do not have the capacity to undergo clonal expansion. Consequently, they must be continuously produced and recruited from the BM. Therefore, the BM must be capable of recognizing the increased demand for myeloid cells during an infection and immediately react with enhanced production, differentiation and mobilization of granulocytes and monocytes. The importance of this so-called ‘emergency myelopoiesis' is best documented in bacterial infections. HSPCs sense the increased demand for myeloid cells by systemic and local danger and inflammatory signals. These include the recognition of conserved microbial products via Toll-like receptor (TLR) activation and of soluble factors such as type I and type II interferons (IFNs).81, 82 TLR signaling has been shown to induce cell cycling and myeloid differentiation in a MyD88-dependent manner in murine HSCs and in human CD34+ progenitor cells.82, 83 Although some TLRs interact with endogenous ligands, no effect of TLR signaling on LSCs has been reported so far.
HSPCs express cytokine receptors and respond to inflammatory signals produced by mature immune cells, such as IFNs.17, 84, 85 IFNs are crucially involved in host protection against various infections and emerged as a major pathway of HSC regulation.86 Type I IFNs (IFNα and β) are synthesized by various cell types, especially plasmacytoid DCs, in response to viral infection and prevent viral replication and thereby viral spread.87 IFNα stimulates dormant HSCs to enter the cell cycle in a signal transducer and activator of transcription 1 (STAT-1)-dependent way.88 In addition, IFN regulatory factor 2, a transcriptional repressor of IFNα signaling, preserves quiescence and multilineage reconstitution capacity of HSCs.89 Thus, acute IFNα production stimulates HSCs to proliferate during viral infection. In contrast, chronic and excessive signaling through this pathway leads to HSC exhaustion.88, 89, 90 A clinically relevant effect of IFNα on CML cells has been well documented. Before the era of the BCR/ABL1-targeting tyrosine kinase inhibitors, IFNα was a standard treatment in CML.91 Clinical and experimental data suggest that IFNα can actually target CML LSCs. The mechanism of action of IFNα in CML is complex, including direct modulation of gene expression in LSCs, induction of apoptosis, anti-proliferative signals and immunomodulatory effects.91 In addition, it has been proposed that IFNα, similar to its effect on HSCs, may induce proliferation of LSCs and render them more susceptible to chemotherapy.92
IFNγ, a type II IFN, is secreted by activated innate and adaptive immune cells, mainly by macrophages and activated T cells. Similar to type I IFNs, IFNγ has activating and suppressive effects on hematopoiesis, probably depending on the timing, duration and amount of secretion. Initially, IFNγ was described as a suppressor of hematopoiesis. This was based on experiments indicating that IFNγ induces differentiation and apoptosis of human and murine HSCs and reduces their colony formation capacity in vitro.93, 94 In analogy, infection of perforin-deficient mice with lymphocytic choriomeningitis virus (LCMV) induced lethal pancytopenia because of the persistence of the virus, leading to prolonged and increased secretion of tumor necrosis factor α (TNFα) and IFNγ by cytotoxic effector CD8+ T cells (CTLs).95 In contrast, accumulating evidence from more physiologic infection models indicates that IFNγ induces an expansion of HSCs and myeloid progenitors and modulates the production of mature myeloid cells.96, 97, 98, 99, 100, 101 Baldridge et al. documented that IFNγ directly increases HSC proliferation through IFNγ receptor 1-STAT-1 signaling during infection with Mycobacterium avium.96 In contrast, we recently demonstrated that IFNγ secreted by activated CTLs during acute LCMV-infection stimulates the expansion of early MPPs and downstream myeloid precursors in the BM.102 Interestingly, IFNγ did not act directly on hematopoietic cells, but stimulated MSCs in the BM to secrete IL-6, which induced proliferation of MPPs and myeloid differentiation. This resulted in elevated myeloid cell counts in the circulation and an increased number of inflammatory monocytes in secondary lymphoid organs that contributed to pathogen clearance. Therefore, IFNγ has an important role in the demand-adapted response to infections and probably regulates HSPC proliferation via direct and indirect mechanisms. However, IFNγ may have comparable effects on LSCs. We found that CTL-secreted IFNγ induces proliferation of LSCs and leukemia progression in a murine CML model (Figure 5).103 Interestingly, this effect was dependent on the amount of secreted IFNγ. If adoptively transferred, activated leukemia-specific CTLs were re-stimulated in vivo by large amounts of antigen, CTL-secreted IFNγ induced LSC proliferation and expansion. The quantity of secreted IFNγ correlated with the leukemia load and with the antigen expression pattern that is, leukemia-specific expression versus expression in, healthy, non-malignant tissue. Similarly, IFNγ increased the colony formation capacity of lin−CD34+ stem/progenitor cells from CML patients in vitro.103
TNFα is another major pro-inflammatory cytokine that is released by activated macrophages, natural killer cells and T cells. Similar to IFNγ, TNFα has been shown to suppress the colony formation capacity of human lin−CD34+ stem/progenitor cells and of murine HSCs in vitro as well as their ability to reconstitute recipient mice.93, 104 Rezzoug et al. documented contradictory results in that TNFα promoted engraftment and colony formation of HSCs resulting in increased numbers of HSPCs.105 A small fraction of CD8+ T-cell receptor-negative cells in the BM was identified as a major source of TNFα in this process. Furthermore, mice deficient of the p55 TNF receptor 1 α (TNFRSF1α−/−) have increased numbers of HSPCs and an increased BM cellularity compared with wild-type mice and mice lacking the p75 TNF receptor 1 β (TNFRSF1β−/−).106 This increase in HSPC numbers is accompanied with a decrease in HSC function. Competitive repopulation assays documented that TNFRSF1α−/− HSCs have an impaired self-renewal capacity. Therefore, TNFα is a major regulator of baseline and demand-adapted hematopoiesis via signaling through the p55 subunit of the TNF receptor (TNFR). However, in analogy to IFNs, prolonged and excessive TNFα signaling is associated with BM failure and myelodysplastic syndrome.107 Early in vitro experiments using AML blasts demonstrated that TNFα has the ability to either support or inhibit cell proliferation, depending on the growth factors present in the culture medium.108, 109 Very recently, two studies highlighted the importance of TNFα for CML and AML stem cell survival and expansion.110, 111 Autocrine TNFα production by LSCs increased nuclear factor κB pathway activation and leukemia progression.
