Using highly sensitive RNAseq to examine the whole transcriptome of enriched aortic hematopoietic stem cells and endothelial cells, the authors find G-protein–coupled receptor, Gpr56, is required to generate the first HSCs during endothelial to hematopoietic cell transition.
Abstract
Hematopoietic stem cells (HSCs) are generated via a natural transdifferentiation process known as endothelial to hematopoietic cell transition (EHT). Because of small numbers of embryonal arterial cells undergoing EHT and the paucity of markers to enrich for hemogenic endothelial cells (ECs [HECs]), the genetic program driving HSC emergence is largely unknown. Here, we use a highly sensitive RNAseq method to examine the whole transcriptome of small numbers of enriched aortic HSCs, HECs, and ECs. Gpr56, a G-coupled protein receptor, is one of the most highly up-regulated of the 530 differentially expressed genes. Also, highly up-regulated are hematopoietic transcription factors, including the “heptad” complex of factors. We show that Gpr56 (mouse and human) is a target of the heptad complex and is required for hematopoietic cluster formation during EHT. Our results identify the processes and regulators involved in EHT and reveal the surprising requirement for Gpr56 in generating the first HSCs.
Hematopoietic stem cells (HSCs) are responsible for the life-long maintenance and regeneration of the adult vertebrate blood system. HSCs are generated through a natural transdifferentiation process occurring in specialized embryonic vascular cells, known as hemogenic endothelial cells (ECs [HECs]). In mice, the first adult HSCs are generated in the aorta-gonad-mesonephros (AGM) region at embryonic day (E) 10.5 (Müller et al., 1994; Medvinsky and Dzierzak, 1996). The emergence of the definitive hematopoietic system in the mouse embryo correlates with the temporal appearance of clusters of hematopoietic cells (HCs) associated with the aortic endothelium and the major arteries (Garcia-Porrero et al., 1995; North et al., 1999; de Bruijn et al., 2000). Chick embryo dye-marking studies were the first to show that aortic ECs give rise to HCs (Jaffredo et al., 1998). In mammalian embryos, the results of phenotypic and genetic studies, supported by stringent in vivo transplantation studies of enriched cell fractions, demonstrate that HSCs are derived from vascular ECs during a short window of developmental time (de Bruijn et al., 2002; North et al., 2002; Zovein et al., 2008; Chen et al., 2009). This developmental process is known as endothelial to hematopoietic cell transition (EHT).
To facilitate the study of HSC emergence in the mouse embryo, numerous markers have been used individually and/or in combination to identify HSCs and their direct precursors. Immunolocalization of these markers in the AGM highlighted the heterogeneous nature of the cells in the hematopoietic clusters (Ody et al., 1999; Taoudi et al., 2005; Yokomizo and Dzierzak, 2010; Robin et al., 2011). Whereas combinations of these markers allow HSC enrichment, so far no combination of endothelial and/or hematopoietic markers has been able to distinguish hemogenic from nonhemogenic aortic ECs.
The Ly6aGFP (Sca1) mouse model, in which all HSCs throughout development are GFP+ (de Bruijn et al., 2002; Ma et al., 2002), has facilitated the study of EHT. Clear proof of EHT was obtained by real-time imaging of the mouse Ly6aGFP embryonic aorta (Boisset et al., 2010). In the E10.5 aorta, at the time when the number of hematopoietic clusters peak (Yokomizo and Dzierzak, 2010), flat endothelial GFP+ cells were observed to transition to morphologically round GFP+ cells that begin to express other HSC markers (Boisset et al., 2010). Real-time imaging of transgenic zebrafish embryos similarly revealed the transition of aortic ECs to HCs (Bertrand et al., 2010; Kissa and Herbomel, 2010), indicating that EHT is an evolutionarily conserved process by which the definitive hematopoietic system of vertebrates is generated.
To specifically understand the molecular program involved in EHT, we set out in this study to identify key genes and processes that are functionally relevant in mouse aortic HECs as they transit to HSCs. Based on the vital imaging of EHT, the Ly6aGFP reporter is currently the most tractable marker to distinguish and enrich the HECs that are undergoing EHT from other aortic ECs, and also the emerging HSCs from other HCs. Here we present RNA sequencing data obtained from highly enriched small numbers of relevant EHT cells from Ly6aGFP embryos, aortic ECs, HECs, and emerging HSCs. Among the few (530) differentially expressed genes (DEGs) during EHT, Gpr56 is the highest up-regulated gene encoding a cell surface receptor. We show for the first time the functional involvement of Gpr56 in HSC emergence during EHT. In addition, the previously described “heptad” transcription factors (TFs; Wilson et al., 2010) are up-regulated during EHT, bind the Gpr56 enhancer, and regulate its expression. This unique dataset expands our understanding of EHT, identifying the gene networks and processes that are essential for HSC generation in the embryo.
RESULTS
Temporal-spatial and transcriptomic quantitation of aortic hemogenic endothelial and emerging HCs
Ly6aGFP expression marks HCs emerging from hemogenic endothelium at the time of HSC generation in the midgestation mouse aorta. To quantify and localize these cells, we performed confocal imaging of whole and sectioned immunostained E10 Ly6aGFP embryos (Fig. 1, A–D). CD31 marks all ECs and HCs, and cKit marks all HCs. However, Ly6aGFP marks only some ECs and some HCs. High-resolution imaging of transverse sections allowed quantitation of four different Ly6aGFP-expressing aortic cell types (Fig. 1 D): flat ECs, bulging cells in the single layer of endothelium, and two differently positioned round cells within the clusters distinguished by the close attachment to (juxtaposed) or a position distal from the endothelium. The total number of GFP+ cells increased from 287 at early E10 (32 sp [somite pairs]) to 1,592 at late E10 (37 sp; Fig. 1 D). From the small fraction of ECs that express GFP (range 13–19%), most aortic GFP+ cells are flat ECs with only 8% of GFP+ cells in hematopoietic clusters. Although by a random distribution more GFP+ cluster cells would be expected in distal positions (as compared with juxtaposed), we observed 70–88% of the GFP+ HCs localized in a juxtaposed position, most likely because GFP+ HCs are emerging from GFP+ ECs and/or are actively maintained at the juxtaposed position.
Figure 1.
Analysis of EHT cell subsets. (A) Whole-mount image of a 34-sp Ly6aGFP embryo showing expression of CD31 (magenta), cKit (red), and GFP (green). The aorta, vitelline artery, and somatic vasculature are indicated. (B) Four types of aortic cells during EHT in a Ly6aGFP AGM section (36 sp) stained with anti-CD31 (magenta) and anti-cKit antibodies (red). ECs are CD31+cKit−GFP−, HECs are CD31+cKit−GFP+, HSCs are CD31+cKit+GFP+, and HCs are CD31+cKit+GFP−. (C) Transverse section through a 36-sp Ly6aGFP embryo showing expression of CD34 (red) and GFP (green). A hematopoietic cluster with some GFP+ cells is located ventrally. GFP+ ECs are scattered throughout the aorta. (D) Different GFP+ cell types (arrowheads) in an E10.5 Ly6aGFP aorta. (Endothelial) two flat GFP+ ECs; (bulging) rounding-up of a GFP+ EC; (juxtaposed) round HC closely adhering to an EC; (distal) round HC on the distal side of the cluster. The number of cells/aorta is listed below at the 32-, 34-, and 37-sp stages. Bars: (A) 100 µm; (B and D) 10 µm; (C) 50 µm. (E) Scatter plot showing the distribution and sorting gates for EHT subsets EC, HEC, HSC, and HC from E10.5/E11 Ly6aGFP AGMs. (F) Hematopoietic progenitor numbers (total CFU-C [CFU-culture]) per AGM. EHT subsets from E10.5 Ly6aGFP AGMs (34–39 sp) were plated in methylcellulose, and colonies were counted at day 12 (SD is shown; n = 4). (G) HSC long-term repopulating activity in E11 Ly6aGFP AGM EHT subsets (40–49 sp). Irradiated adult recipients (n = 4) were injected with 5–9 ee of ECs, 4–9 ee of HECs, 1–5 ee of HSCs, and 4–8 ee of HCs together with 2 × 105 spleen cells (recipient type). Percentage of donor cell chimerism at 4 mo after injection is shown. Indicated above each bar is the number of repopulated recipients/number of recipients injected. (H) Normalized number of mapped fragments for genes encoding the markers used for sorting EHT fractions. FPKMs of CD31, cKit, and Ly6aGFP per fraction are shown (error bars are SD). (I) Heat map of FPKMs for genes encoding several relevant cell surface molecules: Cdh5, Tek, Esam, Kdr, and Eng in each of the sequenced cell fractions and the frequency of cells in each sorted fraction expressing the corresponding protein. Significant positive correlation is observed between FACS and RNAseq data (r2 = 0.54; **, P = 0.01).
Because all HSCs are GFP+ and rare HCs have been observed to emerge from GFP+ ECs, Ly6aGFP is the best marker for high enrichment of HECs. Hence, we developed an enrichment method using the Ly6aGFP, cKit, and CD31 markers: ECs (CD31+cKit−GFP−), HECs (CD31+cKit−GFP+), HSCs (CD31+cKit+GFP+), and progenitor/differentiated HCs (CD31+cKit+GFP−; Fig. 1 B). The distinct cell types were sorted (Fig. 1 E) and hematopoietic function was assessed. Hematopoietic progenitors were found in the HC (64%) and HSC (33%) fractions as expected, with a majority of the immature progenitors (CFU-GEMM) in the HSC fraction (Fig. 1 F). In vivo transplantation assays revealed that only the CD31+cKit+GFP+ fraction contained HSCs (Fig. 1 G). These HSCs provided long-term high-level multilineage repopulation of adult irradiated recipients (Fig. S1 A; 6 engrafted of 10 injected with 1–5 ee [embryo equivalents]). Despite injection of high embryo equivalents of cells from the other fractions (4–9 ee), no repopulation was found with the ECs, HECs, or HCs.
