Significance
Auxin is an important phytohormone that regulates almost all aspects of plant growth and development. Ribosome proteins serve as translational regulators of auxin response. This work highlights the significance of lipid metabolism in the downstream of ribosome protein function to link auxin-regulated developmental patterning with endomembrane trafficking. Our work sheds light on a novel link between auxin signaling, ribosome function, and lipid metabolism.
Keywords: ribosomal mutants, lipid metabolism, vacuole trafficking, auxin response
Abstract
The vacuole is the most prominent compartment in plant cells and is important for ion and protein storage. In our effort to search for key regulators in the plant vacuole sorting pathway, ribosomal large subunit 4 (rpl4d) was identified as a translational mutant defective in both vacuole trafficking and normal development. Polysome profiling of the rpl4d mutant showed reduction in polysome-bound mRNA compared with wild-type, but no significant change in the general mRNA distribution pattern. Ribsomal profiling data indicated that genes in the lipid metabolism pathways were translationally down-regulated in the rpl4d mutant. Live imaging studies by Nile red staining suggested that both polar and nonpolar lipid accumulation was reduced in meristem tissues of rpl4d mutants. Pharmacological evidence showed that sterol and sphingolipid biosynthetic inhibitors can phenocopy the defects of the rpl4d mutant, including an altered vacuole trafficking pattern. Genetic evidence from lipid biosynthetic mutants indicates that alteration in the metabolism of either sterol or sphingolipid biosynthesis resulted in vacuole trafficking defects, similar to the rpl4d mutant. Tissue-specific complementation with key enzymes from lipid biosynthesis pathways can partially rescue both vacuole trafficking and auxin-related developmental defects in the rpl4d mutant. These results indicate that lipid metabolism modulates auxin-mediated tissue differentiation and endomembrane trafficking pathways downstream of ribosomal protein function.
The vacuole is the most prominent compartment in a plant cell. It occupies about 90% of the volume of a cell and is responsible for various unique processes, including storage of ions and metabolites, and is crucial for detoxification and general cell homeostasis (1). For soluble proteins stored in vacuole, at least two separate delivery routes exist based on the location and peptide sequence of the sorting signal. Amino-terminal propeptide (NTPP) and carboxyl-terminal propeptide (CTPP)-containing proteins are transported to the vacuole by distinct pathways that converge at the prevacuolar compartment (2). The NTPP pathway is believed to be common to plants and yeast, and several components of the machinery involved in the sorting of NTPP-type cargoes have been characterized (3, 4). The CTPP pathway is believed to be unique to plants, and different genetic approaches have identified components that are specific for that pathway (5, 6) The biogenesis of vacuoles and trafficking to this compartment is indispensable for plant viability because plants defective in either pathway result in severe developmental defects or even embryonic lethality (7–9).
Extensive studies have linked hormone regulation with vacuole trafficking (8, 10). Auxin is one of the most widely studied (11–13). Polar transport of auxin is an instrumental process for cell viability. The connection between polar auxin transport and endomembrane trafficking is established with the proper membrane targeting of auxin transporters. The asymmetric localization of auxin influx and efflux carrier proteins depends on the actin-dependent vesicle transport machinery (14, 15). Both auxin efflux and influx components PIN-FORMED (PINs) proteins and AUXIN INFLUX1 (AUX1)/LIKE AUX1(LAXs) undergo constitutive cycling between the plasma membrane and endosomes (12, 16–19). GNOM-dependent and independent pathways have been generally described based on the sensitivity to brefeldin A (BFA) treatment (20, 21). The vacuole targeting pathway of PIN proteins in response to environmental signals, such as gravity, also depends on vesicle trafficking regulators. The retromer components SORTING NEXIN1 (SNX1) and VACUOLAR PROTEIN SORTING 29 (VPS29) both act as important factors to retrieve PIN proteins from a late/prevacuolar compartment back to the recycling pathways, and thus regulate the defined rate of PIN protein translocation from prevacuolar compartment to the vacuole (22). Recent publication has elucidated the vacuole trafficking of PIN proteins depends on auxin-regulated both positive- and negative-feedback pathways (23). This auxin effects require the activity of SKP-Cullin-F-boxTIR/AFB (SCFTIR1/AFB)-dependent auxin signaling pathway (23). However, the molecular mechanism between auxin signaling and vacuole trafficking is still not clear. Intriguingly, lipid metabolism has been linked to auxin regulation with cellular trafficking (24–28).
Lipids play essential roles in the spatiotemporal regulation of membrane trafficking because of their essential function as regulators of membrane dynamics (9, 29, 30). In the endomembrane system of animal cells, glycerophospholipids, sphingomyelin, sphingolipids, and sterols are distributed in gradient concentrations among the organelles, with the endoplasmic reticulum (ER) having a low concentration of sphingolipids and cholesterol, the Golgi having an intermediate concentration, and the plasma membrane containing the highest concentration (31). The variability in lipid content within different endomembrane compartments is related to their cellular functions. The lower level of sphingolipids and cholesterol in the ER and biosynthetic recycling pathway provides a suitable environment for insertion and folding of proteins in the ER bilayer, whereas a high level of sphingolipids and cholesterol makes the plasma membrane an impermeable barrier between cytoplasm and the external environment, and makes it easier to form a curved membrane when endocytosis initiates (32, 33). On the other hand, the intermediate concentration of sterols and sphingolipids in the Golgi apparatus makes it serve as an ideal transition system between the biosynthetic and endocytic recycling pathways (33). It was shown that sterols, glucosylceramides, and unsaturation degree of lipid acyl chains are critical for the formation of lipid microdomains. The sterol structure can affect membrane curvature, which is necessary for budding or fusion events (34). Cholesterol is involved in the formation of secretory vesicles from a post-Golgi compartment (35). Other lipids, such as the phospholipids, lysophospholipids, and diacylglycerol, can also induce Golgi vesiculation because their small polar head or structure favor negative membrane curvature and nonlamellar phases (36, 37). The gradual distribution of lipids over organelles is clearly characterized in animal cells but much less in plant cells. Actually, the exact lipid composition of organelles in Arabidopsis still remain uncharacterized. However, lipid biosynthesis pathways have been elucidated in the regulation of endomembrane trafficking of PIN proteins. In Arabidopsis, sphingolipids and sterols have been shown to be involved in the regulation of PINs and AUX1 to maintain their polar subcellular localization (24, 28, 38, 39). Sterols have been implicated in trafficking of PIN2 in its membrane domain. PIN2 localization is defective in the sterol-biosynthesis mutant cyclopropylsterol isomerase 1–1(cpi1-1). In this mutant, PIN2 endocytosis and root gravitropism was compromised as the result of altered sterol composition (27). Another mutant, cotyledon vascular pattern 1(cvp1), which is a knock-out mutant of STEROL METHYLTRANSFERASE2 (SMT2), a key enzyme in the sterol biosynthetic pathway, displayed abnormal vascular cell polarity and defectiveness in cell elongation (39, 40). The results from ceramide mutants indicate that sphingolipids with very long acyl chains define a trafficking pathway with specific endomembrane compartments and polar auxin transport proteins (25). However, whether those biosynthetic enzymes of lipid pathways are direct targets of auxin regulation or other components are also involved is still not clear.
Our work focused on elucidating the mechanism that regulates the plant-specific vacuole-sorting pathway, the CtVSD sorting machinery. The ribosomal large subunit 4 (rpl4) mutant identified in our Vac2 screen (6) was of particular interest because it belongs to a totally different type of protein compared with previously identified plant trafficking determinants. RPL4 was identified as the first regulatory component controlling different vacuolar trafficking pathways and the dynamics of vacuole targeting vs. secretion, which appears to be mediated by the phytohormone auxin (24, 41, 42). Mutations in independent ribosomal proteins result in auxin-related phenotypes, and recent reports propose that the auxin specificity of the phenotypes is determined by the translational regulation of the auxin response factors (ARFs) ETTIN and MONOPTEROS via translation of short upstream ORFs (uORFs) (43, 44). Indeed, our results strongly suggest that the ribosomal complex is involved in the regulation of the vacuolar trafficking pathways by controlling the translation of uORF-containing mRNAs.
In this article, we characterize the downstream targets of RPL4 protein through RNA sequencing. Our analysis showed that lipid metabolism genes were translationally down-regulated in rpl4d mutants. Further experiments with lipid specific dye confirmed that lipid accumulation was compromised in meristem tissue of rpl4d mutants. Tissue-specific complementation of rpl4d mutants with key enzymes in lipid biosynthetic pathways can partially rescue both developmental and vacuole trafficking defects, indicating lipid metabolism is the link between both pathways and is downstream of ribosome action.