IL-6 was originally identified as a T-cell-derived cytokine that induces B-cell maturation into antibody-producing cells.112 IL-6 is expressed by a variety of normal and transformed cells including T and B cells and has important functions in the regulation of immune responses, acute-phase reaction and hematopoiesis.113, 114 IL-6 regulates hematopoiesis in response to Toxoplasma gondii infection in mice. BM stromal fibroblast-derived IL-6 blocked erythroid development but expanded granulocyte-macrophage progenitors.115 Similarly, as discussed above, LCMV infection increased IL-6 secretion by MSCs because of CTL-secreted IFNγ.102 IL-6 then expanded MPPs and myeloid progenitors leading to increased numbers of myeloid cells in the circulation and in lymphoid organs. Quite comparable to its function during an acute infection, IL-6 directs CML MPPs toward myeloid lineage.116 CML cells were the main source of IL-6 and BCR/ABL1 activity controlled IL-6 expression, establishing a feedback-loop that contributed to disease progression.
Although the immune system mainly interacts with HSPCs via soluble factors, direct cell–cell interactions are also involved in the regulation of HSCs and LSCs. CD27, a member of the TNFR superfamily, is expressed on lymphocytes and on HSCs.117 The cellular effects that are initiated after ligation of CD27 by its unique ligand CD70 have been extensively studied in lymphocytes. CD70-CD27 signaling leads to cell expansion, survival, memory formation and cytokine production.118, 119, 120, 121 CD27 signaling on HSPCs reduces colony formation in vitro and lymphocyte, mainly B cell, differentiation in vivo.122 As CD70 is only expressed on lymphocytes and on subsets of DCs upon immune activation, CD27 signaling on HSPCs may represent an important regulatory mechanism during infection. We recently documented that CD27 is expressed on LSCs in CML123 and AML (unpublished results). In contrast to its inhibitory effect on HSCs, CD27 signaling on LSCs increased LSC proliferation and colony formation. Thus, the activated immune system contributes to leukemia progression by CD27 signaling on LSCs and leukemic progenitors (Figure 5). CD27 signaling in LSCs activated the Wnt pathway via the TRAF2- and NCK-interacting kinase. As the Wnt pathway is crucial for CML and AML stem cells,124, 125, 126 blocking CD27 signaling and reducing Wnt pathway activity resulted in a reduction of LSC numbers and delayed disease progression.