RNA sequencing and validation
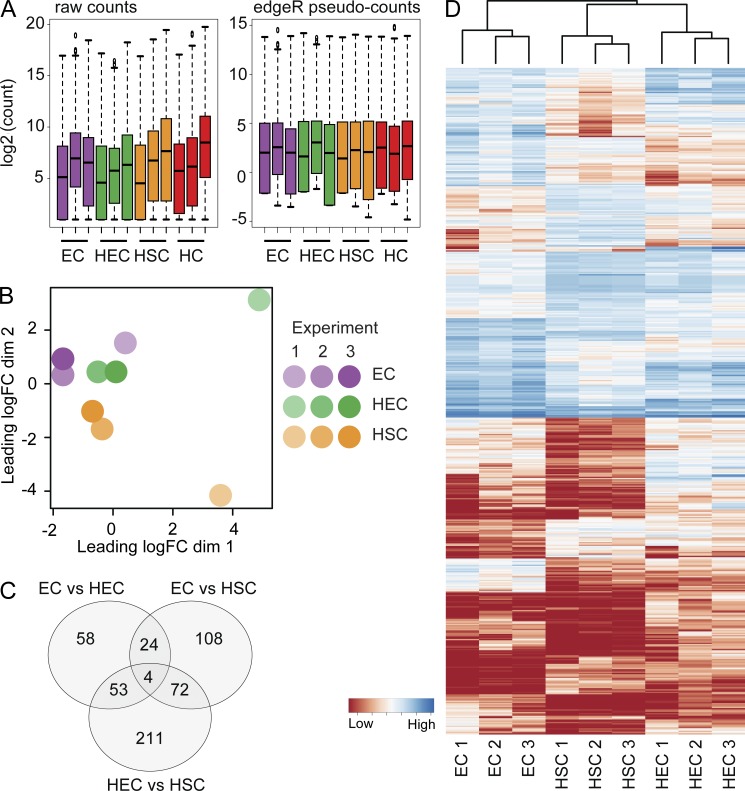
Sorted ECs, HECs, HSCs, and HCs from three independent biological replicates were used for RNA sequencing. As few as 4–14 E10.5 AGM equivalents (34–41 sp) of sorted cells per replicate were obtained, and cDNA was made from as few as 593 sorted cells (see Table S1 for details). The sequence reads of EHT cell fractions were mapped to the mouse genome (NCBI build 37/mm9), and the generated count table (with 7–57 million unique mapped reads to exons per sample) was normalized and analyzed by edgeR (Fig. 2 A; McCarthy et al., 2012).
Figure 2.
RNAseq data analysis. (A) Distribution of raw counts per sample (left) and edgeR internal normalized counts (right). The normalized counts are used in all subsequent analyses. Datasets for three biological replicates are shown for ECs, HECs, HSCs, and HCs. Biological replicate 1 includes two 36-sp and two 37-sp embryos; replicate 2 includes four 34-sp, two 35-sp, three 36-sp, and five 37-sp embryos; replicate 3 includes two 35-sp, one 36-sp, two 38-sp, one 39-sp, one 40-sp, and one 41-sp embryos. (B) BCV in RNAseq samples from three biological replicates of relevant EHT cell fractions: EC, HEC, and HSC. (C) Venn diagram showing numbers of DEGs in comparisons of HECs versus ECs, HSCs versus HECs, and HSCs versus ECs. Total DEGs is 530 (see Table S2 for gene lists). (D) Heat maps showing all 530 DEGs and hierarchical clustering of the genes in each EHT cell fraction from the three biological replicates.
To confirm that the transcriptome analysis was representative of the sorted EHT fractions, we measured the normalized number of fragments (in FPKMs [fragments per kilobase exon reads per million fragments mapped]) of CD31, cKit, and Ly6a (Fig. 1 H). As expected, CD31 transcripts were found in all four subsets (ECs, HECs, HSCs, and HCs), cKit transcripts were found only in HCs and HSCs, and Ly6a transcripts were found in HECs and HSCs.
Gene transcript reads for endothelial genes Cdh5, Tek, Esam, Kdr, and Eng were highest in HECs as compared with ECs and were higher in HSCs than in ECs or HCs. When the four cell fractions were examined by FACS (Fig. 1 I), cell surface expression correlated significantly with transcript levels (r2 = 0.54, P = 0.01). Thus, our datasets reflect a dynamic transcriptional program during EHT.
Global transcriptional differences between the EHT cell subsets
Biological coefficient of variation (BCV) analysis indicates (Fig. 2 B) that EC, HEC, and HSC fractions are closely related but distinct. EC, HEC, and HSC replicate 2 and 3 samples cluster together, whereas replicate 1 EC, HEC and HSC samples show a similar BCV pattern but are further dispersed in the plot. The tighter sample dispersion of replicates 2 and 3 is most likely the result of the higher sequencing depth (Table S1). Hence, distinct transcriptional variation between the EHT fractions is consistent for the three biological replicates.
Dataset comparisons showed a total of 530 DEGs (false discovery rate [FDR] < 0.05; Fig. 2 C and Table S2). The EC to HEC comparison shows 139 DEGs, whereas 340 genes were differentially expressed between HECs and HSCs. Moreover, comparison of ECs with HSCs identified 108 additional genes. MA plots of differential expression analysis show most genes being centered around zero, further confirming the correct normalization of datasets (Fig. S2 A). In the EC to HEC comparison, most DEGs are up-regulated, whereas a majority of the DEGs in HEC to HSC are down-regulated. Hierarchical clustering of DEGs grouped the three biological replicates of each fraction together, suggesting that ECs, HECs, and HSCs have recognizably distinct genetic programs (Fig. 2 D).
Transcriptome analysis reveals processes involved in EHT
DEGs were grouped based on their relative expression levels (H, high; I, intermediate; and L, low expression) into representative patterns for EC genes (HIL, HLL, HHL, and HLI), HEC genes (LHL, IHL, IHI, and LHI), and HSC genes (LHH, LIH, IIH, LLH, and ILH; Fig. 3 and Table S2). Each group was used as input for Gene Ontology (GO), KEGG, and WikiPathways enrichment analysis, and Gene Set Enrichment Analysis (GSEA) was used to detect global shifts of gene sets during each transition.
Figure 3.
GO terms/processes enriched in EHT subsets. (A) DEG patterns that are EC specific are shown (left): high-intermediate-low (HIL), HHL, HLI, and HLL. GO enrichment analysis was performed using WebGestalt, and enriched GO terms are summarized by REVIGO (right). (B) DEG patterns that are HEC specific are shown (left): LHL, IHL, IHI, and LHI. GO enrichment analysis and GO terms are summarized by REVIGO (right). (C) DEG patterns that are HSC specific are shown (left): LHH, LIH, IIH, LLH, and ILH. GO enrichment and GO terms are summarized by REVIGO. Rectangle size represents the number of DEGs in the accompanying GO term. See Table S3 for enriched ontology terms.
EC genes show overrepresentation of “focal adhesion,” “ECM-receptor interaction,” “protein digestion and absorption,” “oxidative stress,” and “chemokine signaling” terms (Fig. 3 A and Table S3), consistent with EC function (Rajendran et al., 2013). Significant enrichment of “inflammatory response” and “TGFbeta signaling” terms suggests ECs to be activated. Whether this is caused by activated endothelium in an actual inflammatory response or by the activation of inflammatory genes that are involved in other signaling pathways in development (Orelio and Dzierzak, 2003; Orelio et al., 2009) requires further study.
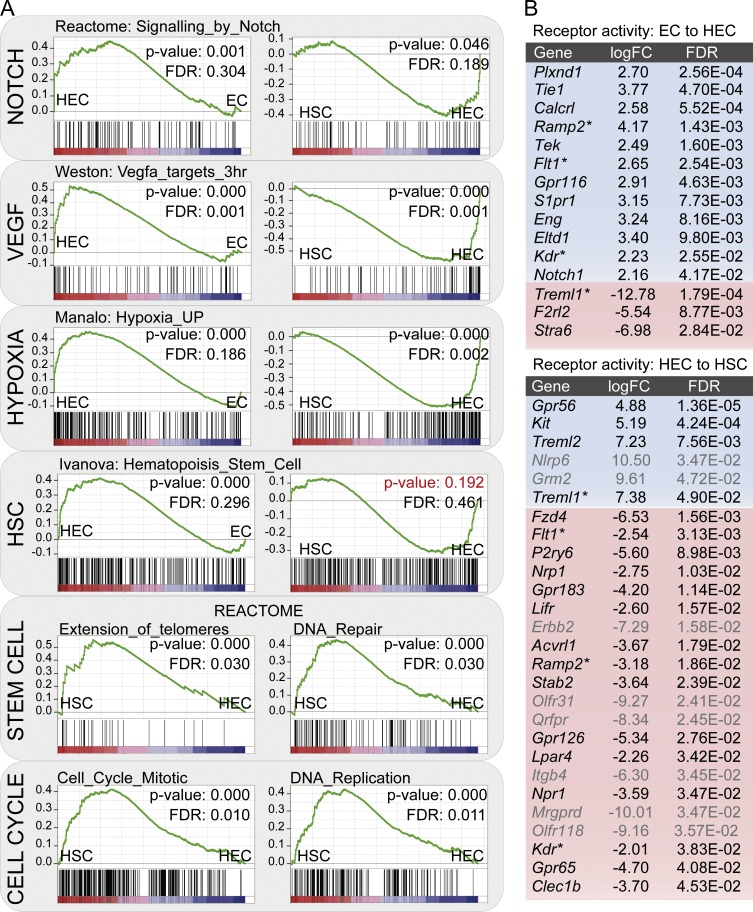
HEC genes are most enriched in “cell adhesion” and “migration” gene sets, consistent with changes required for HECs to move out of the endothelial layer and form clusters of HCs (Fig. 3 B and Table S3). Like ECs, 169 out of 191 genes within the “angiogenesis” GO term (GO:0001525) were present in HECs. HEC genes also showed enrichment in “cardiovascular system development,” most likely because of the presence of many angiogenic and vascular development genes. Indeed, only 3 (Jag1, Sox17, and Fbn1) out of the 76 published essential cardiovascular genes (Van Handel et al., 2012) are present in HEC genes. “Delta-Notch” and “Notch” pathways known to be important for HSC generation and cluster formation (Kumano et al., 2003) were enriched, and whole transcriptome comparisons using GSEA also show Notch pathway gene sets as up-regulated in HECs (Fig. 4 A and Fig. S2 B). In addition, multiple “VEGF” and “hypoxia-regulated” gene sets were enriched in HECs as compared with HSCs and ECs (Fig. 4 A), including Hif1α, which was recently shown to be an important factor for HSC generation (Imanirad et al., 2014). Surprisingly, several previously published HSC gene sets (from MSigDB database v4.0) are significantly enriched in HECs as compared with ECs, whereas no significant enrichment of these genes was found in HSCs as compared with HECs, indicating that the hematopoietic program is already activated in HECs (Fig. 4 A).
