Abstract
Meiotic recombination involves the formation and repair of programmed DNA double-strand breaks (DSBs) catalyzed by the conserved Spo11 protein. This review summarizes recent studies pertaining to the formation of meiotic DSBs, including the mechanism of DNA cleavage by Spo11, proteins required for break formation, and mechanisms that control the location, timing, and number of DSBs. Where appropriate, findings in different organisms are discussed to highlight evolutionary conservation or divergence.
Meiotic recombination is initiated by DNA double-strand breaks, which are catalyzed by Spo11. Recent studies have shed light on Spo11-mediated break formation and how the number, location, and timing of breaks are controlled.
Meiosis is a specialized cell division that generates haploid cells—spores in yeasts, gametes in multicellular organisms—from diploid progenitors. Halving the genetic complement occurs through one round of replication followed by two rounds of chromosome segregation separating homologs (meiosis I), then sister chromatids (meiosis II). A prominent feature of meiosis I is recombination, comprising the formation and repair of programmed DNA double-strand breaks (DSBs). Recombination supports faithful homolog segregation and reshuffles maternal and paternal alleles, thereby increasing genetic diversity in progeny (Handel and Schimenti 2010; Székvölgyi and Nicolas 2010).
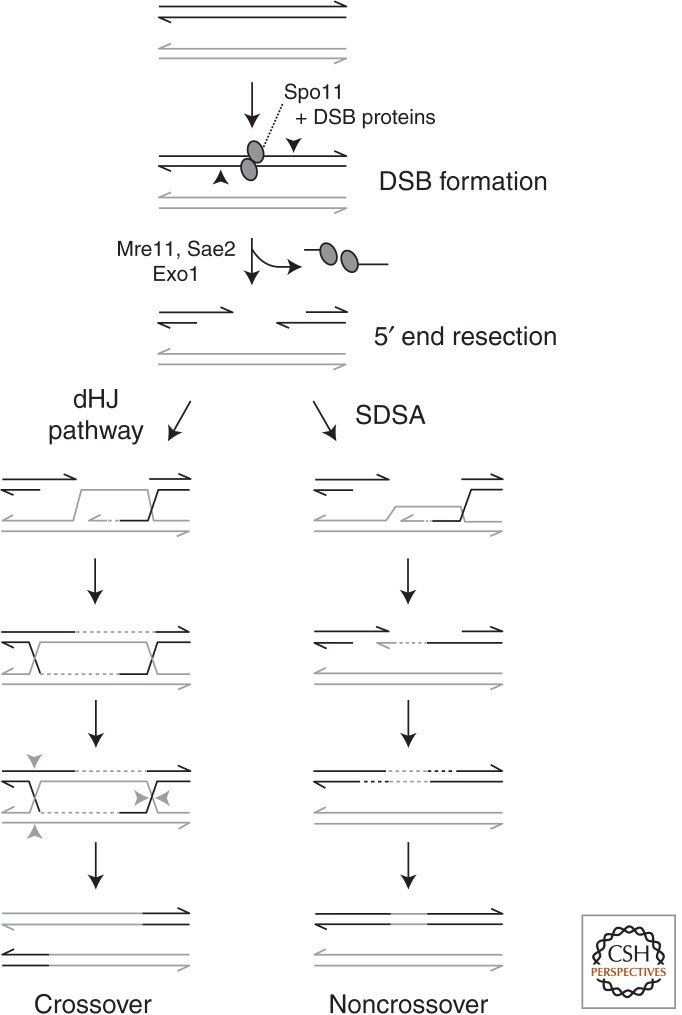
DSBs are formed in prophase I by the conserved Spo11 protein (Fig. 1) (Szostak et al. 1983; Sun et al. 1989; Cao et al. 1990; Bergerat et al. 1997; Keeney et al. 1997). Spo11 remains covalently linked to the 5′ terminus of each broken DNA strand, but is eventually released by nearby endonucleolytic cleavage, likely Mre11 endonuclease and/or Sae2, followed by 3′ to 5′ resection toward the DSB by Mre11 exonuclease activity (de Massy et al. 1995; Keeney and Kleckner 1995; Liu et al. 1995; Neale et al. 2005; Zakharyevich et al. 2010; Garcia et al. 2011). DNA ends are then resected 5′ to 3′ by Exo1 exonuclease to expose 3′ single-stranded tails (Sun et al. 1991; Zakharyevich et al. 2010). Members of the RecA family of strand exchange proteins (Dmc1, Rad51) bind these tails, forming nucleoprotein filaments that catalyze strand invasion into homologous duplex DNA (Chen et al. 2008; San Filippo et al. 2008). In meiosis, some DSBs are repaired via the sister chromatid but recombination occurs most often between homologs, in keeping with the importance of recombination for promoting homolog pairing and segregation (Schwacha and Kleckner 1994; Goldfarb and Lichten 2010).
Figure 1.
The meiotic recombination pathway. A segment of one sister chromatid from each homolog (black, gray) is shown. Spo11 (ovals) catalyzes DSB formation, in association with partner proteins. Endonucleolytic cleavage on either side of the DSB (black arrowheads) releases Spo11 covalently attached to a short oligonucleotide. The DNA ends undergo 5′-to-3′ resection. A 3′ ssDNA tail invades a homologous duplex DNA and initiates repair synthesis. Repair can proceed by either a double Holliday junction (dHJ) pathway, or synthesis-dependent strand annealing (SDSA). In the dHJ pathway, the second end of the DSB is captured to form a dHJ, and its resolution primarily gives rise to crossover recombinants. Only one cleavage pattern for dHJ resolution is shown (gray arrowheads). In SDSA, the invading 3′ strand is displaced after DNA synthesis and reanneals to the other 3′ end of the DSB, followed by further DNA synthesis and nick ligation, ultimately giving rise to noncrossover recombinant products.
Recombination can yield reciprocal exchange of chromosome arms flanking the DSB site (crossovers, COs), or no exchange (noncrossovers, NCO) (Hunter 2007; Serrentino and Borde 2012). Most COs are thought to arise through a double Holliday junction (dHJ) intermediate, whereas most NCOs are formed primarily by synthesis-dependent strand annealing (SDSA) (Fig. 1) (Allers and Lichten 2001; Cromie and Smith 2007b; McMahill et al. 2007). In the dHJ pathway, initial strand invasion is followed by capture of the second DSB end, forming a double Holliday junction that is resolved to generate primarily COs. In SDSA, the invading strand is extended by DNA synthesis, but is then displaced and anneals to the other DSB end.