Immune Responses to Leukemia
It is assumed that HSCs reside in an immune-privileged environment, supported by BM-resident Tregs. In addition, HSCs are resistant to most infectious pathogens with only few exceptions such as the human polyomavirus 2 (JC virus).85 Therefore, during homeostasis and infection, HSPCs are probably not direct targets of CTLs or antibodies. This situation may be different in leukemia. Leukemic cells express antigens that are immunogenic and can be recognized by CTLs.127 Some leukemia antigens originate directly from the oncogenic event and are therefore leukemia-specific, such as BCR/ABL1 in CML128, 129, 130 and DEK/nuceloporin 214,129 promyelocytic leukemia/retinoic acid receptor-α,131, 132 fms-like tyrosine kinase 3—internal tandem duplication (FLT3-ITD)133, 134 and mutated nucleophosmin 1135 in AML. However, apart from BCR/ABL1 and FLT3-ITD, these leukemia-specific antigens are only expressed in a minority of patients. In addition, the vast majority of the more than 200 known leukemia-specific chromosomal translocations does not give rise to antigenic proteins.127 Other antigens are not leukemia specific but are overexpressed by leukemic cells (leukemia-associated antigens), such as Wilms tumor protein, proteinase 3, baculoviral IAP repeat-containing gene 5/survivin, telomerase reverse transcriptase and others. Leukemic cells including LSCs express the molecular repertoire to interact with T cells, that is major histocompatibility molecules and co-stimulatory ligands.103, 136, 137 Clinical and experimental studies have documented immune responses to leukemia.3, 127 CTLs directed against leukemia antigens have been detected in chronic phase CML and in AML patients.18, 138 In an experimental model of CML, depletion of CD8+ T cells by mAbs led to rapid disease progression, documenting an important role of CTLs in the immunosurveillance of leukemia.139 The role of CD4+ T cells in leukemia is less clear and CD4+ T cells have been shown to be dysfunctional in vivo.140 Therefore, there is ample evidence that antigen-specific immune responses toward the leukemia are elicited. However, whether activated T cells can interact with and eliminate LSCs is under debate. It is reasonable to assume that similar to HSCs, LSCs are at least partially protected in an immune-suppressive environment because of the high frequency of Tregs in the BM. In addition, we found in a murine CML model that LSCs express the inhibitory molecule programmed death ligand 1 and that its expression is further upregulated in response to IFNγ. In contrast, leukemia-specific effector CTLs were able to eliminate LSCs in vitro and in vivo in a setting with minimal leukemia load.103 Furthermore, allogeneic HSC transplantation can lead to cure of the leukemia, an effect that is mediated most likely by donor-derived CTLs that eliminate residual LSCs.141, 142 This indicates that human LSCs may also be targeted by donor-derived allo-reactive CTLs in vivo.
Conclusions
Besides its main role as a hematopoietic organ, the BM executes functions of a primary and secondary lymphoid organ.64 Memory CD4+ and CD8+ T cells and antibody-secreting plasma cells are maintained long-term in the BM by cytokines such as IL-7 and IL-15. Up to one-third of all CD4+ T cells in the BM are Tregs. During homeostasis, especially CD4+ T cells contribute to the cytokine milieu in the BM and influence quiescence, proliferation and differentiation of HSCs. During acute infections, the immune system regulates the expansion and differentiation of HSPCs by cell–cell interactions and by the secretion of cytokines. These feedback mechanisms evolved to ensure a concerted action of lymphoid and myeloid cells in response to an infection. Although these mechanisms are beneficial to fight an infection, they may be detrimental in the case of leukemia. LSCs share many characteristics with normal HSCs and inflammatory cytokines and signaling via cell contact-dependent receptors such as the TNFR CD27 may induce their expansion. Cure from leukemia implies the elimination of LSCs, and a better understanding of the ‘immune niche' and its function in the BM microenvironment may help to develop specific therapies targeting leukemia at the level of the LSC.
Glossary
- AML
acute myeloid leukemia
- BCR/ABL1
break point cluster region/Abelson murine leukemia viral oncogene homolog 1
- BM
bone marrow
- CLPs
common lymphoid progenitors
- CML
chronic myeloid leukemia
- CMPs
common myeloid progenitors
- CSCs
cancer stem cells
- CTLs
cytotoxic effector CD8+ T cells
- CXCL12
C-X-C motif chemokine 12
- CXCR4
C-X-C motif chemokine receptor 4
- DCs
dendritic cells
- ECs
endothelial cells
- FLT3-ITD
fms-like tyrosine kinase 3—internal tandem duplication
- GM-CSF
granulocyte-macrophage colony stimulating factor
- HPCs
hematopoietic progenitor cells
- HSCs
hematopoietic stem cells
- HSPCs
hematopoietic stem and progenitor cells
- IFNs
interferons
- IL
interleukin
- JC virus
human polyomavirus 2
- LCMV
lymphocytic choriomeningitis virus
- LEPR
leptin receptor
- LMPPs
lymphoid-primed MPPs
- LRPs
lineage-restricted progenitors
- LSCs
leukemic stem cells
- mAb
monoclonal antibody
- MEP
megakaryocyte-erythrocyte progenitor
- MPPs
multipotent progenitors
- MSCs
mesenchymal stem/stromal cells
- OBs
osteoblasts
- OCs
osteoclasts
- OLPs
osteolineage progenitor cells
- STAT-1
signal transducer and activator of transcription 1
- TGF-β
transforming growth factor β
- TKIs
tyrosine kinase inhibitors
- TLR
Toll-like receptor
- TNFα
tumor necrosis factor α
- TNFR
TNF receptor
- TNFRSF1α
p55 TNF receptor 1 α
- Tregs
CD4+CD25+FOXP3+ regulatory T cells
- VCAM-1
vascular cell adhesion molecule-1
The authors declare no conflict of interest.