Figure 4.
Changing processes and cell surface molecules during EHT. (A) GSEA for VEGF, Notch, Hypoxia up-regulated genes, genes specifically expressed in HSCs, stem cell function–related gene sets like “telomere lengthening” and “DNA repair,” cell cycle–related gene sets, and early hematopoietic progenitor–specific genes. (B) Receptor-related genes with significant expression changes in EC to HEC and HEC to HSC transitions. Blue, increased expression; red, decreased expression; gray font, genes with low overall expression levels as defined by edgeR-calculated logCPM of <3 (and higher probability of being false positive); asterisks, genes differentially expressed during both transitions; logFC, log fold change.
HSC genes showed clear overrepresentation of “hematopoietic processes,” “cell cycle,” and “histone methylation” related genes (Fig. 3 C). Significant enrichment of “cell cycle progression,” “DNA replication,” and “hematopoietic progenitor” sets was also detected by GSEA (Fig. 4 A and Fig. S2 C). “Hematopoietic progenitor” gene sets are enriched in HSCs as compared with HECs, and detection of “acute myeloid leukemia” from the KEGG database and “pluripotency network” from WikiPathways is in agreement with the acquisition of hematopoietic fate and self-renewal capacity in HSCs (Table S3). This is further supported by significant enrichment of gene sets characteristic of stem cells, such as “telomere lengthening” and “DNA repair,” in the HSC fraction by GSEA (Fig. 4 A; Yui et al., 1998; Rossi et al., 2005).
TF expression in cells undergoing EHT
The genetic program directing cell identity is coordinated by TFs, and thus we focused our attention on these genes in our EHT datasets. As compared with ECs, significant up-regulated expression of Mecom, Notch1 and 4, Gfi1, Sox17, Ets2, and Elk3 was found in HECs (Table 1), with Sox7, Sox18, Runx1, Hhex, and Lmo2 among the top up-regulated HEC TFs (Table S4).
Table 1.
Differentially expressed TFs
| HECs versus ECs | HSCs versus HECs | HSCs versus ECs | ||||||
| Gene | logFC | FDR | Gene | logFC | FDR | Gene | logFC | FDR |
| (A) Top 25 up-regulated genes | ||||||||
| Elk3 | 2.42 | 1.1E-02 | Myb | 4.23 | 1.7E-06 | Myb | 4.55 | 7.1E-08 |
| Mecom | 3.21 | 1.4E-02 | Gfi1b | 5.48 | 7.7E-05 | Ikzf2 | 3.13 | 2.5E-04 |
| Notch4 | 5.41 | 2.9E-02 | Hlf | 7.03 | 8.9E-05 | Hlf | 5.99 | 6.7E-04 |
| Notch1 | 2.16 | 4.2E-02 | Meis1 | 2.20 | 8.1E-04 | Runx1 | 2.68 | 1.8E-03 |
| Rab11a | 2.03 | 5.2E-02 | Zfp106 | 2.43 | 3.5E-03 | Myc | 2.27 | 9.5E-03 |
| Gfi1 | 7.69 | 6.8E-02 | Ncoa4 | 2.05 | 8.8E-03 | Dnmt3a | 1.73 | 2.2E-02 |
| Wwtr1 | 3.20 | 6.8E-02 | Nop2 | 2.35 | 1.9E-02 | Chd4 | 1.62 | 2.9E-02 |
| Junb | 4.90 | 6.8E-02 | Elf1 | 2.51 | 2.0E-02 | Dnmt1 | 1.58 | 3.2E-02 |
| Ets2 | 2.50 | 6.9E-02 | Zfp445 | 2.05 | 2.3E-02 | Gfi1 | 8.33 | 3.3E-02 |
| Mapk3 | 1.62 | 7.3E-02 | Nfe2 | 5.05 | 4.1E-02 | Zfp445 | 1.91 | 4.6E-02 |
| Nkx2-3 | 5.95 | 7.4E-02 | Zfp763 | 4.29 | 4.2E-02 | Setbp1 | 2.78 | 5.9E-02 |
| Ldb2 | 2.30 | 7.5E-02 | Ikzf2 | 1.94 | 5.2E-02 | Ikzf1 | 2.60 | 6.8E-02 |
| Hdac7 | 2.20 | 7.6E-02 | Huwe1 | 1.55 | 5.7E-02 | Bcor | 2.21 | 8.4E-02 |
| Sox17 | 5.14 | 8.4E-02 | Orc2 | 2.22 | 6.5E-02 | Trp53bp1 | 1.51 | 8.5E-02 |
| Ctnnb1 | 1.28 | 9.5E-02 | Mpl | 2.84 | 6.6E-02 | Mycn | 2.57 | 9.9E-02 |
| Hey1 | 4.24 | 1.1E-01 | Etv6 | 1.99 | 6.9E-02 | Zfp106 | 1.67 | 1.0E-01 |
| Epas1 | 3.10 | 1.2E-01 | Lmo1 | 8.87 | 7.5E-02 | Suz12 | 1.57 | 1.1E-01 |
| Hey2 | 4.35 | 1.2E-01 | Paxip1 | 1.93 | 7.6E-02 | Cbfa2t3 | 2.63 | 1.2E-01 |
| Sox7 | 4.27 | 1.5E-01 | Cpsf6 | 1.28 | 8.1E-02 | Paxip1 | 1.83 | 1.4E-01 |
| Sox18 | 4.05 | 1.5E-01 | Zfp748 | 2.68 | 8.3E-02 | Notch1 | 1.72 | 1.4E-01 |
| Tsc22d1 | 1.46 | 1.6E-01 | Polr1a | 2.41 | 8.3E-02 | Etv6 | 1.82 | 1.5E-01 |
| Nrarp | 3.76 | 1.7E-01 | Trp53bp1 | 1.42 | 8.9E-02 | Kdm5a | 1.38 | 1.5E-01 |
| Nfic | 2.07 | 1.8E-01 | Dnmt1 | 1.29 | 9.4E-02 | Meis1 | 1.22 | 1.8E-01 |
| Pdlim1 | 1.96 | 2.1E-01 | Med23 | 1.84 | 9.8E-02 | Rreb1 | 1.49 | 1.8E-01 |
| Hmg20b | 2.46 | 2.1E-01 | Krr1 | 1.45 | 1.0E-01 | Bptf | 1.17 | 1.8E-01 |
| (B) Top 25 down-regulated genes | ||||||||
| Pou5f1 | −7.70 | 2.6E-04 | Snai2 | −7.39 | 7.7E-05 | Snai2 | −8.64 | 3.0E-06 |
| Utf1 | −8.22 | 1.0E-02 | Id3 | −3.87 | 5.6E-04 | Rhox6 | −10.09 | 2.9E-05 |
| Gfi1b | −3.77 | 1.3E-02 | Hey2 | −7.55 | 2.3E-03 | Rhox9 | −10.23 | 4.6E-05 |
| Foxd1 | −10.54 | 6.6E-02 | Rhox6 | −6.49 | 6.8E-03 | Tgfb1i1 | −5.76 | 4.6E-05 |
| Lmo1 | −10.20 | 6.7E-02 | Msx2 | −8.54 | 7.4E-03 | Pou5f1 | −7.16 | 2.2E-04 |
| Hand1 | −6.55 | 7.4E-02 | Rhox9 | −6.56 | 9.0E-03 | Etv5 | −3.26 | 6.0E-04 |
| Zfp612 | −7.77 | 1.0E-01 | Etv5 | −2.60 | 1.1E-02 | Utf1 | −8.47 | 3.6E-03 |
| Prrx2 | −7.19 | 1.0E-01 | Isl1 | −4.08 | 3.6E-02 | Msx2 | −8.98 | 5.1E-03 |
| Asb12 | −9.04 | 1.1E-01 | Ebf2 | −7.43 | 4.0E-02 | Id3 | −3.21 | 5.8E-03 |
| Cdc6 | −1.75 | 1.3E-01 | Ebf1 | −5.39 | 4.3E-02 | Prrx2 | −10.99 | 6.9E-03 |
| Ncoa4 | −1.48 | 1.4E-01 | Hey1 | −4.69 | 4.4E-02 | Prss35 | −5.99 | 1.8E-02 |
| Krr1 | −1.47 | 1.4E-01 | Epas1 | −3.42 | 4.6E-02 | Grhl3 | −6.02 | 2.6E-02 |
| Alx4 | −5.93 | 1.5E-01 | Rarb | −3.81 | 5.2E-02 | Foxd1 | −10.54 | 2.8E-02 |
| Wt1 | −4.76 | 2.0E-01 | Hoxd9 | −7.63 | 5.5E-02 | Ripk4 | −9.34 | 3.0E-02 |
| Rhox2e | −3.29 | 2.0E-01 | Sox17 | −5.00 | 5.7E-02 | Ebf2 | −8.08 | 3.0E-02 |
| Pax8 | −8.85 | 2.1E-01 | Creb3l1 | −4.59 | 5.8E-02 | Foxp2 | −3.93 | 3.4E-02 |
| Rhox6 | −3.59 | 2.2E-01 | Ugp2 | −1.79 | 6.5E-02 | Zim1 | −7.30 | 3.4E-02 |
| Neurod6 | −6.02 | 2.2E-01 | Nr3c1 | −2.93 | 6.9E-02 | Creb3l1 | −5.25 | 3.5E-02 |
| Hoxc4 | −8.73 | 2.3E-01 | Notch4 | −4.21 | 7.1E-02 | Hoxd9 | −8.66 | 3.6E-02 |
| Rhox9 | −3.68 | 2.3E-01 | Myt1 | −6.02 | 7.6E-02 | Sall4 | −2.66 | 3.8E-02 |
| Six1 | −7.30 | 2.3E-01 | Elf3 | −6.86 | 7.6E-02 | Id2 | −2.69 | 3.9E-02 |
| Tcf21 | −5.63 | 2.3E-01 | Ankrd1 | −4.84 | 7.7E-02 | Hoxc10 | −5.83 | 5.0E-02 |
| Runx1t1 | −1.79 | 2.4E-01 | Onecut3 | −8.32 | 8.3E-02 | Myt1 | −6.64 | 6.7E-02 |
| Klf1 | −6.56 | 2.6E-01 | Hivep3 | −5.19 | 8.4E-02 | Elf3 | −7.56 | 6.9E-02 |
| Aff3 | −3.18 | 3.1E-01 | Zfp36l1 | −1.90 | 8.6E-02 | Isl1 | −3.79 | 7.0E-02 |
(A and B) Top 25 up-regulated TFs (A) and top 25 down-regulated TFs (B) in HEC versus EC, HSC versus HEC, and HSC versus EC comparisons. FDR, FDR corrected p-value; logFC, log fold change. All genes with FDR < 0.05 except genes with underlining.