In many species, the homology search accompanying recombination promotes recognition and pairing of homologs (Burgess 2002; Bhalla and Dernburg 2008). Those events that become COs then provide physical linkages between homologs, which, combined with sister chromatid cohesion, ensure correct homolog orientation on the meiotic spindle and proper segregation in meiosis I. Depending on the species, absence of recombination or COs results in randomized chromosome segregation, gamete aneuploidy, meiotic arrest, and/or apoptosis (Székvölgyi and Nicolas 2010).
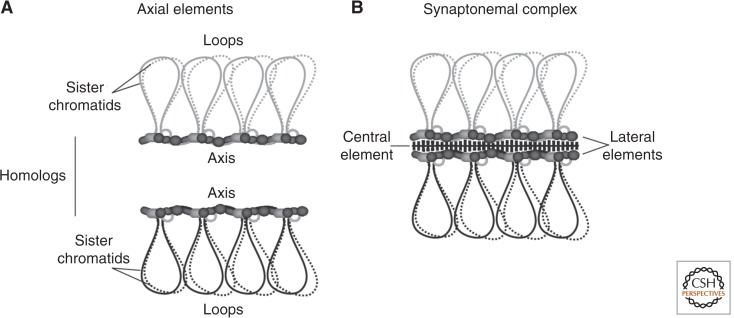
DSB formation and recombination are tightly integrated with higher-order chromosome structure. Pairs of sister chromatids are organized into a series of loops (∼10–20 kb in budding yeast) anchored at their bases along a structural axis called the axial element (Fig. 2A) (Kleckner 1996, 2006; Zickler and Kleckner 1999). At the pachytene stage, homologous chromosomes are held together along their lengths by a tripartite structure called the synaptonemal complex (SC) (Fig. 2B). The SC comprises two lateral elements (formerly the axial element of each homolog) held together by transverse filaments (Zickler and Kleckner 1999). Axial elements are enriched with several protein components, which in budding yeast include Red1, Hop1, and cohesin complexes (Smith and Roeder 1997; Klein et al. 1999; Panizza et al. 2011). Axis proteins are required for normal levels of meiotic DSBs, with DSBs reduced to ∼5%–25% of wild-type levels in hop1 and red1 mutant Saccharomyces cerevisiae (Mao-Draayer et al. 1996; Woltering et al. 2000; Blat et al. 2002; Niu et al. 2005; Carballo et al. 2008).
Figure 2.
Meiotic chromosome organization. (A) Meiotic chromosomes are organized into a series of chromatin loops anchored at their bases by proteinaceous axial elements. (B) At the zygotene stage of prophase I, homologs start to synapse, with the homologous axial elements coming together to form the lateral elements of the synaptonemal complex (SC). The lateral elements are held together by transverse filaments, which, together with central element proteins, make up the central region of the SC. SC formation is completed by the pachytene stage.
MEIOTIC DSB FORMATION BY SPO11
DSB formation by Spo11 orthologs appears to be a universal feature of meiotic recombination initiation in fungi, invertebrates, mammals, and plants (Keeney 2001, 2007). A single gene encodes Spo11 in most organisms, but many plants have multiple SPO11 homologs, of which at least one functions in meiotic recombination (Edlinger and Schlögelhofer 2011).
Biochemical studies of Spo11 have been hampered by difficulties in isolating soluble protein, and appropriate catalytic activity (transesterase, not nuclease) was lacking in reported instances of Spo11 purification (Wu et al. 2004; Shingu et al. 2010). However, Spo11 is homologous with Top6A, the catalytic subunit of archaeal topoisomerase VI, a type II DNA topoisomerase (Bergerat et al. 1997; Keeney et al. 1997). Two structural domains are presumed to be part of the active site of type II topoisomerases: a 5Y-CAP motif containing the catalytic tyrosine and a Toprim domain with a metal-binding pocket (Berger et al. 1998; Nichols et al. 1999; Keeney 2001, 2007). Top6A acts as a dimer with two hybrid active sites for DNA cleavage, each comprising the 5Y-CAP and Toprim domains of separate monomers (Nichols et al. 1999; Keeney 2001).
Structure–function analyses in S. cerevisiae, Schizosaccharomyces pombe (Rec12), and Arabidopsis thaliana (SPO11-1), motivated by the crystal structure of Top6A from Methanococcus jannaschii, strongly support the hypothesis that Spo11 catalyzes meiotic DSB formation via a topoisomerase II–like mechanism (Nichols et al. 1999; Diaz et al. 2002; Kan et al. 2010; Shingu et al. 2010). Thus, Spo11 likely dimerizes and cleaves DNA in a transesterification reaction, resulting in phosphodiester links between the active site tyrosines of the Spo11 protomers with the 5′ DSB ends. DNA cleavage yields a two-nucleotide 5′ overhang (Liu et al. 1995).
OTHER PROTEINS REQUIRED FOR DSB FORMATION
Spo11 is not sufficient for DSB formation in vivo, as essential partners (referred to as DSB proteins) have been identified in many organisms (Table 1). In S. cerevisiae, this cast includes nine proteins that interact directly or indirectly with Spo11 (Keeney 2001, 2007). Null mutants in any of these fail to form DSBs and show reduced sporulation and severely reduced spore viability as a consequence of chromosome missegregation. Some DSB proteins are conserved across phyla, whereas others appear to be unique to species within a narrowly defined clade, or their sequences have diverged to the point of concealing obvious homology. Most meiotic proteins undergo rapid evolutionary divergence (Richard et al. 2005; Keeney 2007); this hinders homology detection, exemplified by Mei4 and Rec114 orthologs not being recognized outside of ascomycetes until two decades after their initial discovery in S. cerevisiae (Kumar et al. 2010).
Table 1.
List of proteins required for meiotic DSB formation in different organisms
| S. cerevisiae | S. pombe | Mus musculus | C. elegans | D. melanogaster | A. thaliana |
|---|---|---|---|---|---|
| Mei4 | Rec24 | MEI4 | PRD2 | ||
| Mer2 | Rec15 | ||||
| Mre11 | Rad32a | MRE11b | MRE-11 | MRE11a | |
| Rad50 | Rad50a | RAD50b | RAD-50a | RAD50a | |
| Rec102 | |||||
| Rec104 | |||||
| Rec114 | Rec7 | REC114 | PHS1a | ||
| Ski8 | Rec14 | WDR61b | SKI8/VIP3a | ||
| Spo11 | Rec12 | SPO11 | SPO11 | Mei-W68 | SPO11-1, SPO11-2, SPO11-3a |
| Xrs2 | Nbs1a | NBS1b | |||
| Mde2 | |||||
| Rec6 | |||||
| MEI1 | PRD1 | ||||
| DSB-1 | |||||
| DSB-2a | |||||
| Mei-P22 | |||||
| Trem | |||||
| PRD3 | |||||
| SWI1 | |||||
| DFO | |||||
| Red1a | Rec10 | ||||
| Hop1a | Hop1a | HORMAD1a | HIM-3a HTP-1a HTP-2a HTP-3a |
ASY1a |
aNot essential for meiotic DSB formation.
bRole in DSB formation is not known.