Results
rpl4d Mutant Is Defective in Lipid Pathway.
The rpl4d mutant was obtained in our previous screen for vacuole trafficking defects (Fig. 1A) in the vac2 background (6). RPL4 was identified as a general regulator of different vacuolar trafficking pathways (42). Further work showed that rpl4d mutants displayed auxin-related developmental defects through translational regulation of ARF transcription factors with uORFs (24, 42).
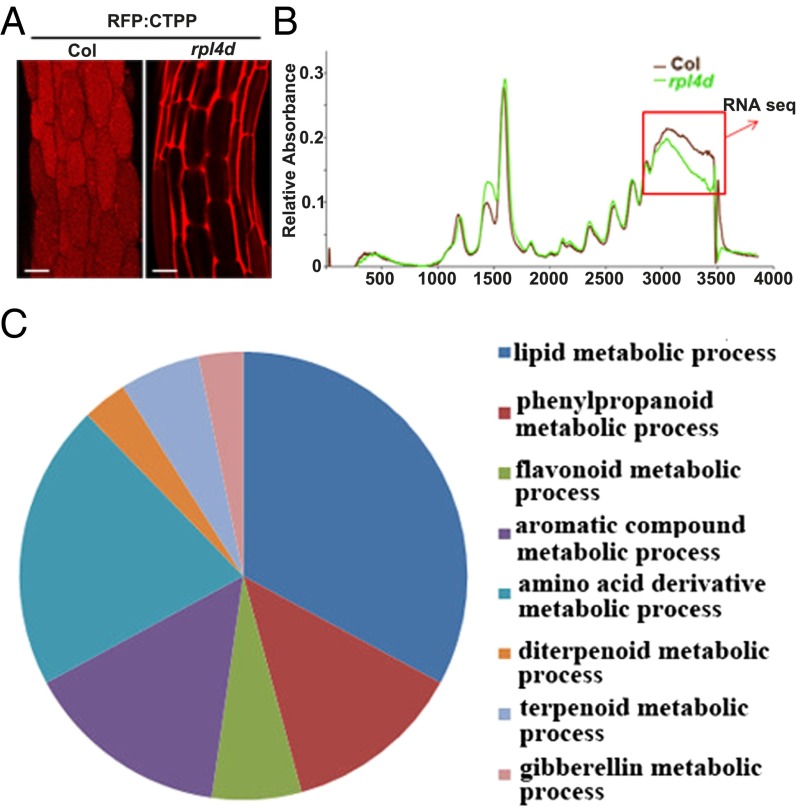
Fig. 1.
Lipid metabolism genes were translational down-regulated in rpl4d mutants. (A) Images of hypocotyl cells from RFP:CTPP lines in the background of Col or rpl4d mutant. (Scale bars, 10 μm.) (B) A254 absorption profiles of total polysomes from 7-d-old seedlings of Col and rpl4d mutants. Larger fraction numbers correspond to higher sucrose concentrations. Red frame indicates sucrose gradient corresponding to polysome bound mRNA. (C) Metabolism catalogs of pathways according to GO classification.
To understand broadly what pathways might be under the regulatory influence of the RPL4 protein and to link auxin pathways with endomembrane trafficking, we first performed polysome profiling to detect actively translated mRNAs and compared the rpl4d mutant with wild-type Col seedlings. Our results showed no general difference in polysome profiling patterns (Fig. 1B), which indicated that the secretion of the vacuole targeting signal and developmental defects were not caused by a general translational deficiency. However, there was a significant reduction of mRNA abundance in a polysome-enriched sucrose fraction (Fig. 1B, red box), suggesting that some specific targets may be responsible for both the vacuole trafficking and developmental defects in this fraction. For this scenario, RNA sequencing was performed by comparing transcript abundance between mutants and wild-type seedlings in both polysome-bound mRNA and total mRNA. The proposed candidate targets were those that were down-regulated in polysome bound mRNA but not in total mRNA. Data analysis of RNA sequencing showed that about 1,800 genes were down-regulated in polysome-bound mRNA when a set criteria P value was 0.001 and the log2 ratio was 0.5 (Dataset S1). These genes were further classified into eight metabolic pathways according to Gene Ontology (GO) catalogs (Materials and Methods and Fig. 1C). Among all of the pathways, the most enrichment of genes was in lipid metabolism, in which about 45 genes were involved in either lipid biosynthesis or desaturation (Dataset S2). Beyond lipid metabolism, we found a few genes involved in endomembrane trafficking pathways (Dataset S3), although much less abundant than in the lipid pathways.
To validate the results from RNA sequencing, real-time PCR was performed to quantify the expression level of genes down-regulated in both lipid metabolism and vesicle trafficking pathways with independent samples collected from polysome-bound mRNA compared with total mRNA (see primers in Dataset S4). About 85% of the results (Materials and Methods and Dataset S2) were consistent with RNA sequencing, indicating that these genes were translationally down-regulated in polysome-bound mRNA but remained unchanged in total mRNA. The ones that did not correlate with the RNA sequencing data either did not show reduced expression of polysome-bound RNA or the transcription in polysome-bound and total RNA was reduced (Dataset S2).
Based on the RNA sequencing results, we concluded that lipid metabolism defects could be responsible for both the vacuole trafficking and developmental defects in rpl4d mutants.
RPL4 Mutation Resulted in Reduced Lipid Accumulation in Meristem Tissue.
Because polysome profiling showed enrichment of genes in lipid metabolism, lipid profiling was first performed with plant tissues extracted from whole 7-d-old seedlings of wild-type Col and rpl4d. However, only minor reductions of polar lipid, sphingolipid, and sterol levels could be detected in rpl4d mutants (Dataset S5). We reasoned that, whereas polysome profiling is a sensitive measure of overall active translation in seedlings, the lipid profiling of seedlings was not sensitive enough to detect tissue-specific changes in lipid accumulation in the rpl4d mutant. To test the hypothesis, physiological experiments were performed by using the lipid dye Nile red. Nile red has been reported to be used as a general lipid dye for both polar and nonpolar lipid in different excitation ranges (45). As shown in Fig. 2A, the red channel represents polar lipids whereas the yellow channel represents nonpolar lipids. Our results showed that wild-type seedlings displayed intense Nile red staining of both polar and nonpolar lipids in the root meristem, and to a lesser extent in the elongation region, whereas rpl4d mutants had significantly reduced signal intensity in the meristematic region in particular (Fig. 2B compared with Fig. 2A, and Fig. 2G). This finding indicates that lipid accumulation was reduced in the meristematic region in rpl4d mutants, which is consistent with the polysome profiling data. However, there was not much difference in signal intensity in the root elongation zone (Fig. 2E compared with Fig. 2D, and Fig. 2H). The tissue differentiation of lipid accumulation is correlated with the RPL4 expression pattern, which is mainly expressed in active dividing meristem tissues (42). This finding indicated that lipid changes in rpl4d mutants were correlated with cell differentiation state and also provided and explained only a minor difference in lipid profiling from whole seedling extracts.
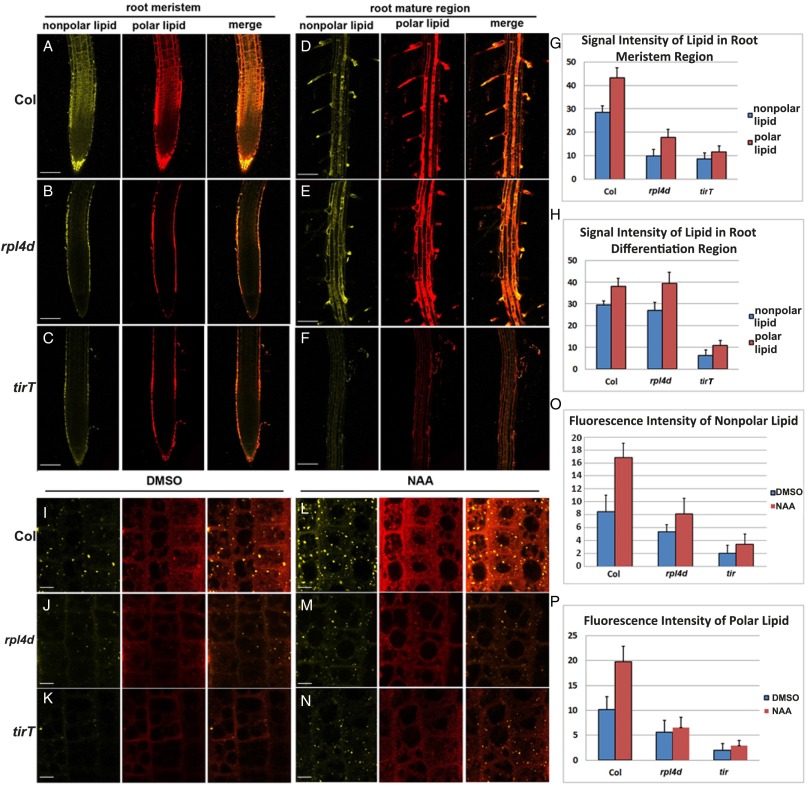
Fig. 2.