In the HSC fraction, many TFs with known roles in HSC development, including Etv6, Gfi1, Gfi1b, Myb, Myc, Hlf, Meis1, Hhex, Runx1, Mpl, and Ikzf1 and 2 (Table 1 and Table S4), were found to be significantly up-regulated as compared with ECs or HECs. We identified several novel TFs not previously reported to be involved in embryonic HSC generation such as zinc-finger proteins Zfp106, Zfp445, Zfp748, and Zfp763, megakaryocyte factor Nfe2, transcriptional corepressor Bcor, and Cbfa2t3 (Eto2). Also present in the top hits were chromatin-remodeling factors Suz12, Paxip1, Kdm5a, Smarca4 (Brg1), Ezh1, Bptf, and Hdac1 and de novo DNA methylation genes Dnmt3a/Dnmt3b and Dnmt1. The down-regulation of several Hox, Tbx, and Fox genes was observed in the EC to HEC and HEC to HSC transition, whereas only Hoxa9, Hhex, and Foxk1 were up-regulated in HSCs as compared with ECs or HECs (Table S4).
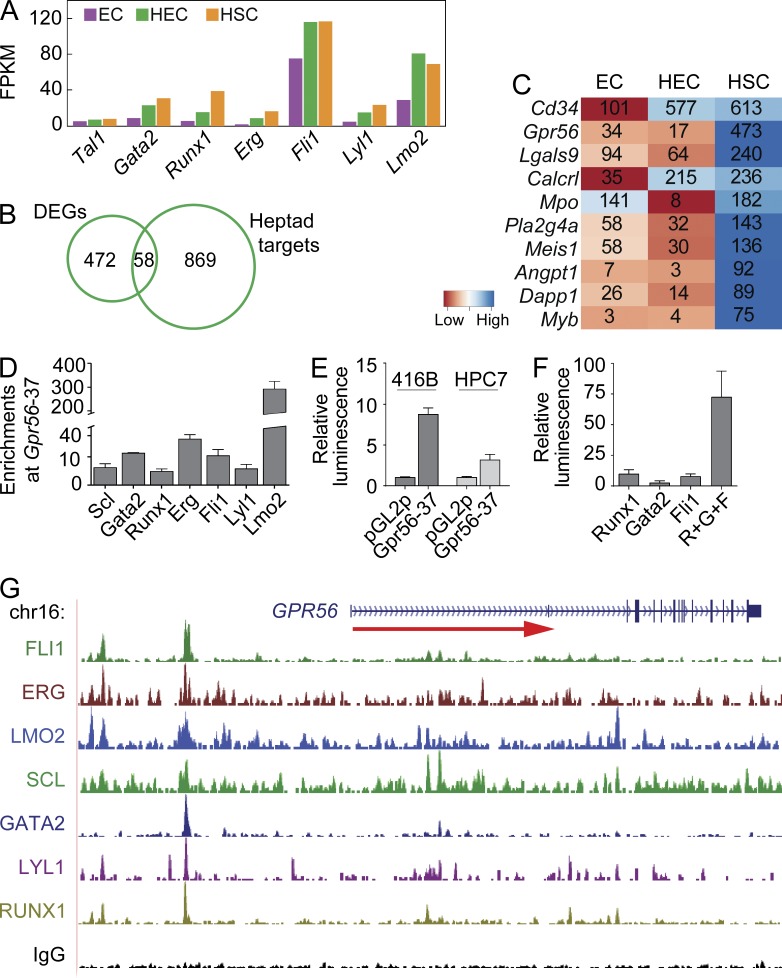
It has been shown that a pivotal (heptad) group of TFs work together in transcriptional regulatory complexes to regulate the expression of downstream target genes in hematopoietic progenitor cell lines (Wilson et al., 2010). The heptad TFs could act as one of the transcriptional hubs for the regulation of EHT. Our RNAseq datasets reveal that all heptad TFs increase during EHT (Fig. 5 A). To identify genes encoding novel EHT and embryonic HSC surface markers, the 530 DEGs were compared with the 927 heptad TF targets identified by chromatin immunoprecipitation (ChIP)seq analyses in HPC7 cells. 58 DEGs were found to be targets of heptad TFs, with CD34 and Gpr56 as the top hits (Fig. 5, B and C). Interestingly, also in the whole transcriptome analysis of EHT, Gpr56 was identified as the top differentially expressed receptor gene in the HEC to HSC transition, followed by cKit (Fig. 4 B). Because both CD34 and cKit function has been studied in HSCs and these markers are used extensively for HSC isolation (Sánchez et al., 1996), we focused on Gpr56.
Figure 5.
Gpr56 is a heptad target in mouse and human blood progenitors. (A) Mean FPKM values of heptad factors in EC, HEC, and HSC fractions. (B) A Venn diagram showing the overlap between sites with combinatorial binding of Scl, Gata2, Runx1, Erg, Fli1, Lyl1, and Lmo2 in HPC7 cells (Heptad targets) and 530 DEGs during EHT. (C) Heat map of top 10 heptad target DEGs based on highest expression in HSCs and with respective mean FPKM values inside heat map. (D) qPCR for TF enrichment at Gpr56-37 as compared with IgG and control in HPC7 mouse myeloid progenitor cells (n = 4). (E) Transfection assays in 416B and HPC7 mouse progenitors show enhancer activity of Gpr56-37 (n = 3). (F) Transactivation assays in Cos7 cells showing synergistic responsiveness of the Gpr56-37 element to Runx1, Gata2, and Fli1 (n = 4). (D–F) Error bars show SD. (G) TF binding at HsGPR56-48 (corresponding region to MmGpr56-37) in primary human CD34 HSCs.
The Gpr56 heptad consensus region in the mouse is located 37 kb upstream of the translational start site. We identified this region as the Gpr56-37 enhancer. Enrichment of heptad factors at the Gpr56-37 element was found in mouse HPC7 cell line by quantitative PCR (qPCR; Fig. 5 D). Transactivation assays in hematopoietic progenitor cell lines showed significant activation of Gpr56-37 enhancer, whereas overexpression of three of the heptad factors (Gata2, Runx1, and Fli1) showed synergistic activation of the Gpr56-37 enhancer (Fig. 5, E and F). Moreover, we identified a homologous element 48 kb upstream of the human GPR56 gene. In human CD34+ HSC-enriched cells, we found binding of all seven heptad TFs to the human GPR56-48 element (Fig. 5 G; Chacon et al., 2014). These data suggest that the heptad TFs and their downstream target Gpr56 are important in HSC generation during EHT, as well as in healthy and leukemic human HCs. Because nothing is known concerning Gpr56 in embryonic hematopoietic development, we examined its regulation and role during EHT.
Gpr56 is required during EHT for HSC generation
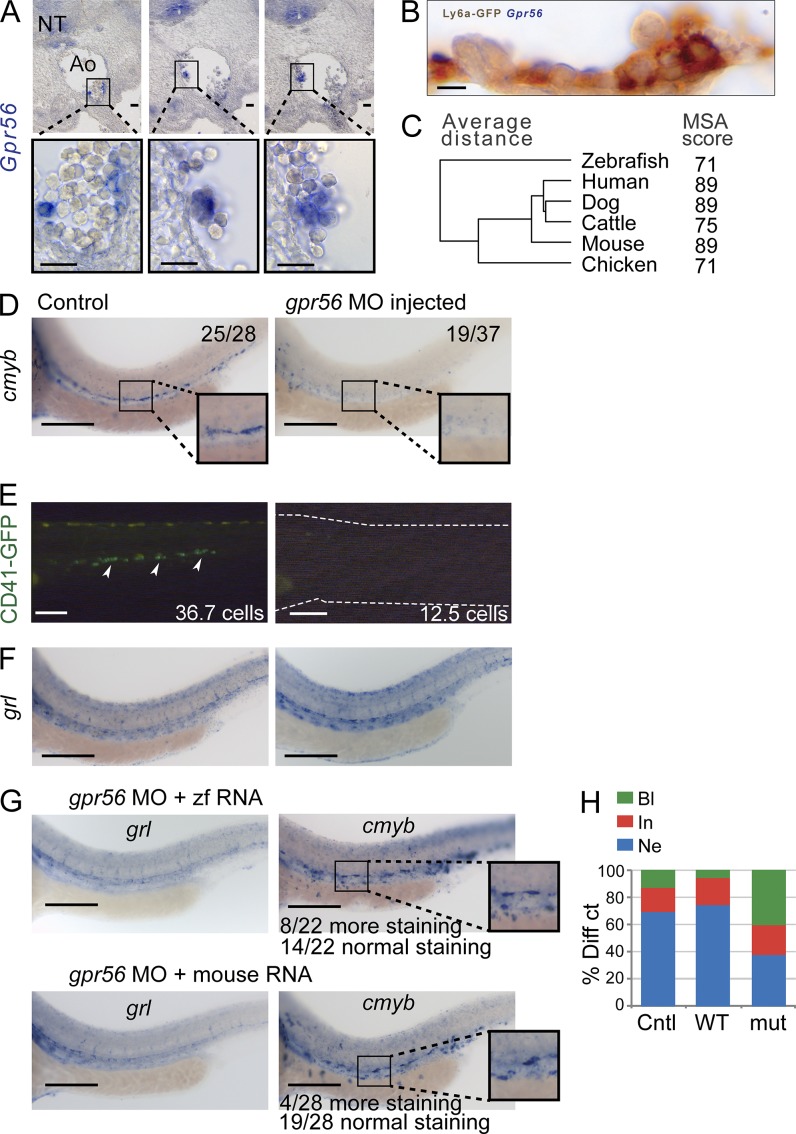
To confirm localized expression of Gpr56 in cells undergoing EHT, we performed in situ hybridization (ISH) analysis of the E10.5 AGM. High-level Gpr56 expression was observed in some aortic HCs (Fig. 6 A), and no/low expression was observed in aortic ECs. An overlap of Gpr56 expression with some GFP+ HCs was found by ISH of Ly6aGFP E10.5 AGM sections (Fig. 6 B). Thus, the localized expression of Gpr56 is consistent with FPKM values derived from RNAseq datasets and strongly suggests a role in HSC generation.