Footnotes
Edited by H-U Simon
References
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- Schürch CM, Riether C, Ochsenbein AF. Dendritic cell-based immunotherapy for myeloid leukemias. Front Immunol. 2013;4:496. doi: 10.3389/fimmu.2013.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman ML, Allan JN. Leukemia stem cells in personalized medicine. Stem Cells. 2013;32:844–851. doi: 10.1002/stem.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Tabe Y, Konopleva M. Advances in understanding the leukaemia microenvironment. Br J Haematol. 2014;164:767–778. doi: 10.1111/bjh.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, Daley GQ. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Xu H, Wang L, Kryczek I, Wu K, Hu Y, et al. Bone marrow and the control of immunity. Cell Mol Immunol. 2012;9:11–19. doi: 10.1038/cmi.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JP, Benjamin A, Costa ES, Barcinski MA, Bonomo A. Normal hematopoiesis is maintained by activated bone marrow CD4+ T cells. Blood. 2005;105:1484–1491. doi: 10.1182/blood-2004-07-2856. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- Molldrem JJ, Lee PP, Wang C, Felio K, Kantarjian HM, Champlin RE, et al. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–1023. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Martin PJ, Najfeld V, Hansen JA, Penfold GK, Jacobson RJ, Fialkow PJ. Involvement of the B-lymphoid system in chronic myelogenous leukaemia. Nature. 1980;287:49–50. doi: 10.1038/287049a0. [DOI] [PubMed] [Google Scholar]
- Jonas D, Lubbert M, Kawasaki ES, Henke M, Bross KJ, Mertelsmann R, et al. Clonal analysis of bcr-abl rearrangement in T lymphocytes from patients with chronic myelogenous leukemia. Blood. 1992;79:1017–1023. [PubMed] [Google Scholar]
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2R gamma c-deficient mice. J Clin Invest. 2011;121:384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- Mansour A, Abou-Ezzi G, Sitnicka E, Jacobsen SE, Wakkach A, Blin-Wakkach C. Osteoclasts promote the formation of hematopoietic stem cell niches in the bone marrow. J Exp Med. 2012;209:537–549. doi: 10.1084/jem.20110994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper AT, Butler JM, Nolan DJ, Kranz A, Iida K, Kobayashi M, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Butler JM, O'Donnell R, Kobayashi M, Ding BS, Bonner B, et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol. 2010;12:1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Malhotra S, Kincade PW. Canonical Wnt pathway signaling suppresses VCAM-1 expression by marrow stromal and hematopoietic cells. Exp Hematol. 2009;37:19–30. doi: 10.1016/j.exphem.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi S, Eto K, Ema H, Nakauchi H. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood. 2009;113:1250–1256. doi: 10.1182/blood-2008-04-146480. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- Tavor S, Petit I, Porozov S, Avigdor A, Dar A, Leider-Trejo L, et al. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 2004;64:2817–2824. doi: 10.1158/0008-5472.can-03-3693. [DOI] [PubMed] [Google Scholar]
- Shen W, Bendall LJ, Gottlieb DJ, Bradstock KF. The chemokine receptor CXCR4 enhances integrin-mediated in vitro adhesion and facilitates engraftment of leukemic precursor-B cells in the bone marrow. Exp Hematol. 2001;29:1439–1447. doi: 10.1016/s0301-472x(01)00741-x. [DOI] [PubMed] [Google Scholar]
- Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood. 2004;104:550–557. doi: 10.1182/blood-2004-02-0566. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Samudio IJ, Munsell M, An J, Huang Z, Estey E, et al. Inhibition of CXCR4 with the novel RCP168 peptide overcomes stroma-mediated chemoresistance in chronic and acute leukemias. Mol Cancer Ther. 2006;5:3113–3121. doi: 10.1158/1535-7163.MCT-06-0228. [DOI] [PubMed] [Google Scholar]
- Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne MR, Mulvey T, Belanger B, Chen S, Pan C, Chong C, et al. BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin Cancer Res. 2013;19:357–366. doi: 10.1158/1078-0432.CCR-12-2333. [DOI] [PubMed] [Google Scholar]
- Jin L, Tabe Y, Konoplev S, Xu Y, Leysath CE, Lu H, et al. CXCR4 up-regulation by imatinib induces chronic myelogenous leukemia (CML) cell migration to bone marrow stroma and promotes survival of quiescent CML cells. Mol Cancer Ther. 2008;7:48–58. doi: 10.1158/1535-7163.MCT-07-0042. [DOI] [PubMed] [Google Scholar]