Lipid accumulation reduced in rpl4d mutants. (A–C) Nile red staining of nonpolar lipid and polar lipid in root meristem region of Col (A), rpl4d (B), and tirT mutants (C). (Scale bars, 100 μm.) (D–F) Nile red staining of polar lipid and nonpolar lipid in root mature region of Col (C), rpl4d (D), and tirT mutants (E). (Scale bars, 100 μm.) (G) Quantification of signal intensity of nonpolar and polar lipid after Nile red staining in root meristem region of Col, rpl4d, and tirT mutants. (H) Quantification of signal intensity of nonpolar and polar lipid after Nile red staining in root mature region of Col, rpl4d, and tirT mutants. (I–K) Nile red staining of nonpolar lipid and polar lipid in root transition zone of Col (I), rpl4d (J), and tirT mutants (K) after DMSO treatment. (Scale bars, 10 μm.) (L–N) Nile red staining of polar lipid and nonpolar lipid in root transition zone of Col (L), rpl4d (M), and tirT mutants (N) after 5-h NAA (100 nM) treatment. (Scale bars, 10 μm.) (O) Quantification of signal intensity of nonpolar lipid after Nile red staining in root transition zone of Col, rpl4d, and tirT mutants before and after NAA treatment. (P) Quantification of signal intensity of polar lipid after Nile red staining in root transition zone of Col, rpl4d, and tirT mutants before and after NAA treatment. For G, H, O, and P, 20 seedlings were imaged for data quantification. Error bars represent SD. Images A to F represent maximum-intensity projections of a stack of confocal optical sections. Images I to N were taken from a single scan in the middle section of the transition region in the root. For all of the figures from A to F and I to N, the Left is nonpolar lipid, Center is polar lipid, and Right is a merged image from the other two. Images represent data from at least three independent experiments.
Because our previous data showed that the rpl4d mutant was defective in an auxin-regulated developmental pathway, we hypothesized that lipid accumulation could also be affected in auxin signal transduction mutants. As shown in Fig. 2C, the auxin receptor triple-mutant tir1 afb1 afb3 (tirT) (46) also displayed significant reduction of signal intensity in both the meristem and mature root region when stained with Nile red (Fig. 2 C and F–H). Because validation of Nile red staining in the meristem tissues by mass spectrometry is difficult because of technique limitations, we applied another commonly used lipid dye, Bodipy 493/503 (47), in the same condition. Our results showed that the signal intensity of lipid droplets stained by Bodipy dye was reduced significantly in rpl4d and tirT mutants, similar to the result from Nile red staining (Fig. S1 A–C and G). This finding indicates both auxin signal transduction and the intact function of the ribosome complex are required for proper lipid accumulation in plant meristem tissue.
Next, we investigated whether lipid accumulation was in response to auxin activation. To our knowledge, there are no reports on Nile red staining in response to auxin, so we first tried 1-naphthaleneacetic acid (NAA) treatment with different time scales. As shown in Fig. S2, 2-h treatment showed no significant change in Nile red signal intensity (Fig. S2B). However, after 5-h treatment, a low concentration of NAA (100 nM) could stimulate Nile red signal in the root meristem and transition region (Fig. S2C), and signal was saturated after 24 h (Fig. S2D). Because NAA is a weak acid, to exclude the possibility that the activation of Nile red is caused by PH changes, the NAA analog 2-NAA, as well as other weak acidic phytohormones, were applied at the same concentration for 5 h. Only NAA could stimulate Nile red signal, whereas 2-NAA produced a weak signal and other hormones, such as 6-BA, JA, GA, and ABA displayed no activation effects (Fig. S3 C–F compared with Fig. S3B). Next, the same condition of NAA treatment was applied to compare lipid response in Col, rpl4d, and tirT mutants. Our results showed that only Col seedlings but not rpl4d or tirT mutants responded to auxin activation in lipid accumulation (Fig. 2 I–P). A similar result was obtained by using the other lipid dye, Bodipy 493/503, in that the signal intensity of lipid droplets was reduced dramatically in the rpl4d and tirT mutants in response to NAA treatment (Fig. S1 D–F and G). This result indicates that lipid metabolism acts downstream of the auxin signal transduction pathway and intact ribosome function is required for lipid accumulation in meristem tissue.
Taking these data together, we find that the RNA sequencing and Nile red staining results indicate that the rpl4 mutation reduced lipid metabolism in meristem tissue, which may explain the defects in vacuole trafficking and the developmental response to auxin regulation.
Lipid Biosynthesis Inhibitors Induced Vacuole Trafficking Defects Similar to Those in rpl4d Mutants.
Because Nile red staining showed a tissue-specific reduction in the meristem region of rpl4d mutants, we tried to search for direct targets downstream of RPL4 regulation for lipid metabolism. T-DNA insertion lines of genes down-regulated in lipid metabolism pathways in polysome-bound mRNA were acquired based on the hypothesis that the mutants should display developmental defects similar to rpl4d mutants. All of the genes chosen for T-DNA insertion mutants contained 5′ uORFs because our previous data showed that RPL4 regulated downstream targets in a uORF-dependent mechanism (48) (genes and T-DNA insertion lines are listed in Dataset S6). Those genes were also among the most down-regulated genes according to our real-time PCR analysis result (Dataset S2). However, no significant phenotypes were observed from any of the mutant lines. When we performed Nile red staining, those mutants also displayed a normal lipid distribution pattern similar to Col wild-type control (Fig. S4). The possible explanation for this is that none of the down-regulated genes are key enzymes in lipid metabolism and some of these genes are from multigene families. Thus, redundant genes may compensate for the defects caused by single-gene mutation.