Figure 6.
In silico and in vivo analysis of Gpr56. (A) ISH of WT mouse E10.5 AGM sections shows specific expression of Gpr56 in some HCs, a few cells lining the aorta (Ao), and the notochord (NT). The top images show low magnification of AGM cross-section, and the bottom images show high magnification of the boxed areas. (B) ISH of E10.5 Ly6aGFP AGM shows coexpression of GFP and Gpr56 in some HCs. (C) Homology relationships of the Gpr56 coding sequence of different vertebrate species. (D–F) Analysis of WT and gpr56 MO zebrafish for the presence of HSCs. (D) ISH with the HSC marker cmyb at 30 hpf. (E) Fluorescent analysis of WT and MO-injected CD41:GFP transgenic embryos at 48 hpf. Numbers in the panels indicate the number of embryos with the depicted phenotype. Arrowheads (left) indicate CD41-expressing HCs in the aorta. The dashed lines (right) indicate the outline of the morphant zebrafish embryo for orientation purposes. (F) ISH with arterial cell marker grl. No vascular or developmental abnormalities can be observed in gpr56 morphant embryos. (G) HSC rescue of gpr56 morphant zebrafish with gpr56 RNA (zebrafish and mouse) as shown by ISH for cmyb. Ectopic cmyb expression in the posterior cardinal vein is clearly visible. No vascular abnormalities can be observed by grl ISH. (D and G) Insets show boxed areas at higher magnification. Bars: (A) 30 µm; (B) 10 µm; (D–G) 100 µm. (H) Effect of human GPR56 activity in neutrophil differentiation of the 32D-CSF3R unipotent stem cell line. 32D-CSF3R cells cultured in medium containing CSF3 efficiently differentiated into neutrophils. Only constitutive active mutant GPR56 (MUT) could block differentiation. Diff ct, differential count; Cntl, empty vector control; WT, WT human GPR56 vector; mut, constitutively active human GPR56 mutant vector; Bl, blast morphology; In, intermediate morphology; Ne, neutrophil morphology.
Gpr56 is highly conserved across different vertebrate species (mean multiple sequencing alignment score = 85%; Fig. 6 C). To validate the involvement of Gpr56 in HSC generation, we used a zebrafish morpholino oligo (MO) knockdown approach. At 30 and 48 h post fertilization (hpf), morphants were assayed by ISH (30 hpf) for cmyb, a marker for emerging HSCs (Jing and Zon, 2011). WT embryos show cmyb-expressing cells along the aorta (Fig. 6 D). In contrast, cmyb-expressing cells are severely reduced in gpr56 MO–injected embryos. To validate that this was a defect in HSC generation, we injected the gpr56 MO into CD41-GFP transgenic embryos (CD41 marks HCs; Lin et al., 2005; Jing and Zon, 2011; Robin et al., 2011). The number of CD41-GFP+ cells in the caudal hematopoietic tissue at 48 hpf is significantly decreased from 36.7 ± 4.0 cells in WT to 12.5 ± 1.8 in gpr56 morphants (Fig. 6 E), suggesting that Gpr56 is important for the emergence of HSCs. No abnormalities in embryo growth or the structure of the vasculature/aorta were found by ISH for arterial endothelial marker gridlock (grl; Fig. 6 F; Zhong et al., 2000). To test whether the gpr56 MO does not show an off-target effect, we performed rescue experiments by injecting gpr56 mRNA. Gpr56 morphants could be rescued with zebrafish gpr56 mRNA, as well as with a mouse Gpr56 mRNA to yield full restoration of aortic cmyb expression (Fig. 6 G). Interestingly, some ectopic expression of cmyb is observed in the region ventral to the aorta. These data indicate that gpr56 is an essential player in the HSC generation program and that its functional domains are maintained between mouse and zebrafish. Ectopic generation of phenotypic HSCs in zebrafish by Gpr56 overexpression further highlights the unexpected function of this molecule in induction of HSC generation.
To further study the function of Gpr56 in HSCs, we used the 32D-CSF3R cell line, a unipotent mouse stem cell differentiation model in which colony-stimulating factor-3 stimulates their differentiation to neutrophils. (Fig. 6 H). When stimulated with CSF3, cells transduced with an empty vector or WT human GPR56 vector lost their blast characteristics and differentiated. However, cells transduced with constitutively active (MUT) human GPR56 resulted in an increase in blast-like cells and in significantly fewer differentiated cells (eight- to fourfold fewer), suggesting that GPR56 is also essential for the maintenance of an undifferentiated cell state.
DISCUSSION
RNA sequencing analyses of EHT and developing HSCs provide a new perspective on the molecules and processes. Whereas previous methods of transcriptome analysis have identified many of the obvious regulators of hematopoietic development, this method provides an accurate accounting of all expressed genes and also small gene expression level changes between the rare, relevant cell types. We have shown here that HECs (the precursors of the earliest emerging HSCs in the midgestation mouse aorta), as distinguished from other ECs by Ly6aGFP expression, have closely related but distinct transcriptional programs. Comparisons between ECs and HECs reveal a developing program indicative of cell migration and changing cell morphology in HECs, while they retain an angiogenic program. The up-regulation of the HSC hematopoiesis program begins in HECs, further distinguishing them from ECs. Of the 530 DEGs, important receptors and TF genes were identified, including Gpr56, which is required for HSC formation. Moreover, the heptad (hematopoietic) TFs were found to be up-regulated during EHT. These factors bind the Gpr56 enhancer and regulate its expression, thus providing a proof of principle for in silico bioinformatical predictions of the combinatory role of the heptad TFs in the emergence of HSCs during EHT. Thus, our datasets are predictive of functionally relevant EHT genes and processes.
RNAseq analysis of small numbers of physiologically relevant cells
The Ly6aGFP transgenic marker in combination with CD31 and c-Kit cell surface markers allowed the high enrichment of HSCs, HECs, and ECs isolated from the aorta at the developmental time when HSCs begin to emerge. Imaging experiments verified the correlation between these markers, the expected cell type, and localization within the AGM region. Moreover, we confirmed that functional adult-repopulating HSCs are exclusively contained within the HSC fraction (0.002% of AGM cells) and that both endothelial fractions (ECs and HECs) do not contain hematopoietic progenitors or HSCs. Thus, the Ly6aGFP marker currently allows the highest level of enrichment for HECs (Fig. S1 B) that will undergo transition to HSCs, as compared with previously used markers.
Previous comparative HSC gene expression profiling (microarray) studies identified several new regulators of AGM HSCs, but the genetic program of HECs was not examined (Mascarenhas et al., 2009; McKinney-Freeman et al., 2012). During preparation of this manuscript, a new microarray study of EHT-relevant populations was performed based on cells expressing the Runx1+23-enhancer marker (Swiers et al., 2013). Runx1+23GFP marks 68% of VE-cadherin+ (endothelial and hematopoietic) cells at E8.5, marking many HECs that are not as yet exhibiting EHT. However, Ly6aGFP expression marks only a small fraction (13–19%) of CD31+ aortic cells and is probably more specifically marking the active HECs at E10.5.
Given the limited number of cells in our enriched aortic EC, HEC, and emerging HSC fractions, RNA sequencing provides the most efficient and sensitive method for analysis of EHT-relevant cells. Only 4–14 embryos (aortas) were used per sequencing experiment to isolate sufficient quantities of total RNA from sorted cell populations. With as few as 593 sorted cells, we successfully applied RNAseq technology with the SMARTER protocol, recently shown to be the best RNAseq method for low numbers of cells (Bhargava et al., 2014). Additionally, RNAseq analysis has the great advantage over microarrays in not only providing the whole transcriptome, but also revealing isoform-specific transcripts in the sequenced samples. For example, Gpr56 expresses two transcript variants. We found variant 1 (GenBank accession no. NM_001198894) of Gpr56 to be expressed exclusively in HSCs, whereas variant 2 (GenBank accession no. NM_018882) was expressed in ECs, HECs, and HSCs. Variant 2 was more highly represented in HSCs (FPKM = 323) as compared with variant 1 (FPKM = 52).
Identification of processes involved in EHT
For the first time, datasets from aortic ECs, HECs, and HSCs provide an overview of the general processes involved during EHT. Quantitative levels of gene expression between EHT-enriched cell fractions show only a small number of significant DEGs: 139 between HECs versus ECs and 340 between HSCs versus HECs. Not surprisingly, the genes with high expression in midgestation aortic ECs are mainly those involved in “general developmental processes.” These and other GO categories related to cell migration and focal adhesion are highly represented in HECs, highlighting the fact that HECs must change their adhesive properties to bulge out of endothelial lining of the aorta, undergo morphological changes as they become HCs, adhere to other HCs within the clusters, and take on hematopoietic identity and function. GO analysis of DEGs with the highest expression in HSCs shows enrichment of “hematopoiesis” and “positive regulation of histone methylation” terms.