In other instances, homologs of S. cerevisiae DSB proteins in other species are not functionally conserved. For example, the Mre11–Rad50–Xrs2 (MRX) complex is required for DSB formation in S. cerevisiae, but not S. pombe, Coprinus cinereus, Drosophila melanogaster, or A. thaliana, although its role in meiotic DSB repair is conserved (Borde 2007; Keeney 2007). In Caenorhabditis elegans, the MRX complex is needed for DSB formation in otherwise wild-type backgrounds, as in yeast (Chin and Villeneuve 2001; Alpi et al. 2003; Goodyer et al. 2008), but not in a rec-8 or smc-3 mutant background (Hayashi et al. 2007; Baudrimont et al. 2011). In a similar vein, Ski8 is essential for DSB formation in S. cerevisiae, S. pombe, and Sordaria macrospora, but not A. thaliana (Evans et al. 1997; Gardiner et al. 1997; Tessé et al. 2003; Arora et al. 2004; Jolivet et al. 2006).
Thus, although the catalytic entity initiating meiotic recombination is highly conserved, the other proteins involved and/or the molecular processes that lead to break formation may be more diverged. The precise molecular function of DSB proteins has been a longstanding question, but recent findings are providing a clearer understanding. Most DSB proteins have been extensively reviewed elsewhere (Keeney 2001, 2007; Hunter 2007; Cole et al. 2010; Edlinger and Schlögelhofer 2011), so we emphasize newer findings below.
DSB Proteins in S. cerevisiae
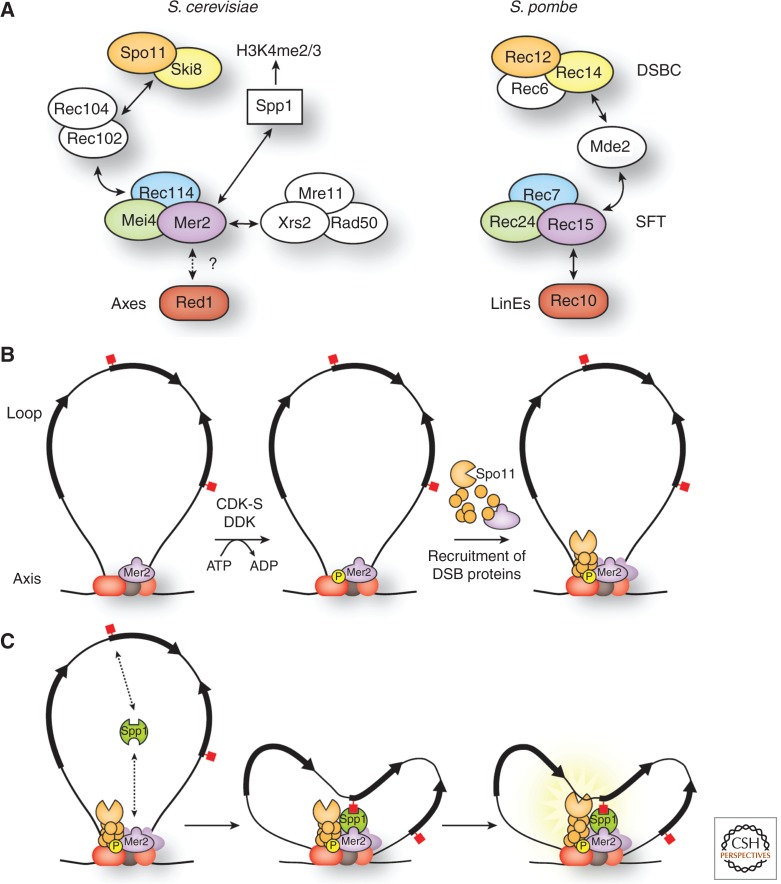
The 10 proteins required for DSB formation in S. cerevisiae behave functionally as four interacting subgroups or subcomplexes (Spo11–Ski8, Rec102–Rec104, Rec114–Mei4–Mer2, and Mre11–Rad50–Xrs2) (Fig. 3A) (Keeney 2007; Maleki et al. 2007). Besides the catalytic role of Spo11 and the post-DSB role of the MRX complex in DNA resection and repair, the biochemical roles of DSB proteins are not clear. Why does the absence of any one of them prevent Spo11 from forming DSBs? Proposed functions include recruiting Spo11 to specific sites, activating Spo11 catalytic activity, and coordinating DSB formation with chromatin and higher-order chromosome structure (Keeney 2007). Recent findings support the latter, and point toward temporal and spatial regulation of Spo11 cleavage by coordinating DSB formation with replication.
Figure 3.
Interaction maps of DSB proteins in S. cerevisiae and S. pombe, and models for roles of DSB proteins in S. cerevisiae. (A) In S. cerevisiae (left), the 10 proteins required for DSB formation form four interacting subcomplexes (see text for details). Mer2 also interacts with Spp1, which recognizes and binds to H3K4me2/me3 marks on chromatin loops. Mer2 and other DSB proteins localize to the chromosome axes, but it is not known whether this is via interaction with the axial element protein Red1 (dashed arrows and question mark), analogous to the interaction of their homologous proteins in S. pombe. In S. pombe (right), the seven proteins required for DSB formation form two subcomplexes (DSBC and SFT) that interact via Mde2. Rec15 interacts with Rec10, a component of linear elements (LinEs) similar to axial elements in S. cerevisiae. Homologous proteins are shown in the same color for the two species; proteins with no known homologs in the other species are shown in white (except for Spp1, whose homolog in S. pombe is not shown, and whose role in DSB formation is not known). (B) Model for DSB regulation via Mer2 phosphorylation (based on data in Henderson et al. 2006; Sasanuma et al. 2008; Wan et al. 2008; Panizza et al. 2011). Mer2 phosphorylation by the replication-associated kinases CDK-S and DDK leads to recruitment of DSB proteins that directly interact with Mer2 (Rec114, Mei4, Xrs2), and perhaps subsequently also other DSB proteins (Rec102, Rec104, Ski8, Spo11, Mre11, and Rad50). Mer2 (purple) is localized at chromosome axes, along with axial element proteins Red1, Hop1, and cohesin Rec8 (red and gray ovals), but is further enriched at axes on phosphorylation by CDK-S. CDK-S primes Mer2 for further phosphorylation by DDK. Arrows on the chromatin loop represent gene open reading frames. Red squares represent H3K4me3 marks. Only one sister chromatid is shown for clarity. (C) Model integrating DSB formation with loop-axis chromosome structure (based on data in Acquaviva et al. 2012; Sommermeyer et al. 2013). Axis-associated Mer2 interacts with Spp1, which binds H3K4me2/me3 marks and thereby tethers a chromatin loop to the axis. The nucleosome-depleted promoter near the tethered loop segment becomes accessible to Spo11, allowing DSB formation. The precise order of events (Mer2 phosphorylation, loop tethering to the axis) is not known. Spp1 interacts with Mer2 independent of Mer2 phosphorylation, so the potential interactions indicated in the left-most panel (H3K4me2/me3–Spp1–Mer2) could also occur in B, but are not shown.
Ski8 is involved in RNA metabolism in vegetative cells but, during meiosis, it relocalizes to the nucleus where it stabilizes the nuclear localization and chromatin association of Spo11 and, to a lesser extent, of Rec102–Rec104 (Arora et al. 2004; Kee et al. 2004; Prieler et al. 2005). Ski8 appears to interact directly with a surface of Spo11 whose amino acid sequence closely matches a motif (Q–R–x–x–Φ) in Ski3, a direct binding partner of Ski8 in mRNA decay (Arora et al. 2004; Halbach et al. 2013). The WD propeller motif in Ski8 is speculated to function as a scaffold for DSB protein complex assembly, but it is not known whether Ski8 contributes in other ways to DSB formation (Keeney 2007).
Rec102 and Rec104 interact with Spo11 and Ski8, and also with Mei4 and Rec114, so one possible role for the Rec102–Rec104 subcomplex is to bridge the Rec114–Mei4–Mer2 subcomplex with the Spo11–Ski8 subcomplex (Fig. 3A) (Arora et al. 2004; Maleki et al. 2007). Rec102 and Rec104 behave as a functional unit, and are required for Spo11 nuclear localization, chromatin association, and binding to hot spots (Kee et al. 2004; Prieler et al. 2005; Sasanuma et al. 2007). Rec102 and Rec104 show a preference for localizing to axis sites, but less prominently than Rec114, Mei4, or Mer2, suggesting a more even distribution genome-wide and possibly association with both loop and axis sequence (Kee et al. 2004; Panizza et al. 2011). Rec104 is phosphorylated, but the function of this modification is unknown (Kee et al. 2004).
Rec114, Mei4, and Mer2 behave as a subcomplex based on two-hybrid, coimmunoprecipitation, and cytological studies (Fig. 3A) (Arora et al. 2004; Henderson et al. 2006; Li et al. 2006; Maleki et al. 2007). Rec114, Mei4, and Mer2 chromosomal-binding sites correlate with axial sites, and exhibit a local anticorrelation with DSB sites (Panizza et al. 2011). Mer2 is recruited to axial sites by axis proteins (Red1, Hop1), but it is not known whether this occurs through direct physical interaction (Panizza et al. 2011).
Mer2 is phosphorylated by the S-phase cyclin-dependent kinase (Cdc28-Clb5/Clb6, also referred to as CDK-S) and the Dbf4-dependent kinase Cdc7 (DDK) (Henderson et al. 2006; Murakami and Keeney 2008; Sasanuma et al. 2008; Wan et al. 2008). CDK-S phosphorylation of Mer2 at S30 primes subsequent phosphorylation on S29 by DDK, with both modifications essential for DSB formation. DDK also independently phosphorylates other Mer2 amino-terminal residues (S11, 15, 19, 22) important for normal DSB levels (Henderson et al. 2006; Sasanuma et al. 2008; Wan et al. 2008). CDK-S also phosphorylates Mer2 at S271, but this is not required for DSB formation (Henderson et al. 2006). Mer2 phosphorylation leads to further enrichment at axes and recruitment of Rec114, Mei4, and Xrs2 (Henderson et al. 2006; Sasanuma et al. 2008; Panizza et al. 2011). Mer2 phosphorylation by replication-associated kinases is thus a critical regulatory step and has been proposed to coordinate premeiotic replication with DSB formation (Fig. 3B) (Murakami and Keeney 2008). A function for Mer2 in coordinating DSB formation with higher-order chromosome structure has recently been identified (see the section Regulation of DSB Timing).
Rec114 phosphorylation is dependent on DSBs and on the DNA damage signal transduction kinases Tel1 and/or Mec1 (Sasanuma et al. 2008; Carballo et al. 2013). Rec114 has eight [S/T]Q motifs, and seven of these are located within two [S/T]Q cluster domains found in many known targets of Tel1 and Mec1 (Carballo et al. 2013). Replacing these eight serines/threonines with nonphosphorylatable alanines (rec114-8A) results in a subtle increase in DSB levels (although not detectable at all sites that were assayed), whereas a phosphomimetic allele (rec114-8D) exhibits reduced DSB levels. The role of Rec114 phosphorylation is not altogether clear, but it has been proposed to mediate a negative feedback loop to inhibit DSB formation (see Concluding Remarks) (Carballo et al. 2013).
Mre11 requires all the other DSB proteins (except Rad50) for association with DSB sites, suggesting the MRX complex is recruited last, once Spo11 is poised to make DSBs (Borde et al. 2004). Recruitment of MRX might occur through Xrs2 interaction with Mer2, dependent on Mer2 phosphorylation by CDK-S (Arora et al. 2004; Henderson et al. 2006). It is speculated that requiring MRX for DSB formation facilitates rapid coordination with repair, ensuring that all breaks are efficiently processed (Borde et al. 2004).
S. pombe
There are six known Rec12 partners (Rec6, Rec7, Rec14, Rec15, Rec24, and Mde2), and a chromosome structure component (Rec10) essential for DSB formation (Fig. 3A) (Cromie and Smith 2007a; Keeney 2007). Other chromosome structure proteins (Rec25, Rec27, and Mug20) are not essential for DSB formation, but determine DSB hot spot location (Fowler et al. 2013). MRX orthologs (Rad32–Rad50–Nbs1) are required for repair of meiotic DSBs but not their formation (Young et al. 2004).
S. pombe does not form synaptonemal complexes, but instead forms structures called linear elements (LinEs) similar to the axial element precursors of the synaptonemal complex in S. cerevisiae (Loidl 2006). Proper LinE formation is dependent on meiotic cohesin subunits Rec8 and Rec11 (Molnar et al. 1995, 2003; Lorenz et al. 2004; Davis et al. 2008; Fowler et al. 2013). The LinE protein Rec10, homologous to S. cerevisiae Red1, is required for DSB formation in S. pombe (Ellermeier and Smith 2005; Lorenz et al. 2006; Bonfils et al. 2011). Rec10 physically interacts with and colocalizes by cytology with the LinE proteins Rec25, Rec27, and Mug20, and these four proteins are interdependent for chromosomal association into cytologically detectable foci (Davis et al. 2008; Spirek et al. 2010; Estreicher et al. 2012; Fowler et al. 2013). Colocalization among LinE proteins is also detected on a population basis by chromatin immunoprecipitation (ChIP), although Rec10 binds uniformly across the genome with only modest enrichment at sites of colocalization (Fowler et al. 2013). Rec25, Rec27, and Mug20 specifically bind to sites of DSB hot spots, even in the absence of DSB formation (Fowler et al. 2013). Moreover, DSBs are significantly reduced at >80% of hot spots in the absence of Rec27, so Rec25, Rec27, and Mug20 are proposed to be determinants of DSB hot spots (discussed below) (Fowler et al. 2013). Unlike for Rec10, mutants lacking Rec25, Rec27, and Mug20 show region-specific reduction in DSB and recombination levels, so these proteins are not essential for DSB formation per se (Davis et al. 2008; Estreicher et al. 2012).
Rec7, Rec15, and Rec24 are orthologs of S. cerevisiae Rec114, Mer2, and Mei4, respectively, and form the SFT (seven-fifteen-twenty-four) subcomplex (Fig. 3A) (Malone et al. 1997; Molnar et al. 2001; Kumar et al. 2010; Miyoshi et al. 2012). As in budding yeast and mouse, Rec7 and Rec24 physically interact and colocalize with LinEs (Steiner et al. 2010; Bonfils et al. 2011; Miyoshi et al. 2012). Rec24 recruits Rec7 to LinEs, and colocalization with LinEs depends on Rec10 but is independent of Rec12 and, thus, DSBs (Lorenz et al. 2006; Bonfils et al. 2011).
Rec15 interacts with both Rec24 and Rec7, and also with itself, Rec10, and Mde2 (Fig. 3A) (Miyoshi et al. 2012). No other Rec12 partner detectably interacts with linear element components, so Rec15 likely recruits Rec24 and Rec7 to LinEs via its interaction with Rec10 (Miyoshi et al. 2012). Rec15 binds to two classes of LinE sites with different genetic dependencies: LinE sites that overlap DSB hot spots (hot-LinE; thought to represent sites of chromatin loops tethered to the axis [see the section Regulation of DSB Timing]), and LinE sites outside of DSB hot spots (Miyoshi et al. 2012). Rec15 binding to hot-LinEs largely depends on Rec24 and Mde2, whereas binding to LinE sites outside of hot spots requires Rec10 (Miyoshi et al. 2012). Rec7 is phosphorylated independently of DSB formation, but the kinase responsible and the role for the modification are unknown (Miyoshi et al. 2012).
Rec6 and Rec14, along with Rec12, form the DSBC (DSB catalytic core) subcomplex (Fig. 3A) (Miyoshi et al. 2012). Rec12–Rec14 interaction can be detected by yeast two-hybrid, but interaction with Rec6 is only detectable in the presence of both Rec12 and Rec14 (Steiner et al. 2010; Miyoshi et al. 2012). Rec14 interacts with Mde2 (Miyoshi et al. 2012). Rec6 has no known ortholog in other organisms, whereas Rec14 is the ortholog of S. cerevisiae Ski8 (Evans et al. 1997).
Mde2 was identified in a screen for genes up-regulated in meiosis, and no orthologs are known (Gregan et al. 2005). Mde2 localizes predominantly to DSB sites, dependent on Rec15, and is required for Rec12 binding to the mbs1 hot spot, suggesting Mde2 recruits Rec12 and the DSBC subcomplex to hot spots (Miyoshi et al. 2012). Mde2 interacts with SFT via Rec15, and with DSBC via Rec14, possibly bridging the two subcomplexes (see the section Regulation of DSB Timing) (Fig. 3A) (Miyoshi et al. 2012). Mde2 expression is regulated by the Rad3–Cds1 meiotic replication checkpoint (Rad3 is the homolog of S. cerevisiae Mec1 and vertebrate ATR kinases), thereby providing a potential mechanism to coordinate replication with DSB formation (Ogino and Masai 2006; Miyoshi et al. 2012).
Taken together, DSB proteins may play a role in recruiting Rec12 to chromatin and/or activating Rec12, whereas hot spot determinants (Rec25, Rec27, and Mug20) likely stabilize or activate Rec12 already bound to chromatin (Miyoshi et al. 2012; Fowler et al. 2013).
M. musculus
Three mouse proteins are known or hypothesized to be required along with SPO11 to generate DSBs (MEI1, MEI4, and REC114). It is not known whether mouse MRX orthologs (MRE11, RAD50, and NBS1) are required for meiotic DSB formation because they are essential for viability (Xiao and Weaver 1997; Luo et al. 1999; Zhu et al. 2001), but their role in repair of meiotic DSBs appears to be conserved (Borde 2007; Cherry et al. 2007; Kumar and de Massy 2010). WDR61 is the Ski8/Rec14 homolog, but whether its meiotic function is conserved is not known (Kumar and de Massy 2010).
Mei1 was identified in an embryonic stem cell-based ethyl methane sulfonate (EMS) chemical mutagenesis screen for fertility mutants (Munroe et al. 2000; Ward et al. 2003). Mei1 mutants display reduced staining for γH2AX (a phosphorylated form of histone H2AX that is a marker of DSBs) and absence of RAD51 foci that can be restored by cisplatin-induced DNA damage (Libby et al. 2002, 2003; Reinholdt and Schimenti 2005). Similar to Spo11−/− mutants (Baudat et al. 2000; Romanienko and Camerini-Otero 2000; Di Giacomo et al. 2005), Mei1−/− mutant spermatocytes exhibit defective chromosome synapsis and prophase I arrest, and epistasis analysis shows that MEI1 acts upstream of DMC1 (Libby et al. 2002; Reinholdt and Schimenti 2005). The biochemical function of MEI1 is unknown. No orthologs have been found in invertebrates (Libby et al. 2003), but MEI1 shares modest homology with plant PRD1 (see the section A. thaliana). It is unknown whether MEI1 physically interacts with SPO11 as for PRD1 (De Muyt et al. 2007).
Mei4 and Rec114 were identified by a phylogenomic approach through conservation of short signature sequence motifs found in their hemiascomycetal orthologs (Maleki et al. 2007; Kumar et al. 2010). MEI4 localizes to discrete foci on chromosome axes independent of SPO11, with focus numbers highest at leptonema and low at pachynema, similar to the spatial organization and kinetics of S. cerevisiae Mei4 and S. pombe Rec24 (Maleki et al. 2007; Kumar et al. 2010; Bonfils et al. 2011). Diagnostic of failure to make DSBs, Mei4−/− spermatocytes exhibit greatly reduced γH2AX; lack of RAD51, DMC1, and RPA foci; synapsis defects; and meiotic prophase arrest (Kumar et al. 2010). MEI4 interacts with REC114 via conserved motifs in the amino-terminal region (Kumar et al. 2010), as also seen in S. cerevisiae and S. pombe orthologs (Maleki et al. 2007; Steiner et al. 2010). It remains to be determined how MEI4 is recruited to the chromosome axes (via a Mer2 ortholog?), whether REC114 function is conserved, and whether orthologs or functional equivalents of other DSB proteins (e.g., Rec102, Rec104, Mer2, Rec6, and Mde2) exist in mouse.
A. thaliana
Several A. thaliana genes required for DSB formation have been identified through genetic screens (PRD1, PRD2, PRD3, SWI1, and DFO) (Edlinger and Schlögelhofer 2011). Some are homologs of DSB proteins in yeasts or mammals (PRD1, PRD2), whereas others have no clear homologs outside the plant kingdom (PRD3, SWI1, and DFO) (Table 1). Besides PRD1, it is not known whether these DSB proteins interact with SPO11 or among themselves in an interaction network similar to those in yeasts. MRX and Ski8 homologs in plants are dispensable for DSB formation (Bleuyard et al. 2004; Pawlowski et al. 2004; Puizina et al. 2004; Jolivet et al. 2006).
PRD1 has modest sequence similarity with human MEI1 and may thus be its functional equivalent (De Muyt et al. 2007). prd1 mutants are asynaptic, exhibit reduced meiotic recombination rates comparable to a strong spo11-2 allele, lack detectable DMC1 foci, and can suppress the chromosome fragmentation defect in DSB repair mutants rad51, scc3, and rec8 (De Muyt et al. 2007). These phenotypes are consistent with a failure to make DSBs. PRD1 interacts with itself and SPO11-1, but whether these interactions are essential for DSB formation is unknown (De Muyt et al. 2007).
PRD2 and PRD3 were identified in a screen for early meiotic recombination defects (De Muyt et al. 2009). prd2 and prd3 mutants lack DMC1 foci; are sterile, asynaptic, and achiasmate; and suppress rad51 or mre11 chromosome fragmentation defects. It is thus concluded that they do not form DSBs (De Muyt et al. 2009). PRD2 is the ortholog of Mei4/Rec24, and PRD3 is the ortholog of PAIR1 in rice, which is required for homolog pairing in meiosis (Nonomura et al. 2004; De Muyt et al. 2009; Kumar et al. 2010).
SWI1/DYAD is involved in sister chromatid cohesion, axial element formation, homolog pairing and synapsis, recombination, and proper histone modification during prophase I (H3 deacetylation and H3K4 dimethylation) (Mercier et al. 2001, 2003; Agashe et al. 2002; Hamant et al. 2006; Boateng et al. 2008). swi1 mutants fail to form DSBs, as seen by the lack of RAD51 foci and suppression of the chromosome fragmentation phenotype of the recombination-defective dif1-1 mutant (Mercier et al. 2003). It is not clear whether SWI1 has separate functions in different meiotic events, or whether a cohesion or axial element defect is responsible for the later meiotic defects.
DFO was identified in a T-DNA insertion mutagenesis screen for sterile mutants (Mercier et al. 2003; Boateng et al. 2008). dfo mutants exhibit asynapsis, severely reduced recombination rates, and impaired DSB formation as shown by the ability to suppress mre11 chromosome fragmentation defects (Zhang et al. 2012). AtDFO is predicted to have three coiled-coil motifs and a helix-turn-helix motif, which might mediate protein–protein interactions and DNA binding, respectively (Zhang et al. 2012).
D. melanogaster
In Drosophila, only two proteins (Mei-P22 and Trem) are currently known to be required for DSB formation besides Mei-W68, the Spo11 ortholog. Mei-P22 was identified as a recombination-defective mutant in a P-element insertion mutagenesis screen for meiotic chromosome missegregation (Sekelsky et al. 1999). mei-P22 mutants are defective for meiotic DSB formation as indicated by the absence of γHis2Av foci (the Drosophila version of γH2AX), and the ability of X-ray treatment to partially rescue defects in crossing over and X chromosome disjunction (Liu et al. 2002; Mehrotra and McKim 2006). No homologs have been identified in other eukaryotes. Mei-P22 forms discrete foci on meiotic chromosomes that partially overlap with γHis2Av foci, and focus formation is dependent on Trem (Mehrotra and McKim 2006; Lake et al. 2011).
Trem (Trade embargo) is a C2H2 zinc finger protein identified in a germline clone screen for mutants with high levels of meiotic nondisjunction (Page et al. 2007). Null trem mutants do not make DSBs and are semisterile; hypomorphic trem alleles yield elevated MI nondisjunction, greatly reduced meiotic recombination frequency, loss of crossover homeostasis, and absence of Mei-P22 foci (Page et al. 2007; Lake et al. 2011). His2Av phosphorylation and MI disjunction, but not the sterility defect, can be partially rescued by X-ray-induced DSBs, suggesting that Trem has a separate role in fertility (Lake et al. 2011). Hawley and colleagues hypothesize that the failure to form DSBs in trem mutants is caused by the failure in Mei-P22 foci formation (Lake et al. 2011). It is not known whether Trem physically interacts with Mei-P22, thereby promoting loading to specific sites, or whether it affects Mei-P22 loading and DSB formation indirectly by altering chromatin structure.
C. elegans
Several C. elegans proteins besides SPO-11 are involved in meiotic recombination initiation (MRE-11, RAD-50, DSB-1, DSB-2, HIM-17, HIM-5, and HTP-3), but it is not yet clear whether their roles are direct or indirect.
DSB-1 is essential for DSB formation, as shown by the absence of RAD-51 foci and chiasmata in dsb-1 germ lines, which can be restored by exogenous DSBs (Stamper et al. 2013). On the other hand, its paralog DSB-2 is not essential but is required for efficient DSB formation (Rosu et al. 2013). DSB-1 and DSB-2 show a similar localization pattern, associating with chromatin from early meiotic prophase to mid-pachytene, which corresponds to the stage of DSB formation (Rosu et al. 2013; Stamper et al. 2013). Chromatin association is independent of DSBs, but dependent on CHK-2 kinase (Rosu et al. 2013; Stamper et al. 2013). DSB-2 chromosome localization and protein levels depend on DSB-1, and DSB-1 localization is partially dependent on DSB-2, yet the two proteins do not colocalize extensively (Rosu et al. 2013; Stamper et al. 2013).
The association of DSB-1 and DSB-2 with chromatin is extended in mutants with CO defects, but not in axial element mutants (despite their CO defect), suggesting that persistence of DSB-1 or DSB-2 requires proper axis organization (Rosu et al. 2013; Stamper et al. 2013). The investigators propose that chromatin association of DSB-1 and DSB-2 indicate a DSB-permissive state, and formation of CO recombination intermediates triggers the removal of DSB-1 and DSB-2 from chromatin, which presumably inactivates DSB formation, thus invoking an obligate CO checkpoint or a negative feedback mechanism (Rosu et al. 2013; Stamper et al. 2013). No homologs of DSB-1 or DSB-2 have been found outside the genus Caenorhabditis. Both DSB-1 and DSB-2 have potential target sites for ATM/ATR family of protein kinases, but it is not known whether they are phosphorylated (Rosu et al. 2013; Stamper et al. 2013).
HIM-17 is required for meiotic DSB formation and proper accumulation of H3K9 methylation on prophase chromosomes (Reddy and Villeneuve 2004). HIM-17 has six C2CH repeat modules seen in zinc-finger DNA-binding motifs (Reddy and Villeneuve 2004). Absence of HIM-17 recapitulates many of the spo-11 mutant phenotypes, such as abolished RAD-51 foci, chromosome missegregation, and defective chiasma formation that can be restored by ionizing radiation-induced DSBs (Reddy and Villeneuve 2004). H3K9me is not a consequence of DSB formation, but it is not known whether the modification is a prerequisite for SPO-11 to cleave DNA, or whether these are two separate roles of HIM-17. Thus, it is possible that the effect of HIM-17 on DSB formation is indirect through histone modifications that alter chromosome structure and make it amenable for SPO-11 catalytic activity.
HIM-5 promotes DSBs specifically on the X chromosome, as seen by severely reduced recombination and elevated frequency of nondisjunction in the X chromosome of him-5 mutants (Broverman and Meneely 1994; Meneely et al. 2012). In him-5 mutants, RAD-51 foci are not detectable on the X chromosome, and exogenous radiation-induced DSBs rescue the X chromosome nondisjunction, premature chromosome desynapsis, and delayed pachytene progression phenotypes (Meneely et al. 2012). On autosomes, him-5 mutants exhibit a redistribution of COs from telomere-proximal regions to chromosome centers, without altering CO levels (Meneely et al. 2012). It has been proposed that HIM-5 targets SPO-11 activity to the X chromosome, either directly as a partner protein, or indirectly by modifying the heterochromatin on the X and distal autosome regions (Meneely et al. 2012).
HTP-3 is a paralog of the axis protein HIM-3 (homolog of S. cerevisiae Hop1) and forms complexes with MRE-11/RAD-50 and HIM-3 (Goodyer et al. 2008). Association of HTP-3 with both MRE-11/RAD-50 and HIM-3 is proposed to link DSB formation with homolog alignment and synapsis, because MRE-11/RAD-50 has been implicated in DSB formation, and HIM-3 functions in homolog alignment, synapsis, and partner choice in recombination (Zetka et al. 1999; Chin and Villeneuve 2001; Alpi et al. 2003; Couteau et al. 2004; Goodyer et al. 2008). HTP-3 associates with chromatin in premeiotic nuclei, is a component of meiotic axes, and is required for HIM-3 localization to axes, homolog alignment, synapsis, and crossing over. RNAi silencing of HTP-3 eliminates RAD-51 and RPA-1 foci formation, and rescues the DSB-dependent diakinesis defects of rad-51 and brc-2 mutants, suggesting that HTP-3 is required for DSB formation (Goodyer et al. 2008). Unlike yeast or mouse, meiotic recombination is not necessary for homolog recognition, presynaptic alignment, and synapsis in C. elegans; therefore, the role of HTP-3 in DSB formation and downstream recombination events likely reflect separate functions in recombination (Dernburg et al. 1998; McKim et al. 1998; Goodyer et al. 2008). It is notable that HORMA domain-containing proteins related to Hop1 promote DSB formation in almost all organisms examined (e.g., budding and fission yeasts Hop1, C. elegans HTP-1 and HTP-3, and mouse HORMAD1; but not A. thaliana ASY1) (Mao-Draayer et al. 1996; Couteau and Zetka 2005; Martinez-Perez and Villeneuve 2005; Sanchez-Moran et al. 2007; Goodyer et al. 2008; Latypov et al. 2010).
SPATIAL DISTRIBUTION OF DSBs
The location of DSBs and subsequent recombination is important for genome integrity. DSBs in repetitive DNA sequence (e.g., rDNA, transposable elements) are at risk of genome rearrangement if repaired using nonallelic homologous sequences as template (Sasaki et al. 2010). Crossovers near centromeres cause an elevated frequency of precocious separation of sister chromatids at meiosis I, resulting in aneuploidy and spore inviability in yeast (Rockmill et al. 2006). Nonetheless, where Spo11 generates a DSB (or not) is not determined by a single factor, but rather by a combination of factors operating over different size scales. This will be discussed in more detail for S. cerevisiae, in which it is best understood, but emerging evidence suggests that multiple layers govern the spatial distribution of DSBs in other organisms as well.
S. cerevisiae
Meiotic DSBs are not randomly distributed along chromosomes, but instead display multiple levels of spatial organization that interact hierarchically with one another (Lichten and Goldman 1995; Petes 2001; Kauppi et al. 2004; Lichten and de Massy 2011; Pan et al. 2011). At the chromosome level, DSBs form preferentially on chromosome arms, and are less frequent within pericentric and subtelomeric zones (two- to threefold below genome average at ∼5–10 kb around centromeres and 3.5-fold ∼20 kb from telomeres) (Blitzblau et al. 2007; Buhler et al. 2007; Pan et al. 2011). Within interstitial regions there are DSB-rich and DSB-poor domains on the order of ∼100 kb (Baudat and Nicolas 1997; Borde et al. 1999). Within these domains, DSBs preferentially form in GC-rich chromatin loop regions rather than AT-rich axis-associated DNA (Fig. 2) (Blat et al. 2002; Kleckner 2006). Locally, there are narrow regions (typically ∼200 bp) in which Spo11 cleaves preferentially, referred to as DSB hot spots. Most hot spots in S. cerevisiae are influenced by chromatin accessibility, and 88% are within nucleosome-depleted regions (NDRs) in gene promoters (Ohta et al. 1994; Wu and Lichten 1994; Berchowitz et al. 2009; Pan et al. 2011). However, chromatin accessibility is not sufficient, that is, not all nucleosome-depleted regions are DSB hot spots. Moreover, a substantial fraction of DSBs (>10%) forms outside of hot spots (Pan et al. 2011).
Posttranslational histone modification, in the form of H3K4 methylation, influences DSB sites. H3K4 trimethylation (H3K4me3) is enriched at the 5′ end of genes, and is a histone mark associated with active transcription (Pokholok et al. 2005; Dehé and Géli 2006). In S. cerevisiae, Set1 is the catalytic subunit of the COMPASS complex responsible for all H3K4me, and DSB levels are reduced in the set1Δ mutant, with sites of high H3K4me3 affected the most, and some novel DSB hot spots arising (Sollier et al. 2004; Dehé and Géli 2006; Borde et al. 2009; Acquaviva et al. 2012; Sommermeyer et al. 2013). However, the spatial correlation between H3K4me3 and DSB levels is weak genome-wide, and H3K4me3 abundance is a poor predictor of DSB hot spot location or heat (Tischfield and Keeney 2012). Recent findings (discussed in the section Regulation of DSB Timing) provide a clearer explanation for the role of H3K4me3 in DSB site selection (Acquaviva et al. 2012; Sommermeyer et al. 2013).
S. pombe
Prominent hotpots in S. pombe are usually widely separated (∼50–100 kb apart) and tend to localize in large intergenic regions (IGR) (Cromie et al. 2007). Large IGRs often include clusters of closely spaced NDRs (de Castro et al. 2012), but NDRs are not as predictive of DSB hot spots in S. pombe as they are in S. cerevisiae (Fowler et al. 2014). Some hot spots are dependent on transcription factor binding (e.g., ade6-M26 hot spot bound by Atf1-Pcr1 transcription factor), whereas others are independent of known transcription factors (e.g., mbs1) (Wahls and Smith 1994; Kon et al. 1997; Cromie et al. 2005; Hirota et al. 2007). Recent work implicates linear element components (Rec25, Rec27, and Mug20) as hot spot determinants (see the section Regulation of DSB Timing) (Fowler et al. 2013).
Unlike S. cerevisiae or mouse, the distribution of DSB hot spots differs from the distribution of crossovers (Young et al. 2002; Cromie and Smith 2007b). Regions with few DSBs have crossover frequencies similar to regions with prominent DSB hot spots, a phenomenon known as crossover invariance (Hyppa and Smith 2010; Fowler et al. 2014). At the heart of crossover invariance is variation in the choice of preferred recombination partner; at DSB hot spots, DSB repair is biased toward the sister chromatid, whereas at DSB cold regions, DSB repair is biased toward the homolog. The mechanism behind this phenomenon is not currently understood.
Mouse and Human
Mouse and human recombination hot spots overlap both genic and intergenic regions, although human recombination rates are on average lower within transcribed regions of genes (McVean et al. 2004; Myers et al. 2005; Arnheim et al. 2007; Frazer et al. 2007; Coop et al. 2008; Kong et al. 2010; Smagulova et al. 2011; Lu et al. 2012). In mouse, DSB hot spots overlap a subset of H3K4me3-enriched sites, but unlike budding yeast, this overlap does not generally include the strong H3K4me3-enriched regions around promoters (Buard et al. 2009; Smagulova et al. 2011). Instead, DSB hot spots in mouse and humans are determined by the DNA-binding specificity of PRDM9 methyltransferase, discussed below (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010; Grey et al. 2011; Brick et al. 2012).
INTEGRATING DSB FORMATION WITH THE LOOP-AXIS STRUCTURE OF CHROMOSOMES
Spp1–Mer2 Interactions Influence DSB Locations in S. cerevisiae
DSBs form preferentially in chromatin loops, not the DNA embedded in axes as defined by ChIP enrichment for Rec8 and other axis components (Blat et al. 2002; Glynn et al. 2004; Kugou et al. 2009; Pan et al. 2011; Panizza et al. 2011). However, cytologically detectable recombination complexes containing Rad51 and/or Dmc1 are associated with axial elements and the synaptonemal complex in various organisms examined, so recombination takes place in the context of chromosome axes (Ashley et al. 1995; Anderson et al. 1997; Barlow et al. 1997; Moens et al. 1998; Tarsounas et al. 1999; Blat et al. 2002; Panizza et al. 2011). This apparent paradox—DSBs are in loops but recombination occurs on axes—can be resolved by the “tethered loop–axis complex” (TLAC) model of Kleckner and colleagues, in which DSB sites in loop DNA are recruited to the proximity of the axes (Blat et al. 2002; Kleckner 2006; Panizza et al. 2011). In principle, tethering could occur before or after DSB formation, but the observation that many DSB proteins are themselves enriched at axes supports a pre-DSB tethering model (Kleckner 2006; Panizza et al. 2011). TLAC structures may help ensure that DSBs form in the context of chromosome axes, thereby promoting interhomolog repair and thus accurate segregation of homologous chromosomes (Kim et al. 2010; Panizza et al. 2011). As discussed below, most available data can be interpreted in light of this model, but it is important to note that there is as yet no direct demonstration of TLACs.
Recent findings provide a mechanism for loop tethering through a recently described physical interaction between Mer2 and Spp1 (Acquaviva et al. 2012; Sommermeyer et al. 2013). Spp1 is part of the COMPASS complex (Set1 is the catalytic subunit) and has a PHD finger motif that binds H3K4me2/me3 marks (Dehé et al. 2006; Shi et al. 2007; Murton et al. 2010). Simultaneous interaction of Spp1 with H3K4me2/me3 and with Mer2 via its carboxyl terminus is proposed to tether chromatin loops to DSB proteins localized on the chromosome axes, thereby activating DSB formation in the nucleosome-depleted regions near the tethered portion of the chromatin loop (Fig. 3C).
Consistent with this model, artificially tethering Spp1 to recombination-cold UASGAL regions using a Gal4 DNA-binding domain fusion is sufficient to induce DSB formation in the nearby NDR (Acquaviva et al. 2012). In this scenario, DSB formation now targeted to a normally cold region did not require H3K4me3, but was still dependent on Mer2 and Spo11. In the absence of normal function of this tethering mechanism (e.g., in set1Δ or H3K4R mutants in which there is no H3K4me), novel DSB hot spots appear, mostly at promoters of genes transcriptionally induced in set1Δ or in chromatin loop regions closest to the axes (Sollier et al. 2004; Borde et al. 2009; Acquaviva et al. 2012; Sommermeyer et al. 2013). Thus, the Mer2–Spp1–H3K4me2/3 interaction influences the location of DSBs.
Targeting DSBs in S. pombe
Ohta and colleagues have proposed a different mechanism for TLAC formation in S. pombe (Miyoshi et al. 2012, 2013). As noted above, the SFT complex localizes to LinEs, presumably through Rec15 interaction with Rec10 (Lorenz et al. