Abstract
Fungal pathogens must assimilate local nutrients to establish an infection in their mammalian host. We focus on carbon, nitrogen, and micronutrient assimilation mechanisms, discussing how these influence host–fungus interactions during infection. We highlight several emerging trends based on the available data. First, the perturbation of carbon, nitrogen, or micronutrient assimilation attenuates fungal pathogenicity. Second, the contrasting evolutionary pressures exerted on facultative versus obligatory pathogens have led to contemporary pathogenic fungal species that display differing degrees of metabolic flexibility. The evolutionarily ancient metabolic pathways are conserved in most fungal pathogen, but interesting gaps exist in some species (e.g., Candida glabrata). Third, metabolic flexibility is generally essential for fungal pathogenicity, and in particular, for the adaptation to contrasting host microenvironments such as the gastrointestinal tract, mucosal surfaces, bloodstream, and internal organs. Fourth, this metabolic flexibility relies on complex regulatory networks, some of which are conserved across lineages, whereas others have undergone significant evolutionary rewiring. Fifth, metabolic adaptation affects fungal susceptibility to antifungal drugs and also presents exciting opportunities for the development of novel therapies.
Fungal pathogens must assimilate nutrients to establish an infection in their host. They inhabit diverse niches—from skin or mucosal surfaces to the internal organs—and display an impressive degree of metabolic flexibility.
Nutrient assimilation is a central and fundamental prerequisite for the growth and survival of all living organisms. Pathogenic fungi inhabit dynamic and contrasting niches and must display rapid and effective adaptation to changes in nutrient availability in these microenvironments. To achieve this, they regulate specific nutrient uptake mechanisms and modulate their metabolism, displaying an impressive degree of metabolic flexibility. This metabolic flexibility, which enhances the fitness of the fungus, is often as essential for pathogenicity as virulence factors, thereby representing an attractive target for potential therapeutic intervention.
The major fungal pathogens of humans have evolved in a polyphyletic manner (i.e., pathogenicity has emerged independently in different phylogenetic branches of the fungal kingdom). Furthermore, major fungal pathogens display different lifestyles, and, consequently, their metabolic flexibility has been shaped by different evolutionary pressures. For example, the ascomycete Candida albicans is a facultative pathogen that exists primarily as a commensal of the oral cavity, gastrointestinal tract, and urogenital tract but can also persist within other extracellular microenvironments (blood, tissues) and within intracellular microenvironments (inside damaged epithelial and endothelial cells, and even the phagolysosome of macrophages). During its commensal stage, C. albicans must adapt to the nutrients present on mucosal surfaces and compete with cohabitating microflora, whereas during infection, the fungus exploits an alternative array of nutrients available in host tissues. The apparent lack of a significant environmental niche for C. albicans (Odds 1988) means that, in recent evolutionary time, the metabolic flexibility of this fungus has been tuned to these host niches. In contrast, fungi such as the basidiomycete Cryptococcus neoformans and the ascomycete Histoplasma capsulatum, are opportunistic pathogens that are associated with environmental niches such as pigeon guano, soil, and trees but can cause chronic pulmonary infections and devastating systemic infections in immunocompromised individuals (Sil 2006; Heitman 2011). The metabolic flexibility of these fungal pathogens ensures survival during the saprophytic phases of their life cycle as well as promoting their ability to cause pulmonary and disseminated infections (Kronstad et al. 2012). Therefore, different evolutionary pressures have been imposed on the processes that mediate metabolic adaptation in major fungal pathogens.
In addition, there are significant differences between fungal pathogens regarding the degree of evolutionary adaptation to their mammalian hosts. For example, the filamentous ascomycete, Aspergillus fumigatus is probably an accidental pathogen. This “grass eater” (Tekaia and Latgé 2005) lives mainly as a saprophyte, degrading plant and other organic material in the environment but can cause severe pulmonary infections in immunocompromised patients. At the other end of the spectrum, Pneumocystis species are obligate fungal pathogens that appear to have coevolved with their mammalian hosts to such an extent that they have shed several metabolic pathways and, so far, it has not been possible to culture them in vitro (Cushion 2004; Cushion et al. 2007; Hauser et al. 2010). C. albicans and Candida glabrata lie between these extremes, apparently being obligate parasites of warm-blooded animals (Odds 1988) and yet retaining a high degree of metabolic flexibility in vitro and in vivo (Brown et al. 2007; Wilson et al. 2009). Clearly the metabolic flexibility of fungal pathogens has been influenced by the evolutionary time scales over which environmental selection pressures have been exerted, as well as by the nature of these pressures.
In this review, we focus on the metabolic flexibility of major fungal pathogens during the infection process: commensalism, colonization, and disease progression. We address major fungal pathogens that are highly significant in clinical settings, such as C. albicans, A. fumigatus, and C. neoformans, mentioning other pathogens where appropriate. We concentrate on nitrogen and carbon metabolism, and micronutrient assimilation, which are both critical for pathogenesis and well characterized in pathogenic fungi.
NITROGEN ASSIMILATION
Nitrogen is required for almost all biosynthetic processes and, like carbon, must be assimilated in large quantities. Therefore, the acquisition of both nitrogen and carbon compounds from the host is essential for pathogenic fungi to survive, grow, and persist within a host. The high degree of metabolic flexibility is reflected in the different types of nitrogen sources exploited by fungal pathogens. In the absence of preferred nitrogen sources, such as ammonia or certain amino acids, these fungi can use compounds like proteins or polyamines. Some pathogenic fungi (e.g., pathogenic Aspergillus species) can also reduce nitrate to ammonia (Zhou et al. 2002). However, the pathogenic Candida and Cryptococcus species are unable to use nitrate as a nitrogen source.
Although fungal nitrogen utilization has been extensively studied in vitro, it is less clear which nitrogen sources are used by human pathogenic fungi in vivo. However, transcriptional profiling studies using microarrays or serial analysis of gene expression (SAGE) in ex vivo or in vivo infection models have provided insights into the nitrogen metabolism of pathogenic fungi such as C. albicans (Fradin et al. 2005; Thewes et al. 2007; Zakikhany et al. 2007; Walker et al. 2009; Wilson et al. 2009; Cheng et al. 2013), C. glabrata (Kaur et al. 2007), A. fumigatus (McDonagh et al. 2008), C. neoformans (Fan et al. 2005; Steen et al. 2003), and dermatophytes (Staib et al. 2010) during infection (reviewed by Cairns et al. 2010).
Proteases, Oligopeptide, and Amino Acid Transporters
During infection, C. albicans expresses various secreted aspartic proteases (Saps), one of the best-investigated virulence attributes of this fungus (Naglik et al. 2003). Liberated oligopeptides and amino acids are then taken up by dedicated oligopeptide transporters (Opt1–8) (Reuss and Morschhauser 2006) and a family of 22 predicted amino acid permeases (Sychrova and Souciet 1994; Kraidlova et al. 2011). When expressed in host tissues, these factors enable C. albicans to use a broad range of nitrogen sources over the course of an infection, driving host tissue damage and invasion (Villar et al. 2007; Naglik et al. 2008; Dalle et al. 2010; Cheng et al. 2013). This proteolytic activity might also promote immune evasion, as factors of the immune system, antimicrobial proteins, and peptides may be degraded (Naglik et al. 2003; Gropp et al. 2009; Meiller et al. 2009). Similarly, the aspartic proteases of C. glabrata and Candida parapsilosis have been implicated in survival following phagocytosis by macrophages (Kaur et al. 2007; Horvath et al. 2012).
Extracellular proteases have also been reported to be virulence attributes for the most common airborne pathogenic fungus, A. fumigatus (Monod et al. 1999), which contains multiple protease genes (Nierman et al. 2005). Although an oligopeptide transporter and a key regulator for extracellular proteolysis are required for in vitro growth of A. fumigatus on complex substrates, these proteins are not crucial for survival in vivo (Hartmann et al. 2011). Similarly, C. albicans Sap2, a protease essential for the growth on protein as the sole nitrogen source, is not essential for survival in vivo (Hube et al. 1994, 1997). Furthermore, similar observations have been made for dermatophytes such as Arthroderma benhamiae and Trichophyton rubrum. Proteases have always been considered de facto to be a key virulence attribute of dermatophytes, as these fungi infect keratin-containing tissue and are necessarily keratinolytic. Indeed, their genomes display a clear enrichment in protease genes (Burmester et al. 2011). However, those protease genes most highly expressed during growth on keratin in vitro, and hence for which an important role in virulence would be expected, differ from those genes that are highly up-regulated during infection (Zaugg et al. 2009; Staib et al. 2010). This highlights the fact that in vitro growth conditions often differ significantly from in vivo microenvironments.
Nitrogen Acquisition in Host Niches
Little is known about the nitrogen sources that pathogenic fungi assimilate in host niches. Presumably the types and concentrations of nitrogen source differ significantly between certain niches, such as mucosal surfaces, the gastrointestinal (GI) tract, and the bloodstream. Most information about fungal nitrogen assimilation has been gleaned through fungal transcriptomics using ex vivo infection models combined with limited molecular dissection via targeted gene mutation.
The environment inside the phagosome of phagocytes is thought to be poor in nitrogen. This is reflected in the transcriptional response of C. albicans and C. glabrata after internalization by macrophages (Lorenz et al. 2004; Seider et al. 2010; Roetzer et al. 2011; Brunke and Hube 2013; Mayer et al. 2013; Miramon et al. 2013). For example, C. albicans up-regulates amino acid biosynthetic pathways, suggesting that the fungus at least transiently faces amino acid deprivation. The arginine biosynthetic pathway is the only amino acid biosynthesis pathway significantly up-regulated in C. albicans cells after phagocytosis by macrophages (Lorenz et al. 2004). Engulfment by neutrophils induces the up-regulation of arginine, leucine, lysine, and methionine anabolic pathways as well as GCN4 (Rubin-Bejerano et al. 2003; Fradin et al. 2005), the gene encoding the master regulator of amino acid synthesis (Tripathi et al. 2002). C. glabrata also up-regulates the synthesis of arginine and lysine on internalization by macrophages (Kaur et al. 2007).