HECs are a transcriptionally dynamic cell type at the interface of EHT. Concurrent with the initiation of the hematopoietic program and HC formation, hematopoietic genes become activated in HECs, whereas endothelium-specific cell adhesion molecules and TFs are down-regulated in HSCs. Our RNAseq data are in agreement with the recent single-cell high-throughput qPCR analysis results for 18 known endothelial and hematopoietic genes during EHT (Fig. S2 D; Swiers et al., 2013). We also identified several genes involved in angiogenesis by selection for GO term “receptor activity” in HEC versus EC comparisons (Fig. 4 A). These include Plxnd1, Eltd1, Calcrl, Ramp2, and S1pr1; Plxnd1 and Eltd1 are both induced by VEGF (Kim et al., 2011; Masiero et al., 2013). Calcrl, a GPCR, induces angiogenesis upon association with Ramp2 and Kdr/Vegf-r2, both of which are significantly induced in HECs (Guidolin et al., 2008). Collectively, these findings suggest a role for angiogenesis-related receptors in activation of hematopoietic potential and generation of HECs.
GPR56: a novel EHT regulator
Gpr56, one of the top hits in our HSC versus HEC comparison (30-fold increase) and bound by all heptad TFs, is indeed a novel regulator for emerging HSCs in the embryonic vasculature. Contrary to expectations raised by the lack of HSC defects in mouse Gpr56 KO embryos (generated by deletion of the first two exons [Saito et al. 2013]), our RNAseq data suggested a strong role for Gpr56 in emergence of HSCs. In the E10.5 mouse aorta, we localized Gpr56 expression to a few HCs/HSCs (Ly6aGFP+). Upon gpr56 knockdown, zebrafish embryos showed severe reduction in HSCs (cmyb) and CD41+ hematopoietic stem/progenitor cells, revealing a requirement for Gpr56 in HSC generation. Our rescue experiments in gpr56 morphants show that both zebrafish and mouse Gpr56 RNA can restore aortic hematopoietic stem/progenitor generation. Moreover, Gpr56 overexpression resulted in ectopic hematopoietic progenitor/stem cell formation in the axial vein, suggesting that the Gpr56 signaling axis may be useful for inducing new HSCs.
We propose that the lack of embryonic lethality in Gpr56 KO embryos could be the result of redundancy by other GPCRs or residual Gpr56 activity in the mouse transgenic model. Our RNAseq and RT-qPCR validation (Fig. S2 E) data show an increase in the expression of Gpr114 (77 kb upstream of Gpr56) and Gpr97 (48 kb downstream) during EHT. The ligand binding N-terminal part of Gpr114 has 47% amino acid similarity (and 27% identity) with Gpr56. Gpr114 is present only in mammals. Also, assays testing a human Gpr56 variant missing a large part of the second exon and the complete third exon showed that it partially retains the ability to activate SRE, E2F, NFAT, and iNOS promoters (Kim et al., 2010). Thus, Gpr56 is an unexpected novel EHT regulator essential for HSC generation and maintenance, and its function is conserved between mouse and zebrafish.
How Gpr56 acts in HECs as they transdifferentiate to HSCs is unknown, but it could affect physical properties such as adherence, cluster formation, signal transduction, migration, and/or self-renewal. Some of these features are consistent with findings in neuronal stem cells, BM HSCs, and leukemic cells, in which it has been proposed that Gpr56 functions in cell adhesion, migration, and/or repression of apoptosis (Iguchi et al., 2008; Saito et al., 2013). We found that Gpr56 functions in the maintenance of the undifferentiated state of a unipotential HSC line. The conservation of Gpr56 across species will allow for future high-throughput study of the mechanism by which Gpr56 affects EHT and generation of HSCs.
Our results on heptad TF binding to the Gpr56 enhancer suggest that other heptad targets in the overlapping list are likely to be relevant in EHT. However, not all genes that we identified as highly up-regulated during EHT are targets of the heptad complex, for example cKit. Because EHT regulation is likely to be multilayered, we are using our whole transcriptome dataset as a resource to identify other candidate transcriptional hubs.
In summary, novel and known EC, HEC, and HSC genes were identified in our RNAseq datasets. These comparative quantitative data have high predictive value for identifying functionally important molecules that direct the cellular processes involved in EHT and could instruct methods for de novo HSC generation either by direct somatic cell conversion or pluripotent stem cell differentiation.
MATERIALS AND METHODS
Cell preparation and flow cytometry.
Ly6aGFP and WT mouse embryos were dissected as described previously (Robin and Dzierzak, 2010), and single cells were prepared by collagenase treatment (0.125%, 45 min, 37°C) and washed with PBS, 10% heat-inactivated FCS, and 1% penicillin/streptomycin (PS). Cells were stained with RαMCD31-AF647 (1:400; BioLegend) and RαMcKit-PE (1:1,200; BD) for 30 min at 4°C, washed with PBS/10% FCS/1% PS, and analyzed/sorted on a FACSAria III or SORP FACSAria II (BD).
Mouse embryo immunostaining and imaging.
10-µm cryosections were prepared as described in Ling et al. (2004; except last dehydration steps were omitted), stained with RαMCD34-biotin (1:100; BD) and Streptavidin-Cy5 (1:500; Jackson ImmunoResearch Laboratories, Inc.), and imaged on an SP5 confocal microscope (Leica).
Whole-mount embryos were prepared as described previously (Yokomizo et al., 2012), stained with RαM-cKit (1:500; eBioscience) and αRat–Alexa Fluor 647 (1:5,000; Invitrogen), RαMCD31-biotin (1:500; BD) and Streptavidin–Alexa Fluor 555 (1:500; Invitrogen), RαGFP (1:2,000; MBL) and GαR–Alexa Fluor 488 (1:1,000; Invitrogen), and imaged on an SP5 microscope. 1.48 µm between stacks; 17 stacks (23.7 µm) merged.
For ISH, embryos (36 sp) were fixed and rotated overnight, 4°C in 4% paraformaldehyde, washed three times in PBS at 4°C, and embedded in paraffin, followed by overnight ethanol dehydration and two xylene washes using a Histokinette (Microm HMP110). 10-µm sections were obtained using a microtome. For Gpr56 cRNA probes, the mouse coding sequence (942 bp) was primed from BM cDNA with FW 5′-TTGCAGCAGCTTAGCAGGTA-3′ and RV 5′-GATAGCCGGGCACATAGGTA-3′ oligos, and the fragment was ligated to pGEM-T Easy (Promega) and linearized before sense and α-sense probes synthesis: overnight at room temperature with DIG-dUTP mix (Roche) and SP6/T7 polymerases (Roche). Hybridization was performed as described previously (Ciau-Uitz et al., 2000), without incubation in H2O2. After developing color for several days, sections were washed five times for 30 min in PBS-0.1% Tween (Tw) and mounted in Kaiser’s Glycerol gelatin (Merck). Ly6aGFP sections were incubated overnight at room temperature with rabbit polyclonal anti-GFP (1:1,000; Abcam) in PBSBlock (1% BSA, 0.05% Tw), washed in PBS+ (0.05% Tw), and incubated for 30 min at room temperature with polyclonal biotinylated GantiRIg (1:400; Dako) in PBSBlock, washed, incubated for 30 min at room temperature with Streptavidin-HRP (PK-7100; Vector Laboratories), washed, and color developed at room temperature in the dark by 6-min incubation in 4 ml of 5% (wt/vol) diaminobenzidine (Fluka) and 200 ml PBS. 70 µl of 35% H2O2 was added to start the reaction. Slides were rinsed with tap water, mounted, and imaged on a BX40F4 microscope and Colorview IIIu camera (Olympus). Sense control probes showed no signal.
Hematopoietic assays.
Sorted cells were plated in triplicate in methylcellulose (MethoCult GF; STEMCELL Technologies) with 1% PS and incubated at 37°C, 5% CO2 for 12 d. Hematopoietic colony types were distinguished by morphology and counted with an inverted microscope.
For all transplantations, 9-Gy irradiated (split dose) C57BL/6 female recipients were used. 2 × 105 C57BL/6 spleen cells were coinjected with the sorted cell samples. Chimerism in hematopoietic tissues was assessed by semi-qPCR for the GFP transgene (eGFP FW 5′-AAACGGCCACAAGTTCAGCG-3′ and RV 5′-GGCGGATCTTGAAGTTCACC-3′), normalized to myogenin (Myo FW 5′-TTACGTCCATCGTGGACAGC-3′ and RV 5′-TGGGCTGGGTGTTTAGTCTTA-3′). Control mixes of Ly6aGFP and WT DNA were used to make a standard curve, and the trend line formula was used to calculate the percentage of reconstitution of each sample. Peripheral blood cell donor chimerism was assayed at 1 and 4 mo after injection, and mice were sacrificed for analysis of donor chimerism in all hematopoietic tissues. Recipients considered reconstituted are ≥10% donor chimerism positive. All experiments have been conducted according to Dutch law and have been approved by the animal experiments committee (Stichting DEC consult, Dier Experimenten commissie, protocol numbers 138-11-01 and 138-12-13).
RNA isolation.
Cells (see Table S1) were directly sorted into PBS/10% FCS/1% PS and centrifuged, and supernatant was removed. Cells were lysed, and RNA was isolated using the mirVana miRNA Isolation kit (Ambion) according to the manufacturer’s protocol. RNA quality and quantity were measured by the 2100 Bioanalyzer (Agilent Technologies).
mRNA sequencing analysis.
RNA samples (Table S1) were prepared by SMARTer protocol. Illumina TrueSeq v2 protocol was used on HiSeq2000 with single read of 36 bp + 7 bp index. Reads were aligned to the mouse genome (NCBI37/mm9) using Tophat/Bowtie and mapped to the mouse genome (NCBI37/mm9), and the generated count table was analyzed by R/Bioconductor package edgeR according to McCarthy et al. (2012). Counts were normalized for mRNA abundance, and differential expression analysis was performed using edgeR (Fig. 2 A). B-H method was used for p-value correction with an FDR of 0.05 as statistically significant. Variance stabilized counts were calculated by R/Bioconductor package “DESeq” for all the genes (Anders and Huber, 2010). Heat maps were generated from the log-scaled variance stabilized counts of DEGs. GSEA was performed using the preranked option in combination with log fold change values of each comparison calculated by edgeR. Cufflinks was used to compute transcript abundance estimates in FPKMs (Trapnell et al., 2013). For DEGs, the FPKMs for each gene across all samples were normalized by division with maximum FPKM observed for that gene. Patterns were generated based on normalized FPKM, with expression levels lower than 1/3 assigned as low (L), between 1/3 and 2/3 as intermediate (I), and more than 2/3 as high (H). Patterns were then categorized as ECs, HECs, or HSCs. Genes corresponding to EC, HEC or HSC patterns were separately used for GO, KEGG, and Phenotype ontology enrichment analysis using the WebGestalt web application (Wang et al., 2013). GO terms were summarized using the REVIGO tool (Supek et al., 2011).