- Tabe Y, Shi YX, Zeng Z, Jin L, Shikami M, Hatanaka Y, et al. TGF-beta-neutralizing antibody 1D11 enhances cytarabine-induced apoptosis in AML cells in the bone marrow microenvironment. PLoS One. 2013;8:e62785. doi: 10.1371/journal.pone.0062785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, et al. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Olsen GH, Dworkin S, Fabb SA, Swann J, McArthur GA, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–244. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier FE, Ragu C, Scadden DT. The bone marrow at the crossroads of blood and immunity. Nat Rev Immunol. 2012;12:49–60. doi: 10.1038/nri3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CL, Colson YL, Wren SM, Watkins S, Simmons RL, Ildstad ST. Phenotypic characterization of a novel bone marrow-derived cell that facilitates engraftment of allogeneic bone marrow stem cells. Blood. 1994;84:2436–2446. [PubMed] [Google Scholar]
- Ho VT, Soiffer RJ. The history and future of T-cell depletion as graft-versus-host disease prophylaxis for allogeneic hematopoietic stem cell transplantation. Blood. 2001;98:3192–3204. doi: 10.1182/blood.v98.12.3192. [DOI] [PubMed] [Google Scholar]
- Greinix HT, Ladiges WC, Graham TC, Maslan S, Raff RF, Sandmaier BM, et al. Late failure of autologous marrow grafts in lethally irradiated dogs given anti-class II monoclonal antibody. Blood. 1991;78:2131–2138. [PubMed] [Google Scholar]
- Huss R, Beckham C, Storb R, Deeg HJ. Major histocompatibility complex class II expression is required for posttransplant immunological but not hemopoietic reconstitution in mice. Transplantation. 1994;58:1366–1371. [PubMed] [Google Scholar]
- Zeng D, Hoffmann P, Lan F, Huie P, Higgins J, Strober S. Unique patterns of surface receptors, cytokine secretion, and immune functions distinguish T cells in the bone marrow from those in the periphery: impact on allogeneic bone marrow transplantation. Blood. 2002;99:1449–1457. doi: 10.1182/blood.v99.4.1449. [DOI] [PubMed] [Google Scholar]
- Price PW, Cerny J. Characterization of CD4+ T cells in mouse bone marrow. I. Increased activated/memory phenotype and altered TCR Vbeta repertoire. Eur J Immunol. 1999;29:1051–1056. doi: 10.1002/(SICI)1521-4141(199903)29:03<1051::AID-IMMU1051>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Sharara LI, Andersson A, Guy-Grand D, Fischer A, DiSanto JP. Deregulated TCR alpha beta T cell population provokes extramedullary hematopoiesis in mice deficient in the common gamma chain. Eur J Immunol. 1997;27:990–998. doi: 10.1002/eji.1830270428. [DOI] [PubMed] [Google Scholar]
- Urbieta M, Barao I, Jones M, Jurecic R, Panoskaltsis-Mortari A, Blazar BR, et al. Hematopoietic progenitor cell regulation by CD4+CD25+ T cells. Blood. 2010;115:4934–4943. doi: 10.1182/blood-2009-04-218826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki J, Wu J, Carlson AL, Silberstein L, Putheti P, Larocca R, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011;474:216–219. doi: 10.1038/nature10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Lucas D, Hidalgo A, Mendez-Ferrer S, Hashimoto D, Scheiermann C, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med. 2011;208:261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher MJ, Rao M, Liu F, Woloszynek JR, Link DC. Expression of the G-CSF receptor in monocytic cells is sufficient to mediate hematopoietic progenitor mobilization by G-CSF in mice. J Exp Med. 2011;208:251–260. doi: 10.1084/jem.20101700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler IG, Sims NA, Pettit AR, Barbier V, Nowlan B, Helwani F, et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood. 2010;116:4815–4828. doi: 10.1182/blood-2009-11-253534. [DOI] [PubMed] [Google Scholar]
- Chow A, Huggins M, Ahmed J, Hashimoto D, Lucas D, Kunisaki Y, et al. CD169(+) macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nat Med. 2013;19:429–436. doi: 10.1038/nm.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin A, Itkin T, Gur-Cohen S, Mildner A, Shezen E, Golan K, et al. Monocytes-macrophages that express alpha-smooth muscle actin preserve primitive hematopoietic cells in the bone marrow. Nat Immunol. 2012;13:1072–1082. doi: 10.1038/ni.2408. [DOI] [PubMed] [Google Scholar]
- Casanova-Acebes M, Pitaval C, Weiss LA, Nombela-Arrieta C, Chevre R, AG N, et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 2013;153:1025–1035. doi: 10.1016/j.cell.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnonen J, Cocks BG, Carballido JM, Bennett B, Peterson D, Aversa G, et al. Soluble and membrane-bound forms of signaling lymphocytic activation molecule (SLAM) induce proliferation and Ig synthesis by activated human B lymphocytes. J Exp Med. 1997;185:993–1004. doi: 10.1084/jem.185.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Floisand Y, Forfang L, Lund-Johansen F. Signaling through Toll-like receptor 7/8 induces the differentiation of human bone marrow CD34+ progenitor cells along the myeloid lineage. J Mol Biol. 2006;364:945–954. doi: 10.1016/j.jmb.2006.09.054. [DOI] [PubMed] [Google Scholar]
- Baldridge MT, King KY, Goodell MA. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 2011;32:57–65. doi: 10.1016/j.it.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatman Zaretsky A, Engiles JB, Hunter CA. Infection-induced changes in hematopoiesis. J Immunol. 2014;192:27–33. doi: 10.4049/jimmunol.1302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch CM, Riether C, Ochsenbein AF. Interferons in hematopoiesis and leukemia. Oncoimmunology. 2013;2:e24572. doi: 10.4161/onci.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek MF, Müller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawski LA, Talpaz M. The role of interferon-alpha in the treatment of chronic myeloid leukemia. Cytokine Growth Factor Rev. 2007;18:459–471. doi: 10.1016/j.cytogfr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Essers MA, Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol Oncol. 2010;4:443–450. doi: 10.1016/j.molonc.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri C, Sato T, Anderson S, Young NS, Maciejewski JP. Interferon-gamma and tumor necrosis factor-alpha suppress both early and late stages of hematopoiesis and induce programmed cell death. J Cell Physiol. 1995;165:538–546. doi: 10.1002/jcp.1041650312. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Williams DE, Lu L, Cooper S, Anderson SL, Beyer GS, et al. The suppressive influences of human tumor necrosis factors on bone marrow hematopoietic progenitor cells from normal donors and patients with leukemia: synergism of tumor necrosis factor and interferon-gamma. J Immunol. 1986;136:4487–4495. [PubMed] [Google Scholar]
- Binder D, van den Broek MF, Kagi D, Bluethmann H, Fehr J, Hengartner H, et al. Aplastic anemia rescued by exhaustion of cytokine-secreting CD8+ T cells in persistent infection with lymphocytic choriomeningitis virus. J Exp Med. 1998;187:1903–1920. doi: 10.1084/jem.187.11.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev NN, Brown DE, Diaz AI, Rae A, Jarra W, Thompson J, et al. Induction of an IL7-R(+)c-Kit(hi) myelolymphoid progenitor critically dependent on IFN-gamma signaling during acute malaria. Nat Immunol. 2010;11:477–485. doi: 10.1038/ni.1869. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ren G, Liang L, Ai PZ, Zheng B, Tischfield JA, et al. Brief report: interferon-gamma induces expansion of Lin(−)Sca-1(+)C-Kit(+) Cells. Stem Cells. 2010;28:122–126. doi: 10.1002/stem.252. [DOI] [PubMed] [Google Scholar]
- MacNamara KC, Oduro K, Martin O, Jones DD, McLaughlin M, Choi K, et al. Infection-induced myelopoiesis during intracellular bacterial infection is critically dependent upon IFN-gamma signaling. J Immunol. 2011;186:1032–1043. doi: 10.4049/jimmunol.1001893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin AM, Libregts SF, Valkhof M, Boon L, Touw IP, Nolte MA. IFNgamma induces monopoiesis and inhibits neutrophil development during inflammation. Blood. 2012;119:1543–1554. doi: 10.1182/blood-2011-07-367706. [DOI] [PubMed] [Google Scholar]
- MacNamara KC, Jones M, Martin O, Winslow GM. Transient activation of hematopoietic stem and progenitor cells by IFNgamma during acute bacterial infection. PLoS One. 2011;6:e28669. doi: 10.1371/journal.pone.0028669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch CM, Riether C, Ochsenbein AF. Cytotoxic CD8+ T cells stimulate hematopoietic progenitors by promoting cytokine release from bone marrow mesenchymal stromal cells. Cell Stem Cell. 2014;14:460–472. doi: 10.1016/j.stem.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Schürch C, Riether C, Amrein MA, Ochsenbein AF. Cytotoxic T cells induce proliferation of chronic myeloid leukemia stem cells by secreting interferon-gamma. J Exp Med. 2013;210:605–621. doi: 10.1084/jem.20121229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybedal I, Bryder D, Fossum A, Rusten LS, Jacobsen SE. Tumor necrosis factor (TNF)-mediated activation of the p55 TNF receptor negatively regulates maintenance of cycling reconstituting human hematopoietic stem cells. Blood. 2001;98:1782–1791. doi: 10.1182/blood.v98.6.1782. [DOI] [PubMed] [Google Scholar]
- Rezzoug F, Huang Y, Tanner MK, Wysoczynski M, Schanie CL, Chilton PM, et al. TNF-alpha is critical to facilitate hemopoietic stem cell engraftment and function. J Immunol. 2008;180:49–57. doi: 10.4049/jimmunol.180.1.49. [DOI] [PubMed] [Google Scholar]
- Rebel VI, Hartnett S, Hill GR, Lazo-Kallanian SB, Ferrara JL, Sieff CA. Essential role for the p55 tumor necrosis factor receptor in regulating hematopoiesis at a stem cell level. J Exp Med. 1999;190:1493–1504. doi: 10.1084/jem.190.10.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Saito I, Kuwata T, Yoshida S, Yamaguchi S, Takahashi M, et al. Overexpression of tumor necrosis factor (TNF)-alpha and interferon (IFN)-gamma by bone marrow cells from patients with myelodysplastic syndromes. Leukemia. 1997;11:2049–2054. doi: 10.1038/sj.leu.2400844. [DOI] [PubMed] [Google Scholar]
- Hoang T, Levy B, Onetto N, Haman A, Rodriguez-Cimadevilla JC. Tumor necrosis factor alpha stimulates the growth of the clonogenic cells of acute myeloblastic leukemia in synergy with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1989;170:15–26. doi: 10.1084/jem.170.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury E, Andre C, Pontvert-Delucq S, Drenou B, Baillou C, Guigon M, et al. Tumor necrosis factor alpha (TNF alpha) downregulates c-kit proto-oncogene product expression in normal and acute myeloid leukemia CD34+ cells via p55 TNF alpha receptors. Blood. 1994;84:2506–2514. [PubMed] [Google Scholar]
- Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S, et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. J Clin Invest. 2014;124:528–542. doi: 10.1172/JCI68101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallipoli P, Pellicano F, Morrison H, Laidlaw K, Allan EK, Bhatia R, et al. Autocrine TNF-alpha production supports CML stem and progenitor cell survival and enhances their proliferation. Blood. 2013;122:3335–3339. doi: 10.1182/blood-2013-02-485607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yasukawa K, Harada H, Taga T, Watanabe Y, Matsuda T, et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature. 1986;324:73–76. doi: 10.1038/324073a0. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Akira S, Taga T. Interleukin-6 and its receptor: a paradigm for cytokines. Science. 1992;258:593–597. doi: 10.1126/science.1411569. [DOI] [PubMed] [Google Scholar]
- Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- Chou DB, Sworder B, Bouladoux N, Roy CN, Uchida AM, Grigg M, et al. Stromal-derived IL-6 alters the balance of myeloerythroid progenitors during Toxoplasma gondii infection. J Leukocyte Biol. 2012;92:123–131. doi: 10.1189/jlb.1011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, et al. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte MA, van Olffen RW, van Gisbergen KP, van Lier RA. Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev. 2009;229:216–231. doi: 10.1111/j.1600-065X.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- Hendriks J, Gravestein LA, Tesselaar K, van Lier RA, Schumacher TN, Borst J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat Immunol. 2000;1:433–440. doi: 10.1038/80877. [DOI] [PubMed] [Google Scholar]
- Ochsenbein AF, Brown M, Baerlocher G, Landsdrop P, Riddell SR, Greenberg PD. CD27 expression is required for long-term survival of effector memory CTL in HIV-infected patients. J Exp Med. 2004;200:1407–1417. doi: 10.1084/jem.20040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter M, Odermatt B, Yagita H, Nuoffer JM, Ochsenbein AF. Elimination of chronic viral infection by blocking CD27 signaling. J Exp Med. 2006;203:2145–2155. doi: 10.1084/jem.20060651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feau S, Garcia Z, Arens R, Yagita H, Borst J, Schoenberger SP. The CD4(+) T-cell help signal is transmitted from APC to CD8(+) T-cells via CD27-CD70 interactions. Nat Commun. 2012;3:948. doi: 10.1038/ncomms1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte MA, Arens R, van Os R, van Oosterwijk M, Hooibrink B, van Lier RA, et al. Immune activation modulates hematopoiesis through interactions between CD27 and CD70. Nat Immunol. 2005;6:412–418. doi: 10.1038/ni1174. [DOI] [PubMed] [Google Scholar]
- Schürch C, Riether C, Matter MS, Tzankov A, Ochsenbein AF. CD27 signaling on chronic myelogenous leukemia stem cells activates Wnt target genes and promotes disease progression. J Clin Invest. 2012;122:624–638. doi: 10.1172/JCI45977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, et al. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguille S, Van Tendeloo VF, Berneman ZN. Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia. 2012;26:2186–2196. doi: 10.1038/leu.2012.145. [DOI] [PubMed] [Google Scholar]
- Yotnda P, Firat H, Garcia-Pons F, Garcia Z, Gourru G, Vernant JP, et al. Cytotoxic T cell response against the chimeric p210 BCR-ABL protein in patients with chronic myelogenous leukemia. J Clin Invest. 1998;101:2290–2296. doi: 10.1172/JCI488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita M, Azuma T, Hamaguchi H, Niiya H, Kojima K, Fujita S, et al. Leukemia-associated fusion proteins, dek-can and bcr-abl, represent immunogenic HLA-DR-restricted epitopes recognized by fusion peptide-specific CD4+ T lymphocytes. Leukemia. 2002;16:2400–2407. doi: 10.1038/sj.leu.2402742. [DOI] [PubMed] [Google Scholar]
- Butt NM, Rojas JM, Wang L, Christmas SE, Abu-Eisha HM, Clark RE. Circulating bcr-abl-specific CD8+ T cells in chronic myeloid leukemia patients and healthy subjects. Haematologica. 2005;90:1315–1323. [PubMed] [Google Scholar]
- Gambacorti-Passerini C, Grignani F, Arienti F, Pandolfi PP, Pelicci PG, Parmiani G. Human CD4 lymphocytes specifically recognize a peptide representing the fusion region of the hybrid protein pml/RAR alpha present in acute promyelocytic leukemia cells. Blood. 1993;81:1369–1375. [PubMed] [Google Scholar]
- Osman Y, Takahashi M, Zheng Z, Toba K, Liu A, Furukawa T, et al. Dendritic cells stimulate the expansion of PML-RAR alpha specific cytotoxic T-lymphocytes: its applicability for antileukemia immunotherapy. J Exp Clin Cancer Res. 1999;18:485–492. [PubMed] [Google Scholar]
- Scholl S, Salzmann S, Kaufmann AM, Hoffken K. Flt3-ITD mutations can generate leukaemia specific neoepitopes: potential role for immunotherapeutic approaches. Leuk Lymphoma. 2006;47:307–312. doi: 10.1080/10428190500301306. [DOI] [PubMed] [Google Scholar]
- Graf C, Heidel F, Tenzer S, Radsak MP, Solem FK, Britten CM, et al. A neoepitope generated by an FLT3 internal tandem duplication (FLT3-ITD) is recognized by leukemia-reactive autologous CD8+ T cells. Blood. 2007;109:2985–2988. doi: 10.1182/blood-2006-07-032839. [DOI] [PubMed] [Google Scholar]
- Greiner J, Ono Y, Hofmann S, Schmitt A, Mehring E, Gotz M, et al. Mutated regions of nucleophosmin 1 elicit both CD4(+) and CD8(+) T-cell responses in patients with acute myeloid leukemia. Blood. 2012;120:1282–1289. doi: 10.1182/blood-2011-11-394395. [DOI] [PubMed] [Google Scholar]
- Whiteway A, Corbett T, Anderson R, Macdonald I, Prentice HG. Expression of co-stimulatory molecules on acute myeloid leukaemia blasts may effect duration of first remission. Br J Haematol. 2003;120:442–451. doi: 10.1046/j.1365-2141.2003.04085.x. [DOI] [PubMed] [Google Scholar]
- Tamura H, Dan K, Tamada K, Nakamura K, Shioi Y, Hyodo H, et al. Expression of functional B7-H2 and B7.2 costimulatory molecules and their prognostic implications in de novo acute myeloid leukemia. Clin Cancer Res. 2005;11:5708–5717. doi: 10.1158/1078-0432.CCR-04-2672. [DOI] [PubMed] [Google Scholar]
- Scheibenbogen C, Letsch A, Thiel E, Schmittel A, Mailaender V, Baerwolf S, et al. CD8 T-cell responses to Wilms tumor gene product WT1 and proteinase 3 in patients with acute myeloid leukemia. Blood. 2002;100:2132–2137. doi: 10.1182/blood-2002-01-0163. [DOI] [PubMed] [Google Scholar]
- Mumprecht S, Schürch C, Scherrer S, Claus C, Ochsenbein AF. Chronic myelogenous leukemia maintains specific CD8(+) T cells through IL-7 signaling. Eur J Immunol. 2010;40:2720–2730. doi: 10.1002/eji.201040404. [DOI] [PubMed] [Google Scholar]
- Kiani A, Habermann I, Schake K, Neubauer A, Rogge L, Ehninger G. Normal intrinsic Th1/Th2 balance in patients with chronic phase chronic myeloid leukemia not treated with interferon-alpha or imatinib. Haematologica. 2003;88:754–761. [PubMed] [Google Scholar]
- Horowitz MM, Gale RP, Sondel PM, Goldman JM, Kersey J, Kolb HJ, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- Weiden PL, Doney K, Storb R, Thomas ED. Antihuman thymocyte globulin for prophylaxis of graft-versus-host disease. A randomized trial in patients with leukemia treated with HLA-identical sibling marrow grafts. Transplantation. 1979;27:227–230. doi: 10.1097/00007890-197904000-00003. [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, et al. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- Bhatia R, Verfaillie CM. The effect of interferon-alpha on beta-1 integrin mediated adhesion and growth regulation in chronic myelogenous leukemia. Leuk Lymphoma. 1998;28:241–254. doi: 10.3109/10428199809092680. [DOI] [PubMed] [Google Scholar]
- Pane F, Mostarda I, Selleri C, Salzano R, Raiola AM, Luciano L, et al. BCR/ABL mRNA and the P210(BCR/ABL) protein are downmodulated by interferon-alpha in chronic myeloid leukemia patients. Blood. 1999;94:2200–2207. [PubMed] [Google Scholar]
- Burchert A, Neubauer A. Interferon alpha and T-cell responses in chronic myeloid leukemia. Leuk Lymphoma. 2005;46:167–175. doi: 10.1080/10428190400012029. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Yang K, Foster SJ, Kondo M, Kelsoe G. Inflammation controls B lymphopoiesis by regulating chemokine CXCL12 expression. J Exp Med. 2004;199:47–58. doi: 10.1084/jem.20031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ikezoe T, Nishioka C, Nobumoto A, Yokoyama A. IL-1beta inhibits self-renewal capacity of dormant CD34(+) /CD38(−) acute myelogenous leukemia cells in vitro and in vivo. Int J Cancer. 2013;133:1967–1981. doi: 10.1002/ijc.28198. [DOI] [PubMed] [Google Scholar]