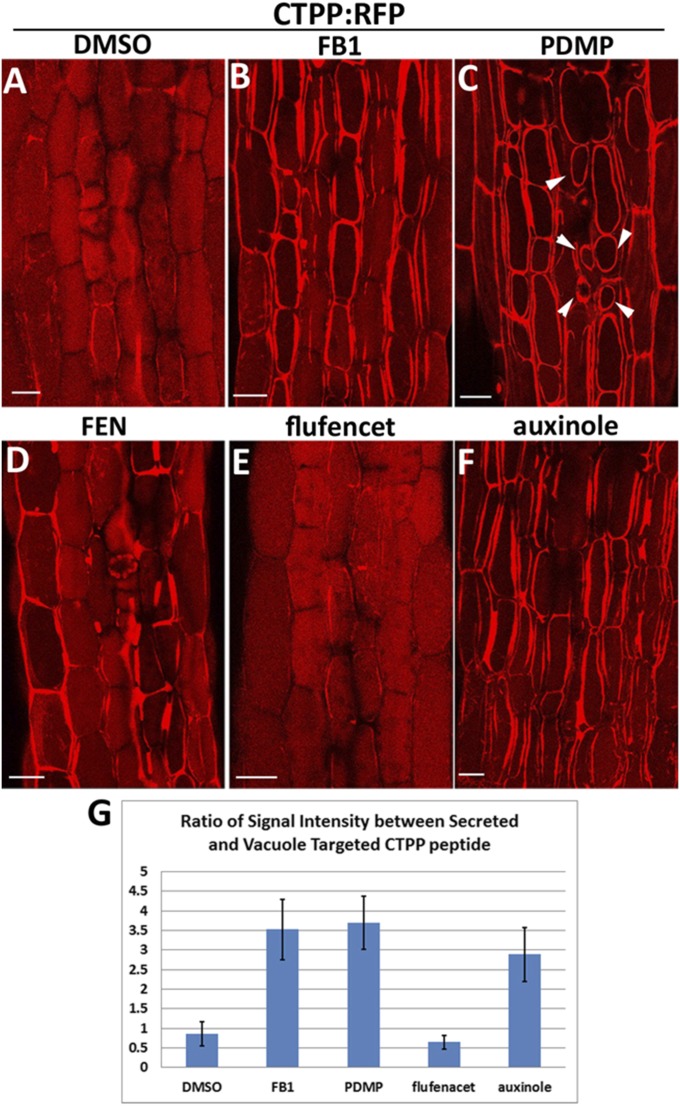
Because single T-DNA insertion mutants showed no detectable phenotypes, we tried to make use of lipid biosynthesis inhibitors. Chemical treatment has its own advantage to circumvent gene redundancy compared with genetic methods. Here we applied three inhibitors that have been published. One is sterol biosynthesis inhibitor fenpropimorph (FEN), which inhibits the C-14 sterol reductase and sterol isomerases, and may inhibit other sterol biosynthetic enzymes in Arabidopsis (49). The other inhibitors were the sphingolipid biosynthetic inhibitor fumonisin B1 (FB1) and 1-phenyl-2-decanoylamino-3- morpholino-1-propanol (PDMP). FB1 specifically inhibits the incorporation of very long-chain fatty acid (VLCFA) into sphingolipids and, thereby, mimicks the disruption of ceramide synthase CERS2 (25, 50). PDMP inhibits glucosylceramide (GluCer) specifically (51). We also applied the fatty acid synthesis inhibitor flufenacet, which specifically inhibits several 3-ketoacyl-CoA synthase and causes a reduction of saturated VLCFAs (52). Treatment of 7-d-old RFP:CTPP seedlings (53) with 100 µM FEN, 5 µM FB1,1 µM PDMP, or 1 µM flufenacet for a short time (2–3 h) did not show any effects. However, long-term treatment (24 h) with FB1 and PDMP resulted in a significant amount of CTPP peptide secreted into the apoplasmic region (Fig. 3 B and C compared with Fig. 3 A and G). PDMP treatment also induced vacuole fragmentation as reported previously (51) (Fig. 3C, arrowheads). Treatment with FEN resulted in variation of secretion phenotypes (Fig. 3D). About 40% of cells (300 cells in 10 seedlings) showed secretion of CTPP peptide, whereas the others remain in the vacuole lumen, compared with 9.5% of cells displayed secretion in control DMSO-treated seedlings. The aberrant secretion of this vacuolar cargo marker is similar to the vacuole trafficking defects in rpl4d mutants (42). However, flufenacet treatment showed no detectable effects (Fig. 3 E and G). A similar secretion phenotype could be detected when treated with auxin signal transduction inhibitor auxinole (54) for the same time period (Fig. 3 F and G). This result suggests that proper trafficking to the vacuole lumen depends on well-organized sterol and sphingolipid biosynthesis but not fatty acid synthesis, and relies on an intact auxin signal transduction pathway. The similarity of vacuole trafficking defects between lipid inhibitors, an auxin signal transduction inhibitor, and the rpl4d mutant, indicates a link between lipid biosynthesis, auxin regulation, and ribosome protein functions.
Fig. 3.
Lipid biosynthesis inhibitor induced secretion of CTPP signal peptide and altered auxin response. (A–F) Images of hypocotyl cells from RFP:CTPP lines after 24-h treatment of DMSO (0.1%), FB1 (5 μM), PDMP (1 μM), FEN (100 μM), flufenacet (1 μM), or auxinole (5 μM). (Scale bars, 10 μm.) (G) Quantification of the ratio between secreted RFP:CTPP signal and vacuole targeting signal; 300 cells from 10 seedlings for each treatment were analyzed. Error bars represent SD.
We also grew rpl4d and Col seedlings on solid medium with different lipid inhibitors or auxinole with the hypothesis that if RPL4 mutation affects certain lipid metabolism pathways, the mutant should be hypersensitive to the lipid inhibitor in that pathway. Our result showed that rpl4d mutants displayed hypersensitivity to root-length inhibition compared with Col control when grown on medium with FB1 (Fig. S5 A, B, H, and F). For FEN, the main root-length inhibition was similar but rpl4d mutants were more sensitive to agravitropism induced by the compound (Fig. S5 C, F, and I). For auxin signaling inhibitor auxinole, rpl4d displayed hypersensitivity to both the root inhibition and the gravitropic response (Fig. S5 E, F, and K). However, rpl4d showed similar sensitivity to fatty acid inhibitor flufenacet in both the root inhibition and the gravitropic response (Fig. S5 D, F, and J). This result indicates that the growth phenotype of rpl4d mutants as well as the defects in vacuole trafficking was correlated with the change of lipid metabolism.
Next, to link lipid metabolism with auxin signaling pathways, lipid inhibitors, auxinole and NAA, were applied together then lipid accumulation was detected with Nile red staining and Bodipy dye. As shown in Fig. S6, FB1, PDMP, FEN, and auxinole treatment reduced both Nile red and Bodipy staining signal in the root transition zone compared with DMSO controls. Cotreatment with either PDMP, FB1, FEN, or auxinole also dramatically inhibited the induction of lipid accumulation by NAA. However, a change of lipid accumulation by either Nile red or Bodipy staining was not observed when treated with fatty acid inhibitor flufenacet (Fig. S6). This result indicates a strong correlation between auxin signal induction and lipid accumulation, which also requires normal RPL4 protein function.
Lipid Biosynthetic Mutants Displayed Vacuole Trafficking Defects.
Because lipid metabolism genes are translationally down-regulated in rpl4d mutants and lipid accumulation was reduced in the mutant, as indicated by Nile red staining (Fig. 2), we raised the hypothesis that proper lipid biosynthesis is indispensable for vacuole trafficking and lipid biosynthetic mutants should display phenotypic defects similar to rpl4d mutants. To test the hypothesis, the recently characterized sterol biosynthetic mutant cpi1-1 was chosen for analysis (27, 28, 38). The ceramide synthase mutant loh1loh2 (25) in the sphingolipid biosynthetic pathway was also tested. The cpi1-1 mutant has been shown to reduce PIN2 polar localization in a sterol-dependent mechanism (27). The loh1loh2 mutant showed a reduction of VLCFA sphingolipid, which was associated with selective aggregation of the plasma membrane auxin transporters AUX1 and PIN1 in the cytosol (25). To detect vacuole trafficking in these mutants, an RFP:CTPP construct (53) was transformed into the mutants to produce stable transgenic lines. F3 homozygous lines were analyzed for vacuole targeting signals in hypocotyls from 7-d-old seedlings. Our results showed that similar to what has been observed in the rpl4d mutant, both sterol and sphingolipid biosynthetic mutants displayed significant aberrant secretion of RFP-labeled CTPP peptide into the apoplasmic region compared with control lines (Fig. 4B compared with Fig. 4A, Fig. 4D compared with Fig. 4C, and Fig. 4E). Based on this result, we concluded that both sterol and sphingolipid biosynthesis pathways play vital roles for proper CtVSD vacuole trafficking.
Fig. 4.
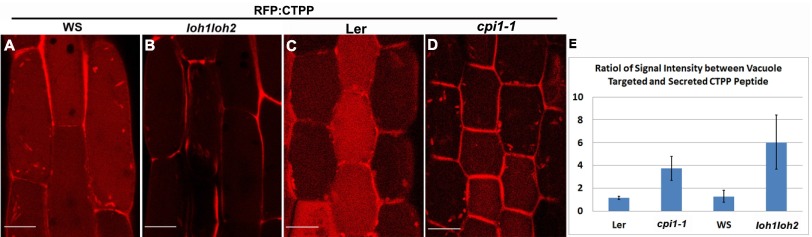
Lipid biosynthesis mutants displayed vacuole trafficking defects. (A–D) Images of hypocotyl cells from RFP:CTPP lines in WS (A), loh1loh2 (B), Ler (C), or cpi1-1 (D) background. (Scale bars, 10 μm.) (E) Quantification of the ratio between secreted RFP:CTPP signal and vacuole targeting signal; 100 cells for each treatment were analyzed. Error bars represent SD.
Lipid Biosynthesis Genes Complement Lipid Accumulation and Auxin-Related Developmental Defects.