Cell lines.
CHOK3 cells transfected with an expression vector for the mouse SCF gene were grown initially in DMEM (Gibco) until they became confluent. They were then grown in Stem Cell Pro media (Gibco) supplemented with 1% PS, l-glutamine, and 0.5% FBS. HPC-7 mouse hematopoietic progenitor cells were grown in IMDM (Gibco) supplemented with 10% CHOK3 conditioned media, 1% PS, 10% FBS, and 1.5 × 10−4 monothioglycerol. See Knezevic et al. (2011) and Wilson et al. (2010) for details.
ChIP.
ChIP assays were performed in HPC-7 cells. 2 × 107 cells per antibody were treated with 0.4% formaldehyde, and cross-linked chromatin was sonicated to fragments of 300–500 bp. Cross-linked, sonicated chromatin was distributed evenly for immunoprecipitation. SYBR Green RT-PCR was performed on a Stratagene Mx3000p and analyzed using the MxPro software. Relative enrichment levels were calculated by normalizing results to the IgG control. For details see Knezevic et al. (2011). CD34 and HPC7 ChIPseq data were downloaded from the BloodChIP database (Beck et al., 2013; Chacon et al., 2014) and from Wilson et al. (2010).
Luciferase and LacZ assays.
Transfection was performed by electroporation of 5–10 × 106 cells with 10 µg vector DNA using a GenePulser Excell (Bio-Rad Laboratories). The luciferase assay was performed using a modified version of the Dual-Luciferase Reporter Assay System (Promega). Transient transfections were cotransfected with the pEFBOS-LacZ vector, and luciferase data were normalized to the lacZ data. For stable transfection assays, cells were cotransfected with pGK Neo and resistant cells were used for luciferase assays as described in Knezevic et al. (2011).
Transactivation.
Cos7 cells were cultured in 6-well plates (5 × 105 cells/well) overnight. Runx1.pcDNA3, CBFb.pcDNA3, Fli1.pcDNA3, and Gata2.pMSCV-PIG or empty vectors were transfected along with the pEFBOS-lacZ control vector and the Gpr56-37.pGL2prom enhancer construct in varying combinations (0.5 µg DNA/well) using the ProFection Mammalian Transfection system (Promega). After 48 h, luciferase and lacZ assays were performed as detailed in Knezevic et al. (2011).
Zebrafish.
Zebrafish (Danio rerio) embryos were raised at 28.5°C (Westerfield, 1995). Heterozygous -6.0itga2b:EGFP embryos (CD41-GFP; Lin et al., 2005) were maintained by crosses with WT zebrafish. For ISH 0.003% 1-phenyl-2-thiourea (PTU)–treated embryos (30 hpf) were fixed over night with 4% PFA in PBS containing 3% sucrose, transferred to MeOH, hybridized with a cmyb probe (gift of R. Patient, University of Oxford, Oxford, England, UK) according to Chocron et al. (2007), and imaged with BX40 (Olympus) and AX10 (Carl Zeiss) fluorescent microscopes with an AxioCam MRm camera (Carl Zeiss).
Human GPR56 expression constructs and differentiation of 32D/G-CSF-R cells.
WT hGPR56 and truncated constitutively active hGPR56 (as described by Paavola et al., 2011) were amplified, and a Kozak sequence and two N-terminal flag tags were added. The WT and constitutively active hGPR56 (wtGPR56/caGPR56) were subcloned into pEGFPN1 to generate an eGFP fusion protein. The wt/caGPR56-eGFP fusion inserts were PCR amplified and cloned into the pLNCX2 retroviral vector (Takara Bio Inc.).
The 32D/G-CSF receptor (32D/G-CSF-R) cell line was cultured at a density <106 cells/ml in RPMI 1640 medium (Life Technologies), supplemented with 1% PS, 10% FCS, and 10 ng/ml mouse IL-3 (CHO conditioned). 106 cells were transduced (RetroNectin; Takara Bio Inc.) with pLNCX2-EGFP, pLNCX2-wtGPR56-EGFP, or pLNCX2-caGPR56-EGFP and selected in 0.8 mg/ml G418 (Life Technologies) 2 d after infection (efficiency 100% at day 4). For the differentiation assay, cells were washed twice in HBSS (Life Technologies), cultured in the same medium, except mIL3 was replaced with 10 ng/ml human CSF3 (Amgen), and placed in 6-well plates (2 × 105 cells/ml). Cell density was adjusted to 2 × 105 cells/ml on a daily basis. Morphology was determined by microscopy on May-Grünwald-Giemsa–stained cytospins at day 7 and scored based on phenotype (blast, intermediate, neutrophil, >100 cells).
Primers.
For cloning: Gpr56-37F, 5′-GAGGATCCTCCATGAGGGACATCTTCAA-3′; Gpr56-37R, 5′-AGTCGACACGGGCTTATCACGAGAAAT-3′. For ChIP-qPCR: Gpr56-37F, 5′-AATGTTATCAACCGTCTGC-3′; Gpr56-37R, 5′-CCTCACCTAATCAAGATATGTC-3′. For RT-qPCR validations: Gpr56 FW, 5′-GCAGAACACCAAAGTCACCA-3′; Gpr56 RV, 5′-TGTCTCTGCTCACTGTCTCG-3′; Gpr97 FW, 5′-CTGGGATATGGCTAAAGGAGAC-3′; Gpr97 RV, 5′-AAGGCGAAGAAGGTCAAGTG-3′; Gpr114 FW, 5′-TCACTGCTCAATAACTATGTCC-3′; Gpr114 RV, 5′-ACTGTATACCCTTCCAGACTC-3′; Ikzf2 FW, 5′-AGCCCTTCAAATGTCCTTTCTG-3′; Ikzf2 RV, 5′-CAGCGTTCCTTGTGTTCCTC-3′; Meis1 FW, 5′-CATCTTTCCCAAAGTAGCCAC-3′; Meis1 RV, 5′-GTAAGTCCTGTATCTTGTGCC-3′; Mpl FW, 5′-TTGGACTTCAGTGCTTTACCT-3′; Mpl RV, 5′-CTCCTCTTCACATTTCTCCCA-3′; Mycn FW, 5′-GGAGAGGATACCTTGAGCGA-3′; Mycn RV, 5′-GGTTACCGCCTTGTTGTTAGAG-3′; Gata2 FW, 5′-CACCCCTAAGCAGAGAAGCAA-3′; Gata2 RV, 5′-TGGCACCACAGTTGACACACT-3′; Gata3 FW, 5′-TGTGGGCTGTACTACAAGCT-3′; Gata3 RV, 5′-TCGATTTGCTAGACATCTTCCG-3′; Runx1 FW, 5′-CAGGTAGCGAGATTCAACGA-3′; Runx1 RV, 5′-TTTGATGGCTCTATGGTAGGTG-3′; Hlf FW, 5′-CGCAAAGTCTTCATTCCCGA-3′; Hlf RV, 5′-GCTCCTTCCTTAAATCAGCCA-3′; Gapdh FW, 5′-GACTTCAACAGCAACTCCCA-3′; Gapdh RV, 5′-GCCGTATTCATTGTCATACCAG-3′.
Online supplemental material.
Fig. S1 provides FACS characterization of CD31+ AGM cells and a representative multilineage repopulation analysis of transplanted mice. Further details of RNAseq analyses and a comparison with the published single-cell qPCR analysis of EHT cells (Swiers et al. 2013) and adult BM HSCs (Riddell et al. 2014) is provided in Fig. S2. Table S1 contains details of the material used for RNAseq analysis. Table S2 contains the 530 DEGs and their expression patterns and groupings. Ontology enrichment analysis results for each group are provided in Table S3. Table S4 contains a list of up- and down-regulated TFs as found by differential expression analysis with edgeR. All sequencing data has been uploaded to GEO repository under accession number GSE63316. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140767/DC1.
Supplementary Material
Acknowledgments
We thank all laboratory members and N. Speck for stimulating discussions, P. Kaimakis for technical assistance, R. Patient for (cmyb and grl) zebrafish ISH probes, and the Experimental Animal Center of Erasmus MC.
This study was supported by a National Institutes of Health grant (R37 DK054077), ZonMw (Netherlands Scientific Research Organization) equipment grant (91109036), ZonMw TOP Award (40-00812-98-11068), NIRM (Netherlands Institute for Regenerative Medicine) FES award, Erasmus MC PhD grant, and Landsteiner Society for Blood Research grant (0407).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AGM
- aorta-gonad-mesonephros
- BCV
- biological coefficient of variation
- ChIP
- chromatin immunoprecipitation
- DEG
- differentially expressed gene
- EC
- endothelial cell
- EHT
- endothelial to hematopoietic cell transition
- FDR
- false discovery rate
- HC
- hematopoietic cell
- HEC
- hemogenic EC
- hpf
- hour post fertilization
- HSC
- hematopoietic stem cell
- ISH
- in situ hybridization
- MO
- morpholino oligo
- qPCR
- quantitative PCR
- TF
- transcription factor
References
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D., Thoms J.A., Perera D., Schütte J., Unnikrishnan A., Knezevic K., Kinston S.J., Wilson N.K., O’Brien T.A., Göttgens B., et al. . 2013. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 122:e12–e22 10.1182/blood-2013-03-490425 [DOI] [PubMed] [Google Scholar]
- Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y.R., and Traver D.. 2010. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 464:108–111 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava V., Head S.R., Ordoukhanian P., Mercola M., and Subramaniam S.. 2014. Technical variations in low-input RNA-seq methodologies. Sci. Rep. 4:3678 10.1038/srep03678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., and Robin C.. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 464:116–120 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Chacon D., Beck D., Perera D., Wong J.W., and Pimanda J.E.. 2014. BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res. 42:D172–D177 10.1093/nar/gkt1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., and Speck N.A.. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 457:887–891 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocron S., Verhoeven M.C., Rentzsch F., Hammerschmidt M., and Bakkers J.. 2007. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev. Biol. 305:577–588 10.1016/j.ydbio.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A., Walmsley M., and Patient R.. 2000. Distinct origins of adult and embryonic blood in Xenopus. Cell. 102:787–796 10.1016/S0092-8674(00)00067-2 [DOI] [PubMed] [Google Scholar]
- de Bruijn M.F., Speck N.A., Peeters M.C., and Dzierzak E.. 2000. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19:2465–2474 10.1093/emboj/19.11.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M.F.T.R., Ma X., Robin C., Ottersbach K., Sanchez M.J., and Dzierzak E.. 2002. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 16:673–683 10.1016/S1074-7613(02)00313-8 [DOI] [PubMed] [Google Scholar]
- Garcia-Porrero J.A., Godin I.E., and Dieterlen-Lièvre F.. 1995. Potential intraembryonic hemogenic sites at pre-liver stages in the mouse. Anat. Embryol. (Berl.). 192:425–435 10.1007/BF00240375 [DOI] [PubMed] [Google Scholar]
- Guidolin D., Albertin G., Spinazzi R., Sorato E., Mascarin A., Cavallo D., Antonello M., and Ribatti D.. 2008. Adrenomedullin stimulates angiogenic response in cultured human vascular endothelial cells: involvement of the vascular endothelial growth factor receptor 2. Peptides. 29:2013–2023 10.1016/j.peptides.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Iguchi T., Sakata K., Yoshizaki K., Tago K., Mizuno N., and Itoh H.. 2008. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a Gα12/13 and Rho pathway. J. Biol. Chem. 283:14469–14478 10.1074/jbc.M708919200 [DOI] [PubMed] [Google Scholar]
- Imanirad P., Solaimani Kartalaei P., Crisan M., Vink C., Yamada-Inagawa T., de Pater E., Kurek D., Kaimakis P., van der Linden R., Speck N., and Dzierzak E.. 2014. HIF1α is a regulator of hematopoietic progenitor and stem cell development in hypoxic sites of the mouse embryo. Stem Cell Res. (Amst.). 12:24–35 10.1016/j.scr.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A., and Dieterlen-Lièvre F.. 1998. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 125:4575–4583. [DOI] [PubMed] [Google Scholar]
- Jing L., and Zon L.I.. 2011. Zebrafish as a model for normal and malignant hematopoiesis. Dis. Model. Mech. 4:433–438 10.1242/dmm.006791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.E., Han J.M., Park C.R., Shin K.J., Ahn C., Seong J.Y., and Hwang J.I.. 2010. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J. Cancer Res. Clin. Oncol. 136:47–53 10.1007/s00432-009-0635-z [DOI] [PubMed] [Google Scholar]
- Kim J., Oh W.J., Gaiano N., Yoshida Y., and Gu C.. 2011. Semaphorin 3E-Plexin-D1 signaling regulates VEGF function in developmental angiogenesis via a feedback mechanism. Genes Dev. 25:1399–1411 10.1101/gad.2042011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K., and Herbomel P.. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 464:112–115 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Knezevic K., Bee T., Wilson N.K., Janes M.E., Kinston S., Polderdijk S., Kolb-Kokocinski A., Ottersbach K., Pencovich N., Groner Y., et al. . 2011. A Runx1-Smad6 rheostat controls Runx1 activity during embryonic hematopoiesis. Mol. Cell. Biol. 31:2817–2826 10.1128/MCB.01305-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K., Chiba S., Kunisato A., Sata M., Saito T., Nakagami-Yamaguchi E., Yamaguchi T., Masuda S., Shimizu K., Takahashi T., et al. . 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 18:699–711 10.1016/S1074-7613(03)00117-1 [DOI] [PubMed] [Google Scholar]
- Lin H.F., Traver D., Zhu H., Dooley K., Paw B.H., Zon L.I., and Handin R.I.. 2005. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 106:3803–3810 10.1182/blood-2005-01-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K.W., Ottersbach K., van Hamburg J.P., Oziemlak A., Tsai F.Y., Orkin S.H., Ploemacher R., Hendriks R.W., and Dzierzak E.. 2004. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200:871–882 10.1084/jem.20031556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Robin C., Ottersbach K., and Dzierzak E.. 2002. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells. 20:514–521 10.1634/stemcells.20-6-514 [DOI] [PubMed] [Google Scholar]
- Mascarenhas M.I., Parker A., Dzierzak E., and Ottersbach K.. 2009. Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood. 114:4645–4653 10.1182/blood-2009-06-230037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero M., Simões F.C., Han H.D., Snell C., Peterkin T., Bridges E., Mangala L.S., Wu S.Y.Y., Pradeep S., Li D., et al. . 2013. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell. 24:229–241 10.1016/j.ccr.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., and Smyth G.K.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman S., Cahan P., Li H., Lacadie S.A., Huang H.T., Curran M., Loewer S., Naveiras O., Kathrein K.L., Konantz M., et al. . 2012. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 11:701–714 10.1016/j.stem.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A., and Dzierzak E.. 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 86:897–906 10.1016/S0092-8674(00)80165-8 [DOI] [PubMed] [Google Scholar]
- Müller A.M., Medvinsky A., Strouboulis J., Grosveld F., and Dzierzak E.. 1994. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1:291–301 10.1016/1074-7613(94)90081-7 [DOI] [PubMed] [Google Scholar]
- North T., Gu T.L., Stacy T., Wang Q., Howard L., Binder M., Marín-Padilla M., and Speck N.A.. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 126:2563–2575. [DOI] [PubMed] [Google Scholar]
- North T.E., de Bruijn M.F., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., and Speck N.A.. 2002. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 16:661–672 10.1016/S1074-7613(02)00296-0 [DOI] [PubMed] [Google Scholar]
- Ody C., Vaigot P., Quéré P., Imhof B.A., and Corbel C.. 1999. Glycoprotein IIb-IIIa is expressed on avian multilineage hematopoietic progenitor cells. Blood. 93:2898–2906. [PubMed] [Google Scholar]
- Orelio C., and Dzierzak E.. 2003. Identification of 2 novel genes developmentally regulated in the mouse aorta-gonad-mesonephros region. Blood. 101:2246–2249 10.1182/blood-2002-07-2260 [DOI] [PubMed] [Google Scholar]
- Orelio C., Peeters M., Haak E., van der Horn K., and Dzierzak E.. 2009. Interleukin-1 regulates hematopoietic progenitor and stem cells in the midgestation mouse fetal liver. Haematologica. 94:462–469 10.3324/haematol.13728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola K.J., Stephenson J.R., Ritter S.L., Alter S.P., and Hall R.A.. 2011. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem. 286:28914–28921 10.1074/jbc.M111.247973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran P., Rengarajan T., Thangavel J., Nishigaki Y., Sakthisekaran D., Sethi G., and Nishigaki I.. 2013. The vascular endothelium and human diseases. Int. J. Biol. Sci. 9:1057–1069 10.7150/ijbs.7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell J., Gazit R., Garrison B.S., Guo G., Saadatpour A., Mandal P.K., Ebina W., Volchkov P., Yuan G.C., Orkin S.H., and Rossi D.J.. 2014. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 157:549–564 10.1016/j.cell.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin C., and Dzierzak E.. 2010. Preparation of hematopoietic stem and progenitor cells from the human placenta. Curr. Protoc. Stem Cell Biol. Chapter 2:Unit 2A.9 10.1002/9780470151808.sc02a09s14 [DOI] [PubMed] [Google Scholar]
- Robin C., Ottersbach K., Boisset J.C., Oziemlak A., and Dzierzak E.. 2011. CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood. 117:5088–5091 10.1182/blood-2011-01-329516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., and Weissman I.L.. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kaneda K., Suekane A., Ichihara E., Nakahata S., Yamakawa N., Nagai K., Mizuno N., Kogawa K., Miura I., et al. . 2013. Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia. 27:1637–1649 10.1038/leu.2013.75 [DOI] [PubMed] [Google Scholar]
- Sánchez M.J., Holmes A., Miles C., and Dzierzak E.. 1996. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 5:513–525 10.1016/S1074-7613(00)80267-8 [DOI] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., and Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 6:e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G., Baumann C., O’Rourke J., Giannoulatou E., Taylor S., Joshi A., Moignard V., Pina C., Bee T., Kokkaliaris K.D., et al. . 2013. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4:2924 10.1038/ncomms3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Morrison A.M., Inoue H., Gribi R., Ure J., and Medvinsky A.. 2005. Progressive divergence of definitive haematopoietic stem cells from the endothelial compartment does not depend on contact with the foetal liver. Development. 132:4179–4191 10.1242/dev.01974 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., and Pachter L.. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B., Montel-Hagen A., Sasidharan R., Nakano H., Ferrari R., Boogerd C.J., Schredelseker J., Wang Y., Hunter S., Org T., et al. . 2012. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 150:590–605 10.1016/j.cell.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Duncan D., Shi Z., and Zhang B.. 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M.1995. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). Third edition University of Oregon Press, Eugene, OR: 385 pp. [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. . 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 7:532–544 10.1016/j.stem.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Yokomizo T., and Dzierzak E.. 2010. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 137:3651–3661 10.1242/dev.051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T., Yamada-Inagawa T., Yzaguirre A.D., Chen M.J., Speck N.A., and Dzierzak E.. 2012. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 7:421–431 10.1038/nprot.2011.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui J., Chiu C.P., and Lansdorp P.M.. 1998. Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood. 91:3255–3262. [PubMed] [Google Scholar]
- Zhong T.P., Rosenberg M., Mohideen M.A.P.K., Weinstein B., and Fishman M.C.. 2000. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 287:1820–1824 10.1126/science.287.5459.1820 [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. . 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 3:625–636 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.