The specific contributions of these amino acids to fungal survival within the phagocyte remain to be tested. However, arginine biosynthetic gene induction might play a critical role in the fungal response to macrophage-derived reactive oxygen species (Jimenez-Lopez et al. 2013). Arginase can convert arginine to urea, which in turn is degraded by urea amidolyase to produce ammonia and CO2. These breakdown products can contribute to neutralization of the acidic pH of the phagolysosome, promoting the intracellular induction of hyphal formation and allowing fungal escape and killing of the host cell (Ghosh et al. 2009). Similarly, the extracellular ureases of C. neoformans and Cryptococcus gattii (Ure1) degrade urea, which is abundant in the cerebrospinal fluid. The liberated CO2 acts as a signal to induce capsule formation, a well-studied virulence factor in this fungus (Frazzitta et al. 2013).
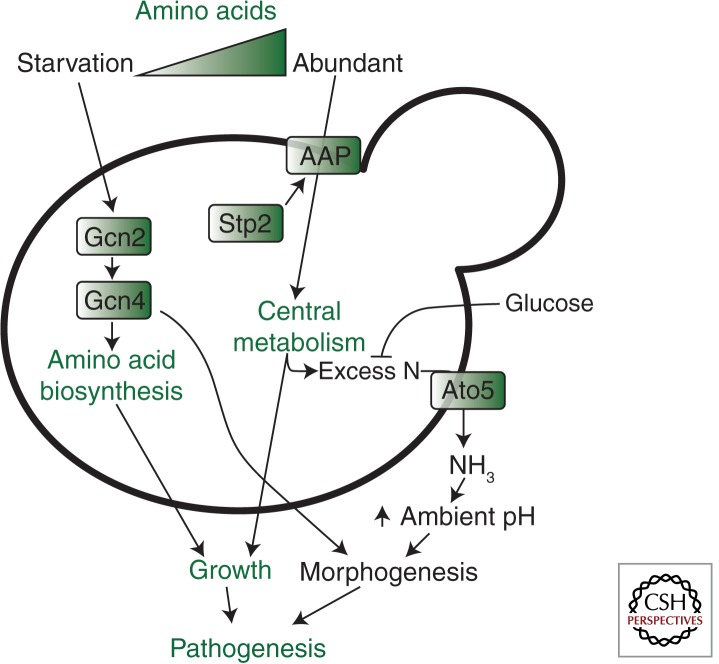
Under sugar-limiting conditions, C. albicans cells can exploit amino acids as a carbon source (Fig. 1). The amino acids are imported via amino acid permeases, the expression of which is up-regulated by the transcription factor Stp2. The amino acids are metabolized in a glucose-repressible manner, and the excess nitrogen is excreted as ammonia via Ato5, a putative ammonia transporter, thereby raising environmental pH and autoinducing hypha formation (Vylkova et al. 2011). Similarly, Dur31 is a sodium/substrate symporter for the polyamine spermidine, and the amino groups of spermidine can potentially be used to generate ammonia. In fact, a dur31Δ mutant is unable to actively increase the local ambient pH. This suggests that Dur31 is an integral component of the mechanisms that promote extracellular alkalinization and hyphal autoinduction. Indeed, DUR31 is required for morphogenesis (Mayer et al. 2012). Interestingly, Dur31 not only transports spermidine but also histatin 5, a highly cytotoxic antimicrobial peptide (Mayer et al. 2012). Therefore, Dur31 appears to have dual functions as a virulence attribute and as an avirulence factor.
Figure 1.
Candida albicans responds to amino acid starvation by activating the GCN pathway, which up-regulates amino acid biosynthesis via Gcn2 and Gcn4 to support the resumption of growth (Tripathi et al. 2002; Tournu et al. 2005). The transcription factor Gcn4 also activates filamentation (Tripathi et al. 2002). When amino acids are abundant, and in the absence of glucose, they are imported via amino acid permeases (AAP), the expression of which is activated by the transcription factor Stp2 (Vylkova et al. 2011). These amino acids are used to sustain central metabolism and the excess nitrogen is exported as ammonia via transporters such as Ato5. This leads to alkalinization of the local microenvironment, which stimulates morphogenesis (Vylkova et al. 2011). Metabolism fuels the growth of C. albicans cells and host colonization, and morphogenesis promotes the virulence of this pathogen (see text).
The response of C. albicans to neutrophils also includes the up-regulation of the ammonium permease genes MEP1 and MEP2 (Fradin et al. 2005). By analogy to the amino acid transporters and the spermidine transporter Dur31, the roles of these ammonium transporters extend beyond the uptake of nitrogen, because Mep2 can activate hyphal formation in response to ammonium limitation. This signaling role appears to be independent of the role in the extracellular alkalinization hyphal autoinduction pathway (Biswas and Morschhauser 2005).
Interestingly, C. glabrata can form pseudohyphae-like structures under certain nitrogen starvation conditions (Csank and Haynes 2000). However, unlike C. albicans, this filamentous morphology is not used as an escape mechanism from macrophages. Instead, C. glabrata continues to replicate as yeasts inside the phagosome until the host cell bursts by an unknown mechanism (Seider et al. 2011). This intracellular survival and replication depends on a specialized form of autophagy (pexophagy) to survive internalization by macrophages (Roetzer et al. 2010).
In general, the recycling of cellular proteins is a common strategy of fungi to overcome nitrogen starvation. For example, C. albicans vacuolar protease genes associated with intracellular protein degradation such as APR1, PRB1, PRB2, or PRC1 are up-regulated in response to phagocytosis by either macrophages (Lorenz et al. 2004) or neutrophils (Fradin et al. 2005). A second, ubiquitin-dependent pathway contributes to protein recycling under nutrient-limiting conditions in C. albicans (Leach et al. 2011). This might explain why autophagy is not essential for this fungus to survive and to form hyphae within macrophages. C. albicans cells with defects in autophagy and cytoplasm-to-vacuole trafficking (atg9Δ) can survive macrophages and retain the ability to kill host cells (Palmer et al. 2007). Meanwhile, C. albicans polyubiquitin (ubi4) mutants display attenuated virulence (Leach et al. 2011). Similarly, A. fumigatus does not require autophagy for full virulence in a neutropenic mouse model (Richie et al. 2007), probably because its hyphae are not taken up by macrophages (Palmer et al. 2008). In contrast, fungi that can reside inside the phagosome for a long time, such as C. glabrata and C. neoformans, seem to require autophagy for full virulence, and the inactivation of the autophagic system reduces the virulence of C. neoformans in mice (Hu et al. 2008).
Regulation of Nitrogen Metabolism
Flexibility in nitrogen assimilation requires sensing of the available nitrogen sources and appropriate regulation of nitrogen assimilation genes. There are strong similarities between C. albicans and the model yeast, Saccharomyces cerevisiae, with respect to the regulation of key nitrogen acquisition pathways. In C. albicans, this occurs mainly via the SPS sensor mechanism, comprising the amino acid receptor Csy1 (a Ssy1 homolog), the scaffold protein Ptr3, and the signaling endopeptidase Ssy5 (Ljungdahl 2009). Similar to S. cerevisiae, the presence of extracellular amino acids is detected by the SPS sensor, leading to the proteolytic processing, activation, and nuclear translocation of the transcription factors Stp1 and Stp2 (Andreasson and Ljungdahl 2002; Martinez and Ljungdahl 2005). In an interesting twist, C. albicans Stp1 and Stp2 each activate a specific subset of nitrogen acquisition-related genes. Stp1 activates the transcription of genes involved in protein utilization, such as the genes encoding the secreted protease Sap2 and the oligopeptide transporter Opt1. On the other hand, Stp2 activates transcription of amino acid permease genes, for example, GAP1, GAP2, and CAN1 (Martinez and Ljungdahl 2005). As Stp1 levels are also strongly reduced in the presence of amino acids, this system allows C. albicans to use free amino acids when they are available, and to obtain nitrogen from extracellular proteins under amino acid-limiting conditions. In the host, this allows optimal utilization of available nitrogen sources, and hence contributes to the pathogenicity potential of C. albicans (Ljungdahl 2009).
Microarray data suggest a key role for the Gpa1-cAMP–PKA pathway in the survival of C. neoformans within macrophages, which includes the up-regulation of amino acid transporters (Fan et al. 2005). This pathway not only regulates the transcription of genes encoding the major virulence attributes of C. neoformans, capsule, and melanin production but also responses to nutrient limitation within phagosomes. Consequently, gpa1Δ and pka1Δ mutants display reduced growth within macrophages (Fan et al. 2005). Microarray experiments also indicate that Paracoccidioides brasiliensis regulates amino acid metabolism during interactions with macrophages (Tavares et al. 2007), and that there is a close link between amino acid assimilation and the infectious and pathogenic states of H. capsulatum as amino acid transporters are differentially regulated in conidia, yeast, and mycelia (Inglis et al. 2013).
Amino acid starvation in C. albicans triggers the induction of genes on essentially all amino acid biosynthetic pathways via general amino acid control (the GCN response) (Fig. 1) (Tripathi et al. 2002; Yin et al. 2004; Tournu et al. 2005). This response is analogous to the GCN response in S. cerevisiae (Hinnebusch 1988; Natarajan et al. 2001; Hinnebusch and Natarajan 2002) and cross-pathway control in Aspergillus and Neurospora species (Paluh et al. 1988; Hoffmann et al. 2001; Krappmann et al. 2004). Briefly, amino acid starvation activates Gcn2, which phosphorylates eIF2α thereby decreasing the activity of this essential translation initiation factor. This reduces the overall rate of mRNA translation while enhancing the translation of GCN4 via upstream open reading frames (uORFs) in the unusually long 5′-leader sequence of this mRNA. Hence, Gcn4 levels increase, leading to the transcriptional activation of amino acid biosynthetic genes via GCN response elements in their promoters. In S. cerevisiae, GCN4 expression is regulated primarily at the translational level (Hinnebusch 1988), whereas in C. albicans Gcn4 synthesis is primarily regulated at the transcriptional level (Tripathi et al. 2002; Tournu et al. 2005). Also, in bakers’ yeast purine biosynthesis is induced by the GCN response, unlike in C. albicans (Yin et al. 2004). Furthermore, in C. albicans Gcn4 interacts with the Ras-cAMP pathway to induce morphogenesis in an Efg1-dependent fashion (Fig. 1) (Tripathi et al. 2002). The GCN response contributes to biofilm formation in C. albicans but is not required for the virulence of this pathogen in the mouse model of systemic infection (Brand et al. 2004; Garcia-Sanchez et al. 2004). In contrast, the corresponding CPC response is required for A. fumigatus virulence in the murine model of pulmonary aspergillosis (Krappmann et al. 2004). These contrasting observations might reflect differential transcriptional outputs of the C. albicans Gcn4 and A. fumigatus CpcA transcription factors, and/or differential amino acid availabilities in the mouse lung and kidney.
CARBON ASSIMILATION
Carbon assimilation is essential for the generation of new biomass, and rapid fungal growth in the host relies on the efficient uptake and metabolism of available carbon sources. These can include fermentable sugars (such as glucose, fructose, and galactose) and nonfermentable carbon sources (such as amino acids and organic acids) (Lorenz and Fink 2001; Lorenz et al. 2004; Piekarska et al. 2006; Vieira et al. 2010; Ueno et al. 2011). Fungal pathogens have evolved different carbon assimilation profiles that presumably reflect their different niches. For example, saprobes that are opportunistic pathogens (such as A. fumigatus) have retained the ability to use a broad range of carbon sources. C. albicans does not display any known auxotrophies, can metabolize a broad range of sugars, and can use all amino acids as sole nitrogen sources (Odds 1988; Kaur et al. 2005). However, C. glabrata lacks certain metabolic pathways that exist in other pathogenic yeasts as a result of gene losses that have occurred during its evolution (Dujon et al. 2004). C. glabrata cannot catabolize galactose (loss of GAL1, 7, 10) or allantoin (DAL1–4, 7), and is auxotrophic for pyridoxine (SNO1, 2, 3), thiamine, and nicotinic acid (BNA1–6) (Dujon et al. 2004; Wong and Wolfe 2005). Presumably, these nutritional restrictions are overcome in the specific host niches that are colonized by C. glabrata. More dramatic gene loss has occurred during the evolution of Pneumocystis species, which appears to have shed significant numbers of metabolic genes, retaining only two of 20 amino acid biosynthetic pathways. Based on gene content, glycolysis, the tricarboxylic acid (TCA) cycle, mitochondrial function, and energy metabolism appear to have remained intact (Cushion 2004; Cushion et al. 2007; Hauser et al. 2010). However, the inability to culture pathogenic Pneumocystis species in vitro has prevented the direct exploration of their metabolic capacity.
Most pathogenic fungi prefer to assimilate sugars because their catabolism via glycolysis and respiration is energetically favorable. During the assimilation of alternative carbon sources, energetically demanding pathways such as gluconeogenesis and the glyoxylate cycle must be invoked to generate the hexose and pentose sugars required for cell wall and nucleotide synthesis, for example. Fungi can be classified into Crabtree-negative and Crabtree-positive species based on their carbon utilization patterns. Crabtree-negative species up-regulate the pyruvate dehydrogenase complex in the presence of glucose, such that most of this carbon flows into the TCA cycle to generate biomass and CO2 (Chambergo et al. 2002; Maeda et al. 2004; Xie et al. 2004). Crabtree-positive fungi ferment most of the glucose to generate ethanol (Klein et al. 1998), which is thought to promote the competitive ability of yeasts within polymicrobial microenvironments owing to the antiseptic nature of ethanol (Crabtree 1928).
Regulatory Rewiring of Central Carbon Metabolism
In general, the central metabolic pathways are conserved across the fungal kingdom. However, S. cerevisiae and C. albicans do display some significant differences in their carbon metabolism gene sets. Indeed, genome-wide comparisons have revealed striking differences, with C. albicans harboring additional genes with roles in respiration and oxidative metabolism (Jones et al. 2004). In addition, there are significant differences between C. albicans and S. cerevisiae with respect to the regulatory networks that control central metabolism. S. cerevisiae is a Crabtree-positive yeast, the balance between fermentation and respiratory metabolism being modulated by glucose concentration, oxygen availability, and growth rate (Gancedo 1998). In contrast, C. albicans has been classified as a Crabtree-negative yeast because it retains respiratory activity even in the presence of glucose (Niimi et al. 1988). Bioinformatic and transcriptomic analyses have revealed significant regulatory rewiring between these yeasts, and close links between pathogenicity and metabolism in C. albicans (Ihmels et al. 2005; Martchenko et al. 2007; Askew et al. 2009; Lavoie et al. 2009). For instance, although galactose utilization genes display a similar syntenic organization in C. albicans and S. cerevisiae, their upstream regulatory regions are completely distinct (Martchenko et al. 2007). In S. cerevisiae the transcription factor Gal4 activates the galactose utilization genes GAL1, GAL7, and GAL10, whereas in C. albicans, Gal4 regulates the balance between respiration and fermentation in a carbon-source-dependent fashion (Askew et al. 2009). This presumably reflects the importance of galactose as a carbon source for the pathogenic fungus (Sabina and Brown 2009), particularly in lactating mothers and their infants. The glycolytic transcriptional circuit has also undergone significant transcriptional rewiring. C. albicans lacks homologs of the S. cerevisiae regulators, Gcr1 and Gcr2, which induce glycolysis regulators in this benign yeast (Askew et al. 2009). Instead Tye7 acts as the key regulator of glycolytic genes in C. albicans with gal4Δ tye7Δ mutants displaying altered glycolytic regulation under hypoxia and attenuated virulence in mouse models of infection (Askew et al. 2009). This highlights the importance of balancing carbon flux between respiration and fermentation in host niches during disease progression.
Recent work has revealed that this regulatory rewiring extends to the posttranscriptional circuitry. Many yeast species, including S. cerevisiae, repress pathways involved in the utilization of alternative carbon sources in the presence of glucose with a view to prioritizing sugar utilization over less favorable carbon sources (Flores et al. 2000). This repression operates at multiple levels. In S. cerevisiae, the transcription of genes involved in alternative carbon source utilization (for example, gluconeogenic and glyoxylate cycle genes) is down-regulated via AMP kinase and cAMP signaling, and the transcriptional repressor Mig1 (Gancedo 1998; Carlson 1999; Johnston 1999; Turcotte et al. 2010). In addition, S. cerevisiae enzymes involved in the assimilation of alternative carbon sources, such as Fbp1, Icl1, and Pck1, are targeted for ubiquitin-mediated degradation (Schork et al. 1995; Gancedo and Gancedo 1997; Schule et al. 2000; Horak et al. 2002; Regelmann et al. 2003). However, in C. albicans the evolutionary rewiring of ubiquitination targets allows these enzymes to persist longer in the cell following glucose exposure (Sandai et al. 2012) even though their genes are subject to strong transcriptional repression in response to glucose (Lorenz et al. 2004; Rodaki et al. 2009). Therefore, unlike S. cerevisiae, C. albicans is able to express glycolytic, gluconeogenic, and glyoxylate cycle enzymes at the same time, allowing it to assimilate glucose and alternative carbon sources simultaneously (Sandai et al. 2012). This has presumably evolved to promote the efficient assimilation of complex mixtures of carbon sources, and hence to enhance the fitness of this pathogen in the host (Lorenz 2013).
Regulation of Central Carbon Metabolism in Host Niches
In some host niches, the assimilation of certain alternative carbon sources is essential for fungal proliferation. This is the case for C. glabrata, which relies on lactate assimilation to survive in the mouse intestine (Ueno et al. 2011). The importance of central carbon metabolism in fungal pathogenesis has been reinforced, for example, by the targeted mutational disruption of specific metabolic functions in C. albicans (Lorenz and Fink 2001; Barelle et al. 2006). Glyoxylate cycle (Icl1), glycolytic (Pyk1), and gluconeogenic enzymes (Pck1) are all required for the full virulence of C. albicans in the murine model of systemic candidiasis (Lorenz and Fink 2001; Barelle et al. 2006). These requirements are paralleled in C. neoformans to a reasonable extent, as mutants with glycolytic defects (pyk1Δ and hxk1Δ hxk2Δ) are severely attenuated in a murine inhalation model of cryptococcosis and display decreased persistence in the central nervous system (Price et al. 2011).
Additional evidence for the significant impact of central carbon metabolism on fungal pathogenicity has been generated by transcript profiling of fungal cells exposed to macrophages. For example, C. albicans glyoxylate cycle and fatty acid β-oxidation genes are induced following phagocytosis by macrophages (Lorenz et al. 2004). Following phagocytosis, C. albicans displays a starvation response, reprogramming its metabolism to generate hexose sugars via lipid catabolism, the glyoxylate cycle, and gluconeogenesis (Lorenz et al. 2004). This reprogramming is absent in the benign yeast S. cerevisiae, suggesting that C. albicans has evolved transcriptional programs to match its pathogenic lifestyle (Lorenz and Fink 2001; Lorenz et al. 2004).
Intriguingly, both glycolytic genes (e.g., PFK2, ENO1, PYK1) and glyoxylate cycle genes (e.g., ICL1, MLS1, MDH1) were shown to be induced in C. albicans cells within 20 min of exposure to human blood (Fradin and Hube 2003). At the time, this was surprising because C. albicans was thought to follow the S. cerevisiae paradigm in which cells do not transcribe glycolytic and gluconeogenic genes at the same time (Yin et al. 2003; Lorenz et al. 2004; Rodaki et al. 2009). Indeed, both species strongly down-regulate gluconeogenic and glyoxylate cycle genes in response to glucose concentrations lower than those found in the bloodstream (about 0.07%) (Yin et al. 2003; Rodaki et al. 2009). Yet the C. albicans glyoxylate cycle is required for virulence during systemic infection (Lorenz and Fink 2001). The paradoxical activation of competing carbon metabolism pathways was further complicated by analyses of fungal cells from different infection models. Both mucosal and intraperitoneal infection models were characterized by the simultaneous induction of glycolytic, gluconeogenic, and TCA cycle genes (Thewes et al. 2007; Wilson et al. 2007; Zakikhany et al. 2007). These observations were thought to reflect the heterogeneous nature of fungal populations in complex host microenvironments. For example, phagocytosed cells that are exposed to carbon starvation activate the glyoxylate cycle, whereas nonphagocytosed cells retain access to glucose and activate glycolysis (Brown et al. 2007; Wilson et al. 2007).
To address this paradox regarding the concomitant roles of opposing central metabolic pathways during C. albicans pathogenesis, single cell profiling was performed with specific GFP (green fluorescent protein) fusions to monitor the activation of the relevant metabolic pathways (PCK1, a gluconeogenic-specific enzyme; ICL1, a glyoxylate cycle enzyme; PFK2 and PYK1, two glycolysis-specific enzymes) (Barelle et al. 2006; Miramon et al. 2012). These studies confirmed that gluconeogenesis and glyoxylate cycle were up-regulated in C. albicans following phagocytosis by macrophages or neutrophils, and that these pathways are indeed repressed by physiologically relevant concentrations of glucose in the majority of fungal cells infecting the kidney. These studies also confirmed the heterogeneity of C. albicans cell populations in the kidney and, hence, the complexity of these microenvironments. However, many of these C. albicans cells expressed both glycolytic and gluconeogenic genes. Subsequently, this apparent paradox was resolved with the discovery that the rewiring of ubiquitin targets in central metabolism permits the relaxation of catabolite repression in C. albicans (Sandai et al. 2012).
In A. fumigatus, the isocitrate lyase Icl1, which is part of the glyoxylate cycle, is not required for full virulence (Schobel et al. 2007; Olivas et al. 2008), although this enzyme is constitutively expressed in conidia during germination within macrophages and not in resting conidia (Ebel et al. 2006). C. neoformans also up-regulates Icl1 after phagocytosis by macrophages, although the ICL1 gene is not necessary for full virulence or growth within macrophages (Rude et al. 2002). In these species, Icl1 has been regarded as a marker for lipid utilization, and, hence, these observations suggest that lipid utilization is not required for infection. Similarly, the C. neoformans malate synthase (Mls1) is dispensable for pathogenicity despite its up-regulation during infection (Idnurm et al. 2007; Kronstad et al. 2012). Nevertheless, peroxisomal and mitochondrial fatty acid β-oxidation affects capsule production and is required for the virulence of C. neoformans (Kretschmer et al. 2012). In contrast, fatty acid catabolism does not appear to be required for the virulence of C. albicans as a fox2Δ mutant, which lacks a key enzyme of fatty acid β-oxidation, displays only a minor defect in virulence in the murine model of systemic candidiasis (Piekarska et al. 2006; Ramirez and Lorenz 2007). Although the glyoxylate cycle and fatty acid catabolism might not be virulence determinants for some species, the gluconeogenic pathway might be crucial during the latter stages of infection and for persistence within infected tissues. Indeed, C. neoformans cells recovered from mouse lungs in the later stages of infection displayed increased levels of enzymes involved in gluconeogenesis, glyoxylate cycle, and β-oxidation, indicative of a glucose-limited environment (Hu et al. 2008). Moreover, a C. neoformans pck1Δ mutant, which has a block in gluconeogenesis, displays a virulence defect (Panepinto et al. 2005).
Impact of Carbon Metabolism on Fungal Virulence Factors
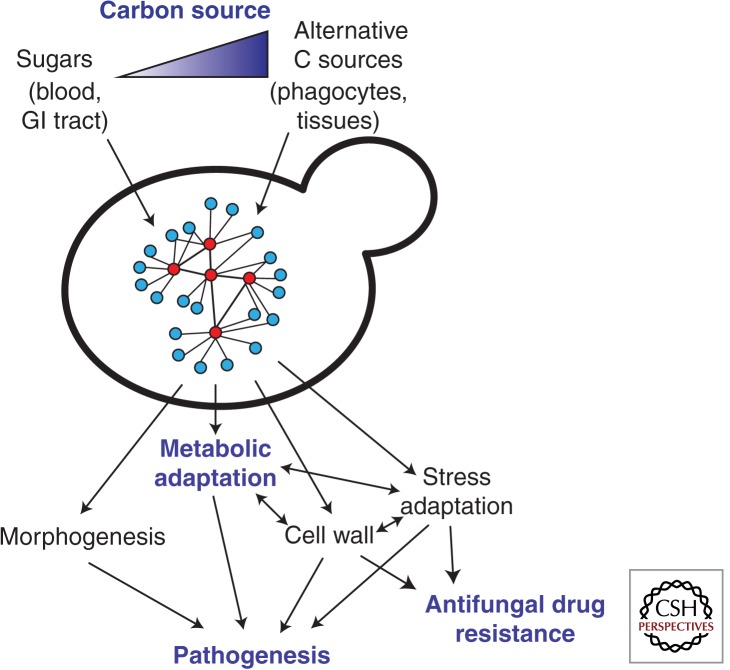
As well as being essential for proliferation in the host, the assimilation of alternative carbon sources can profoundly influence the fitness, physiology, and pathogenicity of fungal pathogens. For example, glucose represses C. neoformans melanization (Zhu and Williamson 2004), and growth on mannitol increases capsule size both in vitro and in vivo, relative to growth on glucose (Guimaraes et al. 2010). Both the capsule and melanization represent important virulence factors in C. neoformans. For C. albicans, growth on lactate rather than glucose modifies the composition and architecture of the cell wall, and, hence, the interaction of C. albicans cells with innate immune cells and host recognition (Ene et al. 2012a,c, 2013). This cell wall remodeling also impacts on adherence, biofilm formation, stress, and drug resistance (Ene et al. 2012a,c). On the other hand, for C. albicans cells grown on lactate, transient exposure to glucose leads to the activation of stress-responsive pathways, increasing their resistance to reactive oxygen species and to an azole antifungal agent (Rodaki et al. 2009). Therefore, changes in carbon source affect many aspects of host–pathogen interactions, and can dramatically impact the virulence of C. albicans and its susceptibility to therapeutic intervention (Fig. 2) (Ene et al. 2012c).
Figure 2.
Candida albicans adapts to changes in carbon sources (C sources) via a complex regulatory network involving sugar signaling and other regulatory modules to tune its metabolism, cell wall architecture, morphology, and stress resistance to the available nutrients (Nantel et al. 2002; Doedt et al. 2004; Hudson et al. 2004; Maidan et al. 2005; Rodaki et al. 2009; Sabina and Brown 2009; Ene et al. 2012c). These changes affect the antifungal drug susceptibility of C. albicans cells and strongly influence their virulence (Ene et al. 2012c) (see text).
An early indication of the intimate links between the regulation of carbon metabolism and virulence was provided by the observation that glycolytic transcript levels are modulated in C. albicans cells undergoing the yeast-hyphal transition, a morphogenetic program that is crucial for the virulence of C. albicans (Swoboda et al. 1994). Also, this morphogenetic transition is influenced by nutritional cues such as glucose and amino acids (Hudson et al. 2004; Maidan et al. 2005; Vylkova et al. 2011). Furthermore, transcript-profiling studies have revealed that, in addition to blocking morphogenesis, the inactivation of the morphogenetic regulator Efg1 has a significant impact on the expression of metabolic genes (Nantel et al. 2002; Doedt et al. 2004). Efg1 induces glycolytic genes while repressing gluconeogenic, TCA cycle, and respiratory functions (Doedt et al. 2004), thereby stimulating fermentative metabolism and repressing respiratory metabolism. Furthermore, a recent report has highlighted the importance of Efg1 during gastrointestinal colonization (Pierce et al. 2013). C. albicans cells that lack Efg1 induce metabolic pathways (carnitine and fatty acid metabolism) that promote hypercolonization of the GI tract in mice (Pierce et al. 2013). Indeed, there is now considerable evidence to suggest that metabolic reprogramming and virulence attributes are integrated by key regulators such as Efg1, Tup1, and Gcn4, which are at the intersections of these regulatory networks (Braun and Johnson 1997; Murad et al. 2001a,b; Tripathi et al. 2002; Doedt et al. 2004).
The white-opaque switch, a developmental program that regulates sexual reproduction in C. albicans, is also accompanied by extensive metabolic rewiring. Glycolytic genes are down-regulated in opaque cells, the mating-competent form of C. albicans, whereas TCA cycle and fatty acid β-oxidation genes are up-regulated (Lan et al. 2002; Tuch et al. 2010). White cells appear to favor fermentative metabolism, whereas opaque cells favor respiratory metabolism, and these programs are presumably associated with their preference for different anatomical niches in the host.
MICRONUTRIENT ASSIMILATION
Microbial survival, persistence, and growth within the host are not only dependent on macronutrients, such as nitrogen and carbon, but also on micronutrients such as trace metals. Among the most important metals are iron, zinc, manganese, and copper, all of which are required for the functionality of many proteins and enzymes. Fungal pathogens must have sufficient access to these essential metals to achieve host colonization, and yet all sterile extracellular niches in humans are essentially devoid of unbound metals. Fungal infections disturb global metal homeostasis in the mammalian host (Potrykus et al. 2013). However, the host has evolved sophisticated mechanisms that restrict microbial access to these metals via a process called “nutritional immunity” (Ganz 2009; Weinberg 2009; Hood and Skaar 2012). Meanwhile, fungal pathogens have evolved elaborate strategies to circumvent this immunity. For example, all studied fungal pathogens can scavenge iron from the host, and this process is essential for their virulence (Almeida et al. 2009; Schrettl and Haas 2011; Kronstad et al. 2012).
The battle over iron during systemic C. albicans infections provides an excellent example of this interplay between pathogen and host (Potrykus et al. 2013). Red blood cell recycling in the spleen is inhibited during systemic candidiasis, leading to the accumulation of heme-associated iron in the kidney. Meanwhile, at a local level, the immune infiltrates surrounding the fungal lesions in the renal cortex impose nutritional immunity thereby limiting the availability of iron to the invading fungus. C. albicans then responds to this host restriction by up-regulating its heme-iron-acquisition mechanisms (Potrykus et al. 2013).
Iron Assimilation
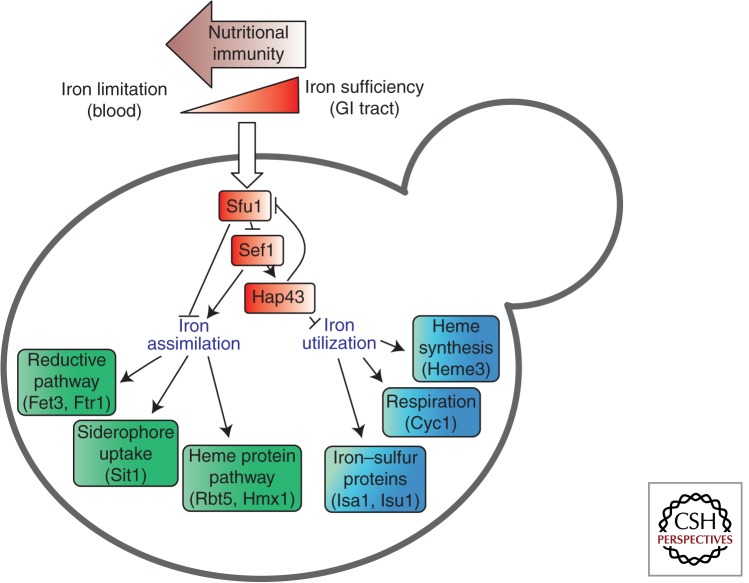
Free iron in human blood is limited to low concentrations (10−24 m), thereby restricting its availability to invading microbes. C. albicans has evolved three main iron-acquisition systems to counteract this restriction (Fig. 3) (Almeida et al. 2009). First, C. albicans does not synthesize siderophores (low molecular mass ferric iron-specific chelators) but it can exploit siderophores synthesized by other microorganisms (xenosiderophores) via the Sit1 transporter—a microbial strategy also described as “iron parasitism” (Heymann et al. 2002). Sit-mediated iron uptake appears universally conserved in the fungal kingdom, having been described in S. cerevisiae, Candida species, Aspergillus species, and C. neoformans (Schrettl and Haas 2011). Second, C. albicans can bind two of the major iron transport and storage proteins of humans: transferrin and ferritin. Transferrin is bound via an unknown receptor (Knight et al. 2005), whereas ferritin is bound via the hypha-associated adhesin and invasin Als3 during invasion into host cells (Almeida et al. 2009). Fungal iron acquisition from host transferrin and ferritin requires the reductive pathway, which is also the main pathway for the uptake of free iron, if available, for example, following host-cell lysis. The reductive pathway requires reductases, oxidases, and iron permeases, each encoded by large gene families (Almeida et al. 2009). These include the high-affinity iron permease gene Ftr1, and the oxidase Fet3. Third, C. albicans can exploit the iron in hemoglobin and other heme proteins. The fungus expresses hemolysins that disrupt red blood cells (Watanabe et al. 1999) and then binds and uses the resultant hemoglobin and heme proteins via the Rbt5/Hmx1 system (Pendrak et al. 2004; Weissman and Kornitzer 2004).
Figure 3.
Candida albicans regulates iron assimilation and iron utilization pathways in response to iron availability within host niches via a regulatory network involving Sfu1, Sef1, and Hap43 (Chen et al. 2011). Host niches differ markedly with respect to iron availability, the local concentrations of free iron within host tissues being reduced by nutritional immunity (Potrykus et al. 2013). The fungus differentially exploits three main iron assimilation pathways depending on the nature of the available iron (Almeida et al. 2009; Potrykus et al. 2013) (see text).
C. glabrata expresses a reductive iron-uptake pathway but is not known to use host ferritin or transferrin as iron sources. Unlike C. albicans, C. glabrata cannot use bacterial siderophores as an iron source (Nevitt and Thiele 2011). However, C. glabrata can use hydroxamate-type xenosiderophores of fungal origin (ferrichrome, ferrirubin, or coprogen) via a Sit1 homolog, and this significantly increases fungal fitness and survival after phagocytosis by macrophages (Nevitt and Thiele 2011).
The macrophage phagosome is characterized by low iron concentrations, and therefore, phagocytosed microbes must rely on internal iron reserves or exploit alternative strategies to scavenge iron and survive within macrophages. H. capsulatum, for example, gains iron via at least three different strategies. First, the fungus inhibits phagosomal acidification thereby promoting the gradual release of iron from transferrin (Eissenberg et al. 1993). Second, H. capsulatum synthesizes and secretes siderophores within macrophages, in contrast to the iron parasitism displayed by C. albicans and C. glabrata (Hwang et al. 2008). Third, H. capsulatum produces surface-bound and secreted enzymes with iron-reducing activity (for example, a secreted γ-glutamyltransferase, Ggt1), which generates an efficient ferric reductant (Zarnowski et al. 2008).
In contrast to the species discussed above, A. fumigatus seems unable to scavenge iron from host proteins. Instead, to acquire iron, this pathogen depends on a low-affinity ferrous iron acquisition system (which exists in other fungi and can also transport zinc) as well as two high-affinity iron uptake systems that involve reductive iron assimilation and siderophore-assisted iron uptake (reviewed in Schrettl and Haas 2011). A. fumigatus excretes two different siderophores, fusarinine C (FsC) and triacetylfusarinine C (TAFC), to mobilize extracellular iron. This pathogen also synthesizes two intracellular siderophores, hyphal ferricrocin (FC) and conidial hydroxyferricrocin (HFC), for the distribution and storage of iron inside the cell (Schrettl et al. 2007; Wallner et al. 2009). In addition, like other fungi, A. fumigatus probably uses vacuolar iron storage, as suggested by the iron-inducible expression of CccA (Schrettl et al. 2008), which is an ortholog of the vacuolar iron importer Ccc1p in S. cerevisiae (Kaplan and Kaplan 2009).
C. neoformans also possesses several complementary iron acquisition strategies. C. neoformans expresses cell surface reductases that reduce ferric iron to its ferrous state, and also exports reductants such as 3-hydroxyanthranilic acid (Nyhus et al. 1997; Jacobson et al. 1998; Nyhus and Jacobson 1999; Kronstad et al. 2012). Cfo1 and Cft1, which are orthologs of Fet3 and Ftr1, are essential for ferric iron uptake and iron acquisition from transferrin. However, they are not required for iron acquisition from heme or the siderophore ferrioxamine. Like C. albicans and C. glabrata, C. neoformans does not produce its own siderophores but expresses a Sit1 transporter that facilitates ferrioxamine B uptake (Tangen et al. 2007). Inactivation of Sit1 does not attenuate the virulence of C. neoformans but inhibits growth under iron-limiting conditions.
Regulation of Fungal Iron Homeostasis
Clearly, evolutionarily diverse fungal pathogens exploit common strategies to scavenge iron from their hosts via parallel pathways that include high-affinity ferroxidase/permease complexes, heme protein utilization, and siderophore uptake systems. However, iron uptake and storage must be tightly regulated because iron is an essential element but excess iron is toxic, generating reactive oxygen species. This is beautifully illustrated by the Sef1-Sfu1 transcriptional circuitry in C. albicans, which differentially controls iron sequestration and iron toxicity in host niches with contrasting iron availabilities. The levels of available iron in the gastrointestinal tract are much higher than in the bloodstream, in which free iron is limited by the host (Martin et al. 1987; Miret et al. 2003). Under iron-limiting conditions (e.g., in the bloodstream), the zinc finger transcription factor Sef1 activates iron assimilation functions and down-regulates the transcriptional repressor Sfu1 (Fig. 3) (Chen et al. 2011). When iron is abundant (e.g., in the gastrointestinal tract), Sfu1 strongly down-regulates Sef1 activity. Sfu1 represses SEF1 expression and inhibits Sef1 functionality by regulating its cellular localization, as well as by promoting Sef1 phosphorylation and destabilization (Chen and Noble 2012). Consequently, Sef1 promotes systemic candidiasis, whereas Sfu1 enhances gastrointestinal commensalism (Chen et al. 2011; Chen and Noble 2012).
An analogous negative-feedback loop regulates iron homeostasis in A. fumigatus. Iron starvation triggers a transcriptional response that is regulated by two interconnected transcription factors, SreA and HapX (Schrettl and Haas 2011). Under conditions of iron sufficiency, SreA represses iron acquisition pathways to limit iron toxicity, and represses the expression of HapX to derepress iron-consuming pathways such as heme biosynthesis, the TCA cycle, and respiration. During iron starvation, HapX represses SreA to up-regulate iron acquisition, and down-regulates iron-consuming pathways such that the available iron is used sparingly. A deficiency in HapX (but not SreA) attenuates the virulence of A. fumigatus, reinforcing the crucial role of iron adaptation mechanisms in the host (Schrettl et al. 2010).
Hap proteins (HapX, Hap3, and Hap5) combine with the Cir1 transcription factor to regulate the response to changing iron levels in C. neoformans (Jung et al. 2006, 2010; Jung and Kronstad 2011). Cir1 is related to C. albicans Sfu1, H. capsulatum Sre1, and Blastomyces dermatitidis SREB (Lan et al. 2004; Chao et al. 2008; Gauthier et al. 2010; Kronstad et al. 2012). Cir1 up-regulates siderophore transport genes via HapX, and down-regulates reductive iron uptake genes. C. neoformans hapX mutants display a minor virulence defect, whereas cir1Δ cells are avirulent (Jung et al. 2006, 2010). The strong virulence defect of C. neoformans cir1Δ mutants could also reflect the roles of Cir1 in the expression of key virulence factors that include growth at 37°C, melanin production, and capsule formation. Cir1 represents a key node in the complex regulatory network, involving HapX, Sre1, Cir1, Tup1, and Gat1, which integrate iron acquisition with critical virulence factors in C. neoformans (Jacobson and Hong 1997; Kronstad et al. 2012). This is further complicated by the influence of ambient pH, which modulates iron homeostasis via the transcriptional regulator Rim101 (Kronstad et al. 2012).
Zinc Assimilation
Zinc is the second most abundant metal in most living organisms and a critical cofactor for many proteins. The mammalian host actively limits zinc availability during bacterial infections via nutritional immunity (Corbin et al. 2008), and the same is probably true during systemic fungal infections. To successfully colonize the host, the fungus must counter this zinc restriction.
C. albicans sequesters host zinc by a “zincophore” system (Citiulo et al. 2012). Analogous to siderophore-mediated iron acquisition, the fungus secretes the zinc-binding protein Pra1 (the “zincophore”), which sequesters the metal from host cells. Pra1 then binds to Zrt1 at the fungal cell surface. Zrt1 is a plasma membrane zinc transporter that mediates the uptake of zinc into the fungal cell. The PRA1 and ZRT1 genes are coregulated in response to zinc availability through their divergent transcription from a common promoter region (Citiulo et al. 2012). The components of this zincophore system, which are essential for C. albicans zinc scavenging during host-cell invasion, are conserved across the fungal kingdom (Wilson et al. 2012). For example, PRA1 and ZRT1 orthologs exist in A. fumigatus (Aspf2 and ZrfC, respectively), and these are organized in an analogous syntenic arrangement, are regulated by environmental zinc levels, and are required for growth under zinc starvation (Amich et al. 2010). However, some fungal pathogens appear to lack a Pra1 orthlog. These species must rely on alternative zinc acquisition systems or rely solely on transporters for zinc uptake (Wilson et al. 2012). For example, C. glabrata apparently lacks both Pra1 and Zrt1. Instead, C. glabrata could acquire zinc via two homologs of the low-affinity S. cerevisiae zinc transporter Zrt2 (Brunke and Hube 2013). Alternatively, other convergently evolved, secreted zinc-binding protein(s) may be deployed. Some fungi may secrete small molecule zinc chelators to sequester this essential metal (Wilson et al. 2012).
Other essential metals such as manganese (Kehl-Fie and Skaar 2010) and copper (Hodgkinson and Petris 2012) may play important roles in host–pathogen interactions. However, their uptake systems in pathogenic fungi remain largely uncharacterized. A putative manganese transporter (Ccc1) and a copper transporter (Ctr1) have been identified in C. albicans (Inglis et al. 2012) but their roles in virulence have not yet been experimentally defined.
CONCLUDING REMARKS
In conclusion, our review has highlighted the fundamental importance of nitrogen, carbon, and micronutrient assimilation for fungal pathogenicity. Clearly fungal pathogens must assimilate the available nutrients within host niches to colonize them. These niches are dynamic, complex, and varied, including the skin, mucosal surfaces, the urogenital and gastrointestinal tracts, the bloodstream, and internal organs. These niches differ with respect to the types and concentrations of available carbon and nitrogen sources, and with regard to micronutrient levels. Fungal pathogens respond by tuning their nutrient acquisition and assimilation mechanisms accordingly (Steen et al. 2003; Lorenz et al. 2004; Fan et al. 2005; Fradin et al. 2005; Kaur et al. 2007; Thewes et al. 2007; Zakikhany et al. 2007; McDonagh et al. 2008; Walker et al. 2009; Wilson et al. 2009; Cairns et al. 2010; Staib et al. 2010; Cheng et al. 2013). In some instances, the host actively reduces the availability of essential nutrients in an attempt to limit fungal colonization. For example, nutritional immunity limits iron availability in the renal cortex during systemic candidiasis, and C. albicans responds by up-regulating its heme-assimilation mechanisms (Potrykus et al. 2013).
Fungal pathogens have evolved to address these metabolic challenges but the evolutionary outcomes often differ for several reasons. First, pathogenicity has evolved independently in different phylogenetic branches of the fungal kingdom. Second, fungal pathogens have been subject to differing evolutionary pressures, possibly over different periods of evolutionary time. Some pathogens, such as A. fumigatus, C. neoformans, and H. capsulatum, have retained significant environmental niches, whereas others, such as C. albicans and Pneumocystis jirovecii, appear to be obligately associated with the host. Third, pathogens differ with respect to their routes of infection. Environmental pathogens such as A. fumigatus, C. neoformans, and H. capsulatum are often inhaled and initially establish pulmonary infections. In contrast, commensal organisms such as C. albicans cause systemic infections when mucosal or gastrointestinal barriers and host immune defenses are compromised. Despite these differences, fungal pathogens display common, evolutionarily ancient strategies to address certain metabolic challenges. These include the tight coordination of iron acquisition and iron-consuming pathways via transcriptional circuitry involving multiple negative-feedback loops, and the global activation of amino acid biosynthesis in response to amino acid starvation via a central transcriptional regulator (Gcn4/CpcA). On the other hand, some conserved metabolic pathways have undergone significant evolutionary rewiring at both transcriptional and posttranscriptional levels during the evolution of fungal pathogens (Ihmels et al. 2005; Martchenko et al. 2007; Askew et al. 2009; Lavoie et al. 2009; Sandai et al. 2012).
The essentiality of nutrient assimilation for fungal pathogenicity represents an opportunity for the development of novel antifungal therapies. Large-scale screens have revealed metabolic targets that might be amenable to pharmaceutical intervention (Roemer et al. 2003), and in some cases, significant differences in assimilation pathways or the catalytic mechanisms of essential mammalian and fungal enzymes enhance their attractiveness as potential antifungal targets (Rodaki et al. 2006; Schrettl and Haas 2011). Recent data have revealed that the susceptibility of fungal pathogens to the currently available antifungal drugs is strongly influenced by nutrient availability. In particular, changes in carbon source affected the resistance of C. albicans cells to azole, polyene, and echinocandin antifungals (Ene et al. 2012c), suggesting that fungal adaptation to different nutrients in host niches affects their susceptibility to therapeutic intervention. Changes in carbon source also affect the stress resistance of C. albicans and exert major effects on the architecture and content of the cell wall, thereby affecting the recognition and killing of C. albicans cells by innate immune cells and the virulence of this pathogen (Rodaki et al. 2009; Ene et al. 2012b,c, 2013).
Although the study of metabolic flexibility and reprogramming has set new paradigms for how microbes adapt to their environment, these processes are fundamental for fungi as they drive both commensalism and infection. These observations highlight the need to investigate the impact of fungal nutrient adaptation on immune recognition and pathogenicity and might help the design of more effective therapeutic strategies.
ACKNOWLEDGMENTS
We are grateful to many colleagues for stimulating discussions, and, in particular, to our friends and colleagues in the European FINSysB Consortium. We apologize to all in the field whose work we were unable to cite because of space limitations. The work of S.B. and B.H. is supported by the European Commission (FINSysB, PITN-GA-2008-214004), the International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS), the excellence graduate school Jena School for Microbial Communication (JSMC), the ERA-NET PathoGenoMics Program (Candicol; BMBF 0315 901 B), the Center for Sepsis Control and Care (CSCC; BMBF 01EO1002), and the Deutsche Forschungsgemeinschaft (DFG Hu 528/15, 16, and 17) including the SPP1580 and the SFB/TR FungiNet. I.V.E. and A.J.P.B. are generously supported by the European Commission (FINSysB, PITN-GA-2008-214004; STRIFE, ERC-2009-AdG-249793), by the UK Biotechnology and Biological Research Council (BBS/B/06679; BB/C510391/1; BB/D009308/1; BB/F000111/1; BB/F010826/1; BB/F00513X/1; BB/K017365/1), and by the Wellcome Trust (080088, 097377).
Footnotes
Editors: Arturo Casadevall, Aaron P. Mitchell, Judith Berman, Kyung J. Kwon-Chung, John R. Perfect, and Joseph Heitman
Additional Perspectives on Human Fungal Pathogens available at www.perspectivesinmedicine.org
REFERENCES
- Almeida RS, Wilson D, Hube B 2009. Candida albicans iron acquisition within the host. FEMS Yeast Res 9: 1000–1012. [DOI] [PubMed] [Google Scholar]
- Amich J, Vicentefranqueira R, Leal F, Calera JA 2010. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot Cell 9: 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson C, Ljungdahl PO 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev 16: 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog 5: e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJP 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol 8: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Morschhauser J 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol 56: 649–669. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJP, Gow NA, Odds FC 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 3: 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105–109. [DOI] [PubMed] [Google Scholar]
- Brown AJP, Odds FC, Gow NA 2007. Infection-related gene expression in Candida albicans. Curr Opin Microbiol 10: 307–313. [DOI] [PubMed] [Google Scholar]
- Brunke S, Hube B 2013. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell Microbiol 15: 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester A, Shelest E, Glockner G, Heddergott C, Schindler S, Staib P, Heidel A, Felder M, Petzold A, Szafranski K, et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol 12: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns T, Minuzzi F, Bignell E 2010. The host-infecting fungal transcriptome. FEMS Microbiol Lett 307: 1–11. [DOI] [PubMed] [Google Scholar]
- Carlson M 1999. Glucose repression in yeast. Curr Opin Microbiol 2: 202–207. [DOI] [PubMed] [Google Scholar]
- Chambergo FS, Bonaccorsi ED, Ferreira AJ, Ramos AS, Ferreira Junior JR, Abrahao-Neto J, Farah JP, El-Dorry H 2002. Elucidation of the metabolic fate of glucose in the filamentous fungus Trichoderma reesei using expressed sequence tag (EST) analysis and cDNA microarrays. J Biol Chem 277: 13983–13988. [DOI] [PubMed] [Google Scholar]
- Chao LY, Marletta MA, Rine J 2008. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 47: 7274–7283. [DOI] [PubMed] [Google Scholar]
- Chen C, Noble SM 2012. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 8: e1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Pande K, French SD, Tuch BB, Noble SM 2011. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10: 118–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Clancy CJ, Xu W, Schneider F, Hao B, Mitchell AP, Nguyen MH 2013. Profiling of Candida albicans gene expression during intra-abdominal candidiasis identifies biologic processes involved in pathogenesis. J Infect Dis 208: 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 8: e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319: 962–965. [DOI] [PubMed] [Google Scholar]
- Crabtree HG 1928. The carbohydrate metabolism of certain pathological overgrowths. Biochem J 22: 1289–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C, Haynes K 2000. Candida glabrata displays pseudohyphal growth. FEMS Microbiol Lett 189: 115–120. [DOI] [PubMed] [Google Scholar]
- Cushion MT 2004. Comparative genomics of Pneumocystis carinii with other protists: Implications for life style. J Eukaryot Microbiol 51: 30–37. [DOI] [PubMed] [Google Scholar]
- Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J 2007. Transcriptome of Pneumocystis carinii during fulminate infection: Carbohydrate metabolism and the concept of a compatible parasite. PLoS ONE 2: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle F, Wachtler B, L’Ollivier C, Holland G, Bannert N, Wilson D, Labruere C, Bonnin A, Hube B 2010. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol 12: 248–271. [DOI] [PubMed] [Google Scholar]
- Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, Brown AJP, Ernst JF 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell 15: 3167–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. 2004. Genome evolution in yeasts. Nature 430: 35–44. [DOI] [PubMed] [Google Scholar]
- Ebel F, Schwienbacher M, Beyer J, Heesemann J, Brakhage AA, Brock M 2006. Analysis of the regulation, expression, and localisation of the isocitrate lyase from Aspergillus fumigatus, a potential target for antifungal drug development. Fungal Genet Biol 43: 476–489. [DOI] [PubMed] [Google Scholar]
- Eissenberg LG, Goldman WE, Schlesinger PH 1993. Histoplasma capsulatum modulates the acidification of phagolysosomes. J Exp Med 177: 1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Cheng SC, Netea MG, Brown AJP 2012a. Growth of Candida albicans cells on the physiologically relevant carbon source, lactate, affects their recognition and phagocytosis by immune cells. Infect Immun 81: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Heilmann CJ, Sorgo AG, Walker LA, de Koster CG, Munro CA, Klis FM, Brown AJP 2012b. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 12: 3164–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Adya AK, Wehmeier S, Brand AC, Maccallum DM, Gow NA, Brown AJP 2012c. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14: 1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Cheng SC, Netea MG, Brown AJP 2013. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun 81: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Kraus PR, Boily MJ, Heitman J 2005. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 4: 1420–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores CL, Rodriguez C, Petit T, Gancedo C 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol Rev 24: 507–529. [DOI] [PubMed] [Google Scholar]
- Fradin C, Hube B 2003. Tissue infection and site-specific gene expression in Candida albicans. Adv Appl Microbiol 53: 271–290. [DOI] [PubMed] [Google Scholar]
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56: 397–415. [DOI] [PubMed] [Google Scholar]
- Frazzitta AE, Vora H, Price MS, Tenor JL, Betancourt-Quiroz M, Toffaletti DL, Cheng N, Perfect JR 2013. Nitrogen source-dependent capsule induction in human-pathogenic Cryptococcus species. Eukaryot Cell 12: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM 1998. Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62: 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM, Gancedo C 1997. Gluconeogenesis and catabolite inactivation. In Yeast sugar metabolism (ed. Zimmermann FK, Entian KD), pp. 359–377 Technomic, Chicago. [Google Scholar]
- Ganz T 2009. Iron in innate immunity: Starve the invaders. Curr Opin Immunol 21: 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C 2004. Candida albicans biofilms: A developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3: 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier GM, Sullivan TD, Gallardo SS, Brandhorst TT, Vanden Wymelenberg AJ, Cuomo CA, Suen G, Currie CR, Klein BS 2010. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog 6: e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW 2009. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun 77: 1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C 2009. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol 47: 465–475. [DOI] [PubMed] [Google Scholar]
- Guimaraes AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD 2010. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol 12: 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, Cairns TC, Olbermann P, Morschhauser J, Bignell EM, Krappmann S 2011. Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol Microbiol 82: 917–935. [DOI] [PubMed] [Google Scholar]
- Hauser PM, Burdet FX, Cisse OH, Keller L, Taffe P, Sanglard D, Pagni M 2010. Comparative genomics suggests that the fungal pathogen Pneumocyctis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS ONE 5: e15152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J 2011. Microbial pathogens in the fungal kingdom. Fungal Biol Rev 25: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF 2002. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun 70: 5246–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev 52: 248–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Natarajan K 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell 1: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson V, Petris MJ 2012. Copper homeostasis at the host-pathogen interface. J Biol Chem 287: 13549–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Valerius O, Andermann M, Braus GH 2001. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol Biol Cell 12: 2846–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Skaar EP 2012. Nutritional immunity: Transition metals at the pathogen-host interface. Nat Rev Microbiol 10: 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Regelmann J, Wolf DH 2002. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem 277: 8248–8254. [DOI] [PubMed] [Google Scholar]
- Horvath P, Nosanchuk JD, Hamari Z, Vagvolgyi C, Gacser A 2012. The identification of gene duplication and the role of secreted aspartyl proteinase 1 in Candida parapsilosis virulence. J Infect Dis 205: 923–933. [DOI] [PubMed] [Google Scholar]
- Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW 2008. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol 69: 1456–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B, Monod M, Schofield DA, Brown AJP, Gow NA 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol 14: 87–99. [DOI] [PubMed] [Google Scholar]
- Hube B, Sanglard D, Odds FC, Hess D, Monod M, Schafer W, Brown AJP, Gow NA 1997. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun 65: 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DA, Sciascia QL, Sanders RJ, Norris GE, Edwards PJ, Sullivan PA, Farley PC 2004. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150: 3041–3049. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Mayfield JA, Rine J, Sil A 2008. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog 4: e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Giles SS, Perfect JR, Heitman J 2007. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot Cell 6: 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309: 938–940. [DOI] [PubMed] [Google Scholar]
- Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G 2012. The Candida genome database incorporates multiple Candida species: Multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40: D667–D674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Voorhies M, Hocking Murray DR, Sil A 2013. Comparative transcriptomics of infectious spores from the fungal pathogen Histoplasma capsulatum reveals a core set of transcripts that specify infectious and pathogenic states. Eukaryot Cell 12: 828–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson ES, Hong JD 1997. Redox buffering by melanin and Fe(II) in Cryptococcus neoformans. J Bacteriol 179: 5340–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson ES, Goodner AP, Nyhus KJ 1998. Ferrous iron uptake in Cryptococcus neoformans. Infect Immun 66: 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Lopez C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, Lorenz MC 2013. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell 12: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet 15: 29–33. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. 2004. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci 101: 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Kronstad JW 2011. Iron influences the abundance of the iron regulatory protein Cir1 in the fungal pathogen Cryptococcus neoformans. FEBS Lett 585: 3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4: e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW 2010. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog 6: e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Kaplan J 2009. Iron acquisition and transcriptional regulation. Chem Rev 109: 4536–4552. [DOI] [PubMed] [Google Scholar]
- Kaur R, Domergue R, Zupancic ML, Cormack BP 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol 8: 378–384. [DOI] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci 104: 7628–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie TE, Skaar EP 2010. Nutritional immunity beyond iron: A role for manganese and zinc. Curr Opin Chem Biol 14: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Olsson L, Nielsen J 1998. Glucose control in Saccharomyces cerevisiae: The role of Mig1 in metabolic functions. Microbiology 144: 13–24. [DOI] [PubMed] [Google Scholar]
- Knight SA, Vilaire G, Lesuisse E, Dancis A 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun 73: 5482–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraidlova L, Van Zeebroeck G, Van Dijck P, Sychrova H 2011. The Candida albicans GAP gene family encodes permeases involved in general and specific amino acid uptake and sensing. Eukaryot Cell 10: 1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S, Bignell EM, Reichard U, Rogers T, Haynes K, Braus GH 2004. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol Microbiol 52: 785–799. [DOI] [PubMed] [Google Scholar]
- Kretschmer M, Wang J, Kronstad JW 2012. Peroxisomal and mitochondrial β-oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 11: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, Geddes JM, Griffiths EJ, Choi J, Cadieux B, et al. 2012. Adaptation of Cryptococcus neoformans to mammalian hosts: Integrated regulation of metabolism and virulence. Eukaryot Cell 11: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N 2002. Metabolic specialization associated with phenotypic switching in Candidaalbicans. Proc Natl Acad Sci 99: 14907–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N 2004. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol 53: 1451–1469. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Whiteway M 2009. Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr Opin Microbiol 12: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Stead DA, Argo E, MacCallum DM, Brown AJP 2011. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol 79: 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl PO 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem Soc Trans 37: 242–247. [DOI] [PubMed] [Google Scholar]
- Lorenz MC 2013. Carbon catabolite control in Candida albicans: New wrinkles in metabolism. MBio 4: e00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR 2001. The glyoxylate cycle is required for fungal virulence. Nature 412: 83–86. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3: 1076–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Sano M, Maruyama Y, Tanno T, Akao T, Totsuka Y, Endo M, Sakurada R, Yamagata Y, Machida M, et al. 2004. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expressed sequence tags. Appl Microbiol Biotechnol 65: 74–83. [DOI] [PubMed] [Google Scholar]
- Maidan MM, Thevelein JM, Van Dijck P 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem Soc Trans 33: 291–293. [DOI] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol 17: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RB, Savory J, Brown S, Bertholf RL, Wills MR 1987. Transferrin binding of Al3+ and Fe3+. Clin Chem 33: 405–407. [PubMed] [Google Scholar]
- Martinez P, Ljungdahl PO 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol Cell Biol 25: 9435–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Jacobsen ID, Miramon P, Grosse K, Hube B 2012. The novel Candida albicans transporter Dur31 is a multi-stage pathogenicity factor. PLoS Pathog 8: e1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Hube B 2013. Candida albicans pathogenicity mechanisms. Virulence 4: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, et al. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiller TF, Hube B, Schild L, Shirtliff ME, Scheper MA, Winkler R, Ton A, Jabra-Rizk MA 2009. A novel immune evasion strategy of Candida albicans: Proteolytic cleavage of a salivary antimicrobial peptide. PLoS ONE 4: e5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramon P, Dunker C, Windecker H, Bohovych IM, Brown AJP, Kurzai O, Hube B 2012. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS ONE 7: e52850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramon P, Kasper L, Hube B 2013. Thriving within the host: Candida spp interactions with phagocytic cells. Med Microbiol Immunol 202: 183–195. [DOI] [PubMed] [Google Scholar]
- Miret S, Simpson RJ, McKie AT 2003. Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr 23: 283–301. [DOI] [PubMed] [Google Scholar]
- Monod M, Jaton-Ogay K, Reichard U 1999. Aspergillus fumigatus-secreted proteases as antigenic molecules and virulence factors. Contrib Microbiol 2: 182–192. [DOI] [PubMed] [Google Scholar]
- Murad AM, d’Enfert C, Gaillardin C, Tournu H, Tekaia F, Talibi D, Marechal D, Marchais V, Cottin J, Brown AJP 2001a. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol Microbiol 42: 981–993. [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, et al. 2001b. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J 20: 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67: 400–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, et al. 2008. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 154: 3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van het Hoog M, Gordon P, et al. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell 13: 3452–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt T, Thiele DJ 2011. Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog 7: e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: 1151–1156. [DOI] [PubMed] [Google Scholar]
- Niimi M, Kamiyama A, Tokunaga M 1988. Respiration of medically important Candida species and Saccharomyces cerevisiae in relation to glucose effect. J Med Vet Mycol 26: 195–198. [PubMed] [Google Scholar]
- Nyhus KJ, Jacobson ES 1999. Genetic and physiologic characterization of ferric/cupric reductase constitutive mutants of Cryptococcus neoformans. Infect Immun 67: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhus KJ, Wilborn AT, Jacobson ES 1997. Ferric iron reduction by Cryptococcus neoformans. Infect Immun 65: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC 1988. Candida and candidosis. Bailliere Tindall, London. [Google Scholar]
- Olivas I, Royuela M, Romero B, Monteiro MC, Minguez JM, Laborda F, De Lucas JR 2008. Ability to grow on lipids accounts for the fully virulent phenotype in neutropenic mice of Aspergillus fumigatus null mutants in the key glyoxylate cycle enzymes. Fungal Genet Biol 45: 45–60. [DOI] [PubMed] [Google Scholar]
- Palmer GE, Kelly MN, Sturtevant JE 2007. Autophagy in the pathogen Candida albicans. Microbiology 153: 51–58. [DOI] [PubMed] [Google Scholar]
- Palmer GE, Askew DS, Williamson PR 2008. The diverse roles of autophagy in medically important fungi. Autophagy 4: 982–988. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Orbach MJ, Legerton TL, Yanofsky C 1988. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci 85: 3728–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Liu L, Ramos J, Zhu X, Valyi-Nagy T, Eksi S, Fu J, Jaffe HA, Wickes B, Williamson PR 2005. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest 115: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendrak ML, Chao MP, Yan SS, Roberts DD 2004. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to α-biliverdin. J Biol Chem 279: 3426–3433. [DOI] [PubMed] [Google Scholar]
- Piekarska K, Mol E, van den Berg M, Hardy G, van den Burg J, van Roermund C, MacCallum D, Odds F, Distel B 2006. Peroxisomal fatty acid β-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell 5: 1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JV, Dignard D, Whiteway M, Kumamoto CA 2013. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot Cell 12: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus J, Stead D, Maccallum DM, Urgast DS, Raab A, van Rooijen N, Feldmann J, Brown AJP 2013. Fungal iron availability during deep seated candidiasis is defined by a complex interplay involving systemic and local events. PLoS Pathog 9: e1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. MBio 2: e00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MA, Lorenz MC 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell 6: 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, Thumm M, Wolf DH 2003. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: A genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell 14: 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Morschhauser J 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol Microbiol 60: 795–812. [DOI] [PubMed] [Google Scholar]
- Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, Rhodes JC, Askew DS 2007. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell 6: 2437–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaki A, Young T, Brown AJP 2006. Effects of depleting the essential central metabolic enzyme fructose-1,6-bisphosphate aldolase on the growth and viability of Candida albicans: Implications for antifungal drug target discovery. Eukaryot Cell 5: 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaki A, Bohovych IM, Enjalbert B, Young T, Odds FC, Gow NA, Brown AJP 2009. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol Biol Cell 20: 4845–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50: 167–181. [DOI] [PubMed] [Google Scholar]
- Roetzer A, Gratz N, Kovarik P, Schuller C 2010. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol 12: 199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A, Gabaldon T, Schuller C 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: Important adaptations for an opportunistic pathogen. FEMS Microbiol Lett 314: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci 100: 11007–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rude TH, Toffaletti DL, Cox GM, Perfect JR 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun 70: 5684–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina J, Brown V 2009. Glucose sensing network in Candida albicans: A sweet spot for fungal morphogenesis. Eukaryot Cell 8: 1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, Bohovych I, Ene IV, Kastora S, Budge S, et al. 2012. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. MBio 3: e00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel F, Ibrahim-Granet O, Ave P, Latgé JP, Brakhage AA, Brock M 2007. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect Immun 75: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork SM, Thumm M, Wolf DH 1995. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem 270: 26446–26450. [DOI] [PubMed] [Google Scholar]
- Schrettl M, Haas H 2011. Iron homeostasis—Achilles’ heel of Aspergillus fumigatus? Curr Opin Microbiol 14: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, Wallner A, Arst HN Jr, Haynes K, Haas H 2007. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, Werner ER, Jacobsen I, Illmer P, Yi H, et al. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol 70: 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, et al. 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 6: e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule T, Rose M, Entian KD, Thumm M, Wolf DH 2000. Ubc8p functions in catabolite degradation of fructose-1, 6-bisphosphatase in yeast. EMBO J 19: 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seider K, Heyken A, Luttich A, Miramon P, Hube B 2010. Interaction of pathogenic yeasts with phagocytes: Survival, persistence and escape. Curr Opin Microbiol 13: 392–400. [DOI] [PubMed] [Google Scholar]
- Seider K, Brunke S, Schild L, Jablonowski N, Wilson D, Majer O, Barz D, Haas A, Kuchler K, Schaller M, et al. 2011. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol 187: 3072–3086. [DOI] [PubMed] [Google Scholar]
- Sil A 2006. Using genomics to study the life cycle of Histoplasma capsulatum. In Fungal genomics—The mycota (ed. Brown AJ), pp. 221–236 Springer, Berlin. [Google Scholar]
- Staib P, Zaugg C, Mignon B, Weber J, Grumbt M, Pradervand S, Harshman K, Monod M 2010. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology 156: 884–895. [DOI] [PubMed] [Google Scholar]
- Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, Perfect JR, Kronstad J 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell 2: 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda RK, Bertram G, Delbruck S, Ernst JF, Gow NA, Gooday GW, Brown AJP 1994. Fluctuations in glycolytic mRNA levels during morphogenesis in Candida albicans reflect underlying changes in growth and are not a response to cellular dimorphism. Mol Microbiol 13: 663–672. [DOI] [PubMed] [Google Scholar]
- Sychrova H, Souciet JL 1994. CAN1, a gene encoding a permease for basic amino acids in Candida albicans. Yeast 10: 1647–1651. [DOI] [PubMed] [Google Scholar]
- Tangen KL, Jung WH, Sham AP, Lian T, Kronstad JW 2007. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 153: 29–41. [DOI] [PubMed] [Google Scholar]
- Tavares AH, Silva SS, Dantas A, Campos EG, Andrade RV, Maranhao AQ, Brigido MM, Passos-Silva DG, Fachin AL, Teixeira SM, et al. 2007. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect 9: 583–590. [DOI] [PubMed] [Google Scholar]
- Tekaia F, Latgé JP 2005. Aspergillus fumigatus: Saprophyte or pathogen? Curr Opin Microbiol 8: 385–392. [DOI] [PubMed] [Google Scholar]
- Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B 2007. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol 63: 1606–1628. [DOI] [PubMed] [Google Scholar]
- Tournu H, Tripathi G, Bertram G, Macaskill S, Mavor A, Walker L, Odds FC, Gow NA, Brown AJP 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot Cell 4: 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJP 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J 21: 5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet 6: e1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 10: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Matsumoto Y, Uno J, Sasamoto K, Sekimizu K, Kinjo Y, Chibana H 2011. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE 6: e24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira N, Casal M, Johansson B, MacCallum DM, Brown AJP, Paiva S 2010. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol Microbiol 75: 1337–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A 2007. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect Immun 75: 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2: e00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LA, Maccallum DM, Bertram G, Gow NA, Odds FC, Brown AJP 2009. Genome-wide analysis of Candida albicans gene expression patterns during infection of the mammalian kidney. Fungal Genet Biol 46: 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner A, Blatzer M, Schrettl M, Sarg B, Lindner H, Haas H 2009. Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus. Appl Environ Microbiol 75: 4194–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takano M, Murakami M, Tanaka H, Matsuhisa A, Nakao N, Mikami T, Suzuki M, Matsumoto T 1999. Characterization of a haemolytic factor from Candida albicans. Microbiology 145: 689–694. [DOI] [PubMed] [Google Scholar]
- Weinberg ED 2009. Iron toxicity: New conditions continue to emerge. Oxid Med Cell Longev 2: 107–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol 53: 1209–1220. [DOI] [PubMed] [Google Scholar]
- Wilson D, Tutulan-Cunita A, Jung W, Hauser NC, Hernandez R, Williamson T, Piekarska K, Rupp S, Young T, Stateva L 2007. Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol Microbiol 65: 841–856. [DOI] [PubMed] [Google Scholar]
- Wilson D, Thewes S, Zakikhany K, Fradin C, Albrecht A, Almeida R, Brunke S, Grosse K, Martin R, Mayer F, et al. 2009. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res 9: 688–700. [DOI] [PubMed] [Google Scholar]
- Wilson D, Citiulo F, Hube B 2012. Zinc exploitation by pathogenic fungi. PLoS Pathog 8: e1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Wolfe KH 2005. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet 37: 777–782. [DOI] [PubMed] [Google Scholar]
- Xie X, Wilkinson HH, Correa A, Lewis ZA, Bell-Pedersen D, Ebbole DJ 2004. Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurosporacrassa. Fungal Genet Biol 41: 1104–1119. [DOI] [PubMed] [Google Scholar]
- Yin Z, Wilson S, Hauser NC, Tournu H, Hoheisel JD, Brown AJP 2003. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol Microbiol 48: 713–724. [DOI] [PubMed] [Google Scholar]
- Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, McLnerney T, Gaskell S, Oliver SG, Cash P, Brown AJP 2004. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics 4: 2425–2436. [DOI] [PubMed] [Google Scholar]
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B 2007. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 9: 2938–2954. [DOI] [PubMed] [Google Scholar]
- Zarnowski R, Cooper KG, Brunold LS, Calaycay J, Woods JP 2008. Histoplasma capsulatum secreted γ-glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol Microbiol 70: 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg C, Monod M, Weber J, Harshman K, Pradervand S, Thomas J, Bueno M, Giddey K, Staib P 2009. Gene expression profiling in the human pathogenic dermatophyte Trichophyton rubrum during growth on proteins. Eukaryot Cell 8: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Takaya N, Nakamura A, Yamaguchi M, Takeo K, Shoun H 2002. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J Biol Chem 277: 1892–1896. [DOI] [PubMed] [Google Scholar]
- Zhu X, Williamson PR 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res 5: 1–10. [DOI] [PubMed] [Google Scholar]