To test the hypothesis that lipid metabolism defects are responsible for both the developmental and vacuole trafficking defects in rpl4d mutants, key enzymes from sterol, sphingolipid, and fatty acid biosynthetic pathways were cloned and expressed in the rpl4d background using the RPL4 promoter. We preferred to use the RPL4 promoter instead of the 35S constitutive promoter because the native promoter should present complementation in tissue-specific regions where RPL4 is normally expressed. For complementation, we chose LCB1 (At4g36480) as a key regulator of sphingolipid biosynthesis. LCB1 is the subunit of heterodimer serine palmitoyltransferase, which catalyzes the first step of sphingolipid biosynthesis. Mutation of LCB1 resulted in embryonic lethality (55). For the sterol biosynthesis pathway, we chose C-24 SMT2 (At1g20330), which encodes a C-24 SMT that catalyzes the reaction that distinguishes the synthesis of sterols from signaling brassinosteroid derivatives. The SMT2 mutant displayed developmental defects, including aberrant cotyledon vein patterning, serrated petals, and reduced stature. The mutants also affected auxin response, demonstrated by reduced auxin sensitivity, enhanced axr1 auxin resistance, and ectopically expressed DR5:GUS signal (40, 56). For the fatty acid pathway, we chose β-Ketoacyl-[acyl carrier protein] synthase I (KASI) (At5g46290), which catalyzes the elongation of de novo fatty acid synthesis. The T-DNA insertion mutant kas1 displayed multiple morphological defects and dramatically reduces fatty acid levels (57).
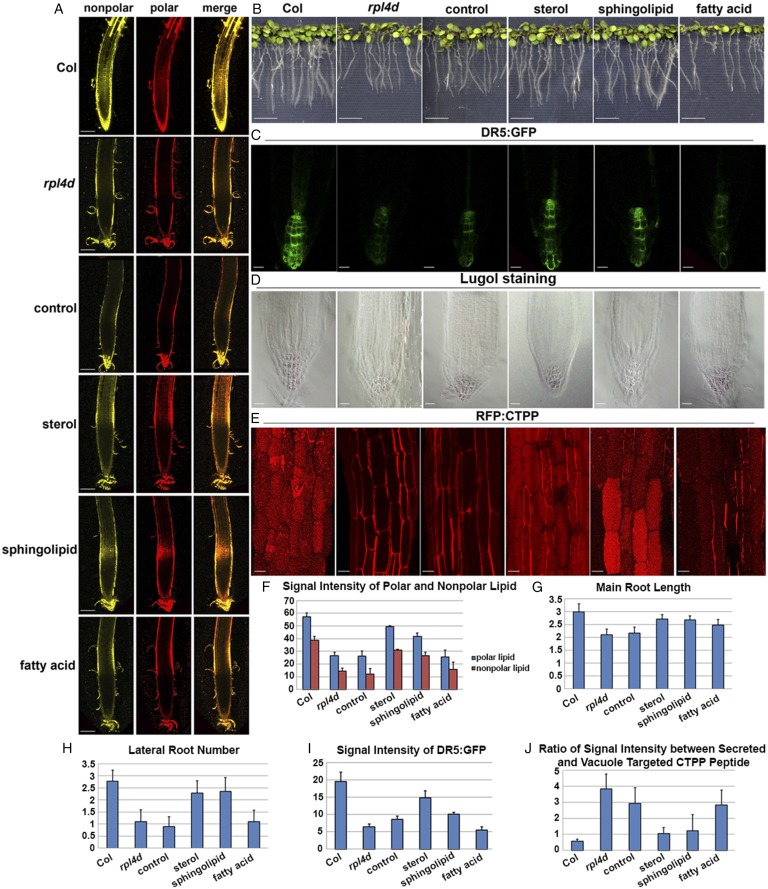
We first asked whether lipid accumulation could be rescued by introducing these lipid biosynthetic genes. As shown in Fig. 5, live imaging of Nile red staining from transgenic lines with sterol and sphingolipid biosynthetic genes, but not fatty acid synthesis genes, could partially rescue lipid accumulation in the root meristem region in rpl4d (Fig. 5 A and F). Next, developmental phenotypes and auxin response were also detected in F3 homozygous transgenic lines compared with parental lines. Consistent with the result for lipid accumulation, only sterol biosynthetic and sphingolipid biosynthetic genes, but not the fatty acid synthesis genes, in complementation lines can partially rescue the defective main root length and reduced lateral root number in rpl4d mutants (Fig. 5 B, G, and H). DR5:GFP signal distribution pattern was also examined in these transgenic lines compared with the rpl4d mutant. As shown in Fig. 5, only sterol biosynthetic and sphingolipid biosynthetic genes, but not the fatty acid synthesis genes, can partially rescue the reduced DR5 signal in rpl4d mutants (Fig. 5 C and I). Because the columella and root cap differentiation pattern is correlated with the proper distribution of auxin maxima in root meristems, lugol staining was performed with F3 homozygous lines. The lugol staining pattern was consistent with the DR5 signal pattern. Only sterol and sphingolipid biosynthesis gene complementation lines can partially rescue the mis-organized collumella cell pattern, whereas fatty acid gene complementation lines were similar to control lines (Fig. 5D). We concluded that mis-regulation of sterol and sphingolipid metabolism is responsible for the developmental defects and altered auxin response in rpl4d mutant.
Fig. 5.
Complementation with sterol and sphingolipid biosynthetic genes, but not fatty acid synthesis genes, can partially rescue both auxin-regulated developmental defects and vacuole trafficking defects in rpl4d mutants. (A) Nile red staining in root meristems of 7-d-old seedlings from Col, rpl4d mutants, or rpl4d mutants complemented with control vector, sterol, sphingolipid, or fatty acid biosynthesis genes. The yellow channel represents nonpolar lipid and the red channel represents polar lipid. Images represent maximum-intensity projections of a stack of confocal optical sections. (Scale bars, 100 μm.) (B) Phenotypes of 10-d-old seedlings on MS medium. From left to right are seedlings in the genotype of Col, rpl4d mutants, or rpl4d mutants complemented with control vector, sterol biosynthesis gene, sphingolipid, or fatty acid biosynthesis gene. (Scale bars, 1 cm.) (C) DR5:GFP signal distribution pattern in primary roots of 7-d-old seedlings. From left to right are represents of DR5:GFP signal from genotype of Col, rpl4d mutants, or rpl4d mutants complemented with vector, sterol biosynthesis gene, sphingolipid, or fatty acid biosynthesis gene. (Scale bars, 20 μm.) (D) Starch deposition pattern after lugol staining from root columella cells of 7-d-old seedlings. From left to right are represents of lugol staining pattern from genotype of Col, rpl4d mutants, or rpl4d mutants complemented with vector, sterol biosynthesis gene, sphingolipid, or fatty acid biosynthesis gene. (Scale bars, 20 μm.) (E) CTPP peptide distribution pattern from hypocotyl cells of 7-d-old seedlings. From left to right are represents of CTPP vacuole targeting phenotypes from genotype of Col, rpl4d mutants or rpl4d mutants complemented with vector, sterol biosynthesis gene, sphingolipid, or fatty acid biosynthesis gene. (Scale bars, 10 μm.) (F) Quantification of signal intensity of both nonpolar and polar lipid after Nile red staining. Twenty seedlings were imaged for data quantification. (G) Statistic record of primary root length of 10-d-old seedlings from different genotypes as is shown in C. Sixty seedlings of each genotype were measured. (H) Quantification of lateral root number of 10-d-old seedlings from different genotypes as is shown in C. Sixty seedlings of each genotype were analyzed. (I) Quantification of signal intensity of DR5 signal in roots from different genotypes as is shown in D. Thirty seedlings of each genotype were measured. (J) Quantification of the ratio between secreted RFP:CTPP signal and vacuole targeting signal from hypocotyl cells in the genotypes as is shown in E. One-hundred cells for each treatment were analyzed. For all of the figures from A to E, images represent data taken from at least three independent experiments. For quantification analysis from F to J, error bars represent SD.
Tissue Specific Complementation with Lipid Biosynthesis Genes Partially Rescued Vacuole Trafficking Defects in rpl4 Mutants.
Vacuole trafficking was also monitored in the rpl4d mutant with the same complementation lines. RFP:CTPP signal was detected in F3 homozygous transgenic lines compared with control lines. Consistent with developmental phenotypes, only sterol and sphingolipid, but not the fatty acid biosynthetic genes, can partially rescue the mis-targeting vacuole phenotypes (Fig. 5 E and J). We concluded that defects in lipid biosynthesis are responsible for both development and vacuole trafficking defects in rpl4 and play a role in regulating vacuole trafficking and auxin responses.
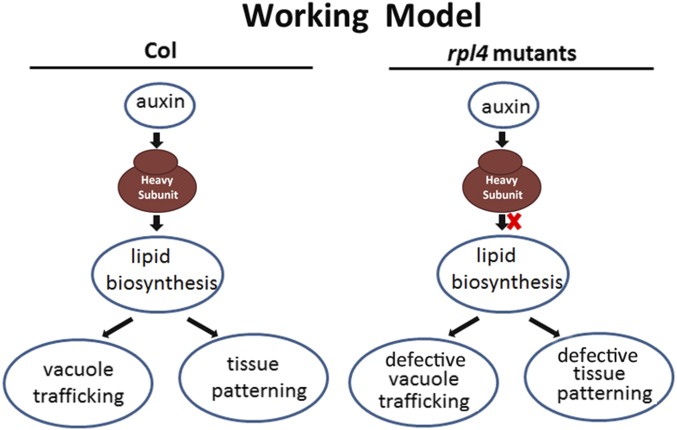
Based on these results, we propose a working model for RPL4 function. RPL4 serves as a central hub linking auxin-regulated development with endomembrane vacuole trafficking, possibly through the translational control of the downstream lipid metabolism pathway (Fig. 6). When lipid accumulation is down-regulated as the result of RPL4 protein deficiency, both the developmental process and endomembrane trafficking pathway were altered in rpl4d mutants (Fig. 6).
Fig. 6.
Working model for ribosome protein function. Ribosomal protein work as the hub to integrate auxin-regulated developmental program and vacuole trafficking through the translational regulation of lipid metabolism. However, under the condition of ribosome mutant, lipid accumulation was compromised, which result in defects of both tissue patterning and mis-localization of vacuole trafficking.
Discussion
In this report we have elucidated a novel link between auxin-regulated development with endomembrane trafficking through mediation of lipid metabolism downstream of ribosome function. Data analysis from RNA sequencing results put forward a new insight into the involvement of lipid metabolism in both auxin-mediated tissue differentiation and vacuole trafficking.
We demonstrated that auxin signaling mutants were defective in lipid accumulation and that auxin specifically stimulated lipid accumulation within 5 h. Although only future experiments using mass spectrometry can link changes in specific lipid species with auxin treatment, our results show a clear correlation between auxin activation and lipid accumulation. Our results also link auxin signal transduction with proper vacuole targeting, because treatment with the auxin signaling inhibitor auxinole resulted in aberrant secretion of CTPP peptide, which was similar to defects observed in the rpl4d mutant. Therefore, we propose that auxin-mediated endomembrane trafficking is achieved at least partially by the regulation of lipid metabolism, and ribosome functions as the hub to link the two pathways together.
Our results clearly demonstrate that lipid accumulation was reduced in meristem tissue of rpl4d mutants compared with wild type Col seedlings. The minor difference in lipid profiling result may be explained in that the RPL4 mutation only caused tissue-specific defects in seedling meristem tissue. Because of technical limitations, we were unable to perform lipid profiling from only meristem tissues. The fact that the rpl4d mutant displayed only reduced lipid accumulation in the meristem but not the fully differentiated tissues is also worth further investigation. The possible explanation could be that actively dividing tissues such as meristems require more involvement of intact ribosomes, so these fast dividing tissues are more likely to show defects when RPL4 loses function. The other explanation could be that lipid metabolism is required during vacuole formation, but to a lesser extent, once vacuoles are already fully formed in differentiation cells.
The mechanism of how membrane lipids regulate vesicle trafficking has been widely demonstrated in mammalian models. Lipid gradients exist along the biosynthetic pathway with increased density of cholesterol and sphingolipids from the ER to the plasma membrane (58). The biophysical nature of different lipid species determines the extent fluidness and curvature during budding (59). For this reason, the ER is enriched in unsaturated lipid, which facilitates protein exit after synthesis, whereas plasma membrane and endosomes contain more cholesterol, which eases membrane curvature (31, 60). In Arabidopsis, mutants defective in sphingolipid synthesis lead to auxin-dependent inhibition of lateral root emergence and selective aggregation of the auxin transporters AUX1 and PIN1 in the cytosol (25). The sterol biosynthesis mutant cpi1-1 indicates that sterol composition affects postcytokinesis acquisition of PIN2 polarity by endocytosis and thus sterol plays an indispensable role for the establishment of polar protein localization (27). In our results, both sphingolipids and sterol biosynthesis mutants displayed vacuole trafficking defects, leading to secretion of RFP:CTPP signal, which resembled the phenotype observed in the rpl4d mutant. Additionally, our results showed tissue-specific complementation with key enzymes from the sterol and sphingolipid biosynthetic pathways, but not fatty acid synthesis, for both vacuole trafficking and developmental defects in rpl4d. This finding suggests that sterol and sphingolipid biosynthesis pathways are indispensible pathways for vacuole trafficking, which is downstream of ribosome function.
Our results clearly indicate that sphingolipid and sterol metabolism are involved in RPL4 regulation. However, in the list of down-regulation genes of lipid-metabolism, only a few genes are directly involved in the sphingolipid and sterol metabolism. Many genes identified are involved in fatty acid and glycerolipid metabolism. This result suggests that acyl modification of sphingolipids and sterols may be important for RPL4 function as the two lipid esters contain acyl groups. Although we have no direct evidence that RPL4 selectively regulates acyl abundance in sphingolipid and sterol lipids, the fact that loh1loh2 double-mutants display greatly reduced VLCFA levels and the mutants phenocopy vacuole trafficking defects (28) similar to rpl4d indicates the possible correlation between acyl accumulation, RPL4 function, and vacuole trafficking. Moreover, there is no clear cut between the change in sphingolipid and other lipid species, such as phospholipids and fatty acids, because reduction in one lipid species always results in compensation by other lipids. Thus, the fact that rpl4d only showed limited changes in sterol and sphingolipid biosynthesis genes could be explained by compensation effects.
It was known that auxin regulates cell proliferation, tissue differentiation, and endomembrane trafficking. Our result shows that lipid synthesis is downstream of auxin-ribosome function and that tissue-specific complementation of lipid synthesis can partially rescue auxin signal intensity, distorted root pattern, and vacuole trafficking defects in rpl4d. This finding indicates that lipid metabolism is the key factor integrating auxin-mediated tissue differentiation and endomembrane trafficking, which requires translational regulation by ribosome proteins. Because the auxin receptor triple-mutant tir1 afb1 afb3 (tirT) also displayed a reduction in lipid accumulation (Fig. 2C), it suggests that lipid metabolism is modulated directly or indirectly by TIR1/AFBs signaling pathways and that ribosome proteins serve as the hub to link the two pathways together.
The molecular mechanism of auxin-regulated vacuole trafficking still remains elusive. A recent publication has elucidated the involvement of the TIR1/AFB signaling pathway in the regulation of gravity induced PIN2 vacuole trafficking (23). The molecular mechanism connecting auxin signaling and vacuole trafficking is still not clear. However, the result from the same group showed that mutation in the ARF2 gene disturbs vacuolar trafficking of PIN2 protein (23). Interestingly, our previous report elucidated that RPL4 regulates translational efficiency of ARF2, ARF3, ARF7, and ARF19 in a uORF-dependent mechanism (48). Based on these facts, it would be plausible to speculate that auxin signaling-regulated lipid metabolism requires active ARF transcriptional factors, which are under translational regulation by ribosome proteins.
In the future, it will be important to begin to understand the mechanism of auxin regulation, ribosomal protein function, lipid metabolism, and endomembrane trafficking. One possible mechanism could involve the conserved mammalian target of rapamycin (mTOR) pathway, which has been well-studied in mammalian cell systems. The TOR complex functions as a center hub to coordinate cell proliferation, nutrient supply, energy sufficiency, stress, hormone activation with metabolism, and translation regulation (61). Interestingly, evidence has emerged in Arabidopsis of the possible involvement of TOR-S6K1 signaling pathway downstream of auxin stimulation to facilitate translational reinitiation of plant mRNAs containing uORFs at their 5′ UTR (62). Intriguingly, TORC1 is a positive regulator of lipid synthesis in mammalian cells that promotes accumulation of lipids, TAGs, and long-chain polyunsaturated fatty acids in Arabidopsis (63). Because ribosomal proteins regulate both development and endomembrane trafficking downstream of auxin signaling through the control of lipid synthesis (present study), we speculate that the TOR pathway may link auxin activation with ribosome-mediated translational regulation.
Taking these data together, our work sheds light on a novel link between translational regulation by ribosome proteins, auxin-mediated tissue differentiation, and endomembrane trafficking through the regulation of lipid metabolism (see model in Fig. 6). It will be a new area of research to study the relationship between auxin signaling, ribosome function, and lipid metabolism.
Materials and Methods
Plant Material and Growth Condition.
The rpl4d (salk_029203c) and RFP:CTPP (53) plants are in Arabidopsis thaliana Columbia ecotype background. cpi1-1 (27) is in the Ler ecotype, and loh1loh2 (25) is in the WS ecotype. Seeds were sterilized in sterilization solution [80% ethanol and 20% (vol/vol) bleach] and vernalized for 3 d at 4 °C, then germinated on solid Murashige-Skoog (MS) medium containing 1% sucrose under long-day conditions (16-h light/8-h dark) before being transferred to soil.
Plasmid Construction.
Plasmids used for complementation were constructed in pMDC123, containing gateway cassettes. RPL4D promoter was obtained by PCR using the specific primers RPL4P-F and RPL4P-R. Primer sequences are given in Dataset S4. The PCR product was digested with XbaI and SacI and ligated into pMDC123 digested with the same restriction enzymes. CDS of LCB1, SMT2, and KAS1 were first introduced into pDONR, and then transferred to modified pMDC123 vector through LR reaction.
Quantitative RT-PCR Conditions.
Quantitative RT-PCR (qRT-PCR) reactions were performed in triplicate in 96-well microtiter plates (Bio-Rad, Cat # MLL9601). The total reaction volume was 20 μL, which contained 10 μL 2× SYBR Green Master Mix reagent (Bio-Rad, Cat # 172–5264), 0.5-μM gene-specific primers, and 2 μL appropriately diluted cDNA. The qRT-PCR reactions were performed using a CFX 96 Real-Time System (Bio-Rad). The following standard thermal profile was used for all PCR reactions: 90 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 58 °C for 30 s. Amplicon dissociation curves (melting curves) were recorded after cycle 40 by heating from 65 °C to 95 °C with a ramp speed of 0.5 °C per 5 s. Data were analyzed by using the Bio-Rad self-contained program followed by Microsoft Office Excel 2007. The PCR (E) efficiency was calculated from the exponential phase of each individual amplification plot according to the following formula: E = 10(−1/slope) − 1, where an efficiency of 1 corresponds to 100%. The e-values corresponding to PCR efficiencies of reference gene Ubiquitin in polysome bound and total mRNA in rpl4d mutants and Col were 0.935, 0.912, 0.941, 0.910, respectively.
Lipid Extraction and Electrospray Ionizaton-MS/MS Analysis.
Lipid extraction, electrospray ionization (ESI)-MS/MS analysis, and quantification was performed as described previously (64, 65). Briefly, 7-d-old seedlings of Col wild-type or rpl4d mutants were harvested and transferred immediately into 3 mL of isopropanol with 0.01% butylated hydroxytoluene at 75 °C. The tissue was extracted with chloroform/methanol (2:1, vol/vol) four times with 1.5 h of agitation each time. The remaining plant tissue was dried in the hood overnight and weighed as “dry weight” of the plants. Lipid samples were analyzed on a triple quadruple MS/MS equipped for ESI. Data processing was performed as previously described (66). The lipids in each class were quantified in comparison with two internal standards of the class. Six replicates of each treatment for each genotype were analyzed. The Q-test was performed on the total amount of lipid in each head group class, and data from discordant samples were removed. Paired values were subjected to the t test to determine statistical significance.
Hormones and Inhibitor Treatment.
Seedlings in different backgrounds were germinated and grown in vertical placed solid MS medium containing 1% sucrose for 7 d before being transferred to liquid medium with different hormones and inhibitors at final concentration of 0.1% for various time scales, as described in Results. For each treatment, per one well of a 24-well cell-culture plate, 7–10 seedlings were incubated in in 2 mL liquid medium (1/2 MS, 1% sucrose, pH 5.8) containing the respective inhibitor or corresponding amounts of solvent. All of the lipid dye, inhibitors, and hormones were first dissolved in DMSO as stock solution. Nile red and Bodipy 493/503 were added from a 1-mg/mL stock, auxinole was added from a 5-mM stock in DMSO, FB1 was added from a 5-mM stock, PDMP was added from a 10-mM stock, FEN was added from a 100-mM stock, and flufenacet was added from a 1-mM stock. Different hormones including NAA, 2-NAA, GA3, JA, and ABA were prepared in 100-μM stock. Hormones and inhibitors were purchased from Sigma. The data presented were reproduced in three to five independent experiments.
Lugol Staining.
Lugol staining of starch deposition in Arabidopsis roots was performed as described previously (67), with minor modification of the protocol. Seedlings were incubated with 1% Lugol’s solution for 5 min. Roots were then washed twice with distilled water and mounted on a tissue clear solution [chloral hydrate: glycerol: water (8:1:2, wt/vol/vol)] for 2 h before being visualized with Zeiss AX10 microscope equipped with DIC optics. Pictures were taken using charged-coupled device camera (ProgRes C5, JENOPTIK). Images were further processed with Photoshop CS5.
Live-Cell Imaging and Image Process.
For confocal microscopy images, the SP5 (Leica) confocal microscope was used. Detection of nonpolar lipid (excitation at 514 nm, emission at 520–560 nm), polar lipid (excitation at 534 nm, emission at 600–700 nm) after Nile red staining was performed as described previously (68). Sequential scanning was used to avoid any interference between fluorescence channels. Fluorescence signal for DR5:GFP (excitation at 488 nm, emission at 490–550 nm) and RFP:CTPP signal (excitation at 534 nm, emission at 600–700 nm) were detected using GFP and RFP settings, respectively.
Hypocotyl or root-tip imaging was performed with seedlings mounted in water, and high magnification images were taken with a 40× objective lens (water immersion) with a line average of 4. Confocal pictures were processed with Photoshop CS5. The absolute fluorescence intensities were measured by using imageJ (NIH; rsb.info.nih.gov/ij). The statistical significance was evaluated with Student’s t test.
Polysome Profiling.
Polysome profiling was performed using freshly ground tissue from 7-d-old seedlings as described in Mustroph et al. (69).
Library Preparation for RNA Sequencing.
Total RNA and polysome-bound mRNA were collected from sucrose gradients as described in polysome profiling (69). RNA was first extracted with trizol RNA extraction solution (invitrogen), and then purified by Plant RNeasy mini kit (Qiagen). Libraries were prepared using mRNA-SEq. 24-sample prep kit (Illumina, Cat. # RS-930-2002). All samples were purified and multiplexed using 12 of the three-nucleotide barcoded adapters, randomly assigned to three different pools with four samples per pool. These pooled libraries were subjected to Illumina HiSEq. 2000 paired-end sequencing at the University of California Riverside Core Facility following the standard Illumina sequencing protocols. The experiment generated ∼50 M–90 M read pairs for each sample.
RNA Sequencing Data Analysis.
Distribution for the FASTQ quality score for each sample was analyzed for sequencing quality assessment. Reads were then aligned against to Arabidopsis Information Resource (TAIR10) version of Arabidopsis genome using TOPHAT v2.00 supplied with The TAIR10 GFF at default setting. Alignment bam index files were generated using SAMtools v0.1.19. Approximately >85% of read pairs aligned to TAIR10 genome. Read counts for each gene were quantified using summarizeOverlaps R function in GenomicFeatures package with the settings mode = union. RPKM value was calculated based on the read counts. Quality control check of the sample reproducibility was done by computing a correlating matrix using RPKM value in an R script. Differential expression gene was determined using edgeR R package v2.0. The cut-off of false-discovery rate was set with <0.001 and log2FC > 0.5 or logFC < −0.5.
GO term enrichment of identified differential expression gene sets was analyzed using an R script with computing a hypergeometric distribution test and returning the corresponding raw and Bonferroni-corrected P values with cut-off 0.001 for TAIR10 full GO or GO slim nodes. Additionally, MapMan (v3.5.2.1R2) software was run for pathway analysis.
Supplementary Material
Acknowledgments
We thank Dr. Lorenzo Frigerio (University of Warwick) for kindly providing the RFP:CTPP vacuolar targeting marker and construct; Dr. Markus Grebe (Umea University) for sharing the material of cpi1-1; Dr. Jean-Denis Faure (Institut Jean-Pierre Bourgin) for the gifts of loh1loh2 double-mutants; Dr. Hiroshi Nozaki (Okayama University of Science) for providing the inhibitor auxinole; J. Brimo (University of California, Riverside) for administrative support; and Wilhelmina van de Ven, Emily Hsu, and Ivan Sotelo for technical support. The lipid analyses described in this work were performed at the Kansas Lipidomics Research Center Analytical Laboratory. The Kansas Lipidomics Research Center was supported by National Science Foundation (EPS 0236913, MCB 0455318, DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20RR16475), and Kansas State University. All microscopy and Illumina sequencing and analysis were performed at the Centralized Research Facilities of the Institute for Integrative Genome Biology and Center for Plant Cell Biology at Univerisity of California, Riverside (genomics.ucr.edu). This research was funded by the Division of Chemical Sciences of the US Department of Energy through Grant DE-FG02-02ER15295 (to N.V.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422656112/-/DCSupplemental.
References
- 1.Marty F. Plant vacuoles. Plant Cell. 1999;11(4):587–600. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao Y, Li KY, Li HY, Yao X, Jiang L. The vacuolar transport of aleurain-GFP and 2S albumin-GFP fusions is mediated by the same pre-vacuolar compartments in tobacco BY-2 and Arabidopsis suspension cultured cells. Plant J. 2008;56(5):824–839. doi: 10.1111/j.1365-313X.2008.03645.x. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed SU, et al. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol. 2000;149(7):1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassham DC, Raikhel NV. Unique features of the plant vacuolar sorting machinery. Curr Opin Cell Biol. 2000;12(4):491–495. doi: 10.1016/s0955-0674(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 5.Sanmartín M, et al. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(9):3645–3650. doi: 10.1073/pnas.0611147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohn EJ, et al. The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc Natl Acad Sci USA. 2007;104(47):18801–18806. doi: 10.1073/pnas.0708236104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surpin M, et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15(12):2885–2899. doi: 10.1105/tpc.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surpin M, Raikhel N. Traffic jams affect plant development and signal transduction. Nat Rev Mol Cell Biol. 2004;5(2):100–109. doi: 10.1038/nrm1311. [DOI] [PubMed] [Google Scholar]
- 9.Jurgens G. Membrane trafficking in plants. Annu Rev Cell Dev Biol. 2004;20:481–504. doi: 10.1146/annurev.cellbio.20.082503.103057. [DOI] [PubMed] [Google Scholar]
- 10.Hicks GR, Raikhel NV. Advances in dissecting endomembrane trafficking with small molecules. Curr Opin Plant Biol. 2010;13(6):706–713. doi: 10.1016/j.pbi.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Norambuena L, Hicks GR, Raikhel NV. The use of chemical genomics to investigate pathways intersecting auxin-dependent responses and endomembrane trafficking in Arabidopsis thaliana. Methods Mol Biol. 2009;495:133–143. doi: 10.1007/978-1-59745-477-3_12. [DOI] [PubMed] [Google Scholar]
- 12.Robert S, et al. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci USA. 2008;105(24):8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413(6854):425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 14.Nagawa S, et al. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 2012;10(4):e1001299. doi: 10.1371/journal.pbio.1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin D, et al. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol. 2012;22(14):1319–1325. doi: 10.1016/j.cub.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhonukshe P, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17(6):520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Kleine-Vehn J, Friml J. Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol. 2008;24:447–473. doi: 10.1146/annurev.cellbio.24.110707.175254. [DOI] [PubMed] [Google Scholar]
- 18.Feraru E, Friml J. PIN polar targeting. Plant Physiol. 2008;147(4):1553–1559. doi: 10.1104/pp.108.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleine-Vehn J, Dhonukshe P, Swarup R, Bennett M, Friml J. Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell. 2006;18(11):3171–3181. doi: 10.1105/tpc.106.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geldner N, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112(2):219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 21.Teh OK, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448(7152):493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 22.Kleine-Vehn J, et al. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA. 2008;105(46):17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baster P, et al. SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 2013;32(2):260–274. doi: 10.1038/emboj.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roudier F, et al. Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell. 2010;22(2):364–375. doi: 10.1105/tpc.109.071209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markham JE, et al. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell. 2011;23(6):2362–2378. doi: 10.1105/tpc.110.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, et al. Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J. 2013;74(1):37–47. doi: 10.1111/tpj.12103. [DOI] [PubMed] [Google Scholar]
- 27.Men S, et al. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol. 2008;10(2):237–244. doi: 10.1038/ncb1686. [DOI] [PubMed] [Google Scholar]
- 28.Pan J, et al. The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell. 2009;21(2):568–580. doi: 10.1105/tpc.108.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville C, Browse J. Plant lipids: Metabolism, mutants, and membranes. Science. 1991;252(5002):80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- 30.Hawes CR, Brandizzi F, Andreeva AV. Endomembranes and vesicle trafficking. Curr Opin Plant Biol. 1999;2(6):454–461. doi: 10.1016/s1369-5266(99)00023-0. [DOI] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz J, Phair RD. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu Rev Biophys. 2010;39:559–578. doi: 10.1146/annurev.biophys.093008.131357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux A, et al. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24(8):1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samsonov AV, Mihalyov I, Cohen FS. Characterization of cholesterol-sphingomyelin domains and their dynamics in bilayer membranes. Biophys J. 2001;81(3):1486–1500. doi: 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 35.Pol A, et al. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. 2005;16(4):2091–2105. doi: 10.1091/mbc.E04-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arouri A, Mouritsen OG. Membrane-perturbing effect of fatty acids and lysolipids. Prog Lipid Res. 2013;52(1):130–140. doi: 10.1016/j.plipres.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Bagatolli LA, Mouritsen OG. Is the fluid mosaic (and the accompanying raft hypothesis) a suitable model to describe fundamental features of biological membranes? What may be missing? Front Plant Sci. 2013;4:457. doi: 10.3389/fpls.2013.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willemsen V, et al. Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell. 2003;15(3):612–625. doi: 10.1105/tpc.008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carland FM, Fujioka S, Takatsuto S, Yoshida S, Nelson T. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell. 2002;14(9):2045–2058. doi: 10.1105/tpc.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carland F, Fujioka S, Nelson T. The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 2010;153(2):741–756. doi: 10.1104/pp.109.152587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosado A, Raikhel NV. Understanding plant vacuolar trafficking from a systems biology perspective. Plant Physiol. 2010;154(2):545–550. doi: 10.1104/pp.110.161091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosado A, et al. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010;22(1):143–158. doi: 10.1105/tpc.109.068320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17(11):2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou F, Roy B, von Arnim AG. Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 2010;10:193. doi: 10.1186/1471-2229-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz G, Melis M, Batetta B, Angius F, Falchi AM. Hydrophobic characterization of intracellular lipids in situ by Nile Red red/yellow emission ratio. Micron. 2008;39(7):819–824. doi: 10.1016/j.micron.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Horn PJ, et al. Visualization of lipid droplet composition by direct organelle mass spectrometry. J Biol Chem. 2011;286(5):3298–3306. doi: 10.1074/jbc.M110.186353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA. 2012;109(48):19537–19544. doi: 10.1073/pnas.1214774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He JX, et al. Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 2003;131(3):1258–1269. doi: 10.1104/pp.014605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abbas HK, et al. Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 1994;106(3):1085–1093. doi: 10.1104/pp.106.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krüger F, et al. PDMP induces rapid changes in vacuole morphology in Arabidopsis root cells. J Exp Bot. 2013;64(2):529–540. doi: 10.1093/jxb/ers345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bach L, et al. Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana. J Cell Sci. 2011;124(Pt 19):3223–3234. doi: 10.1242/jcs.074575. [DOI] [PubMed] [Google Scholar]
- 53.Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 2007;145(4):1371–1382. doi: 10.1104/pp.107.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi K, et al. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol. 2012;7(3):590–598. doi: 10.1021/cb200404c. [DOI] [PubMed] [Google Scholar]
- 55.Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB. The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell. 2006;18(12):3576–3593. doi: 10.1105/tpc.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neelakandan AK, et al. Molecular characterization and functional analysis of Glycine max sterol methyl transferase 2 genes involved in plant membrane sterol biosynthesis. Plant Mol Biol. 2010;74(4-5):503–518. doi: 10.1007/s11103-010-9692-6. [DOI] [PubMed] [Google Scholar]
- 57.Wu GZ, Xue HW. Arabidopsis β-ketoacyl-[acyl carrier protein] synthase i is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell. 2010;22(11):3726–3744. doi: 10.1105/tpc.110.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorre B, et al. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc Natl Acad Sci USA. 2009;106(14):5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manneville JB, et al. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc Natl Acad Sci USA. 2008;105(44):16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiong Y, Sheen J. Moving beyond translation: Glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle. 2013;12(13):1989–1990. doi: 10.4161/cc.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schepetilnikov M, et al. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32(8):1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caldana C, et al. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73(6):897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- 64.Li W, et al. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J Biol Chem. 2008;283(1):461–468. doi: 10.1074/jbc.M706692200. [DOI] [PubMed] [Google Scholar]
- 65.Welti R, et al. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277(35):31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 66.Devaiah SP, et al. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry. 2006;67(17):1907–1924. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA. 2010;107(26):12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenspan P, Mayer EP, Fowler SD. Nile red: A selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100(3):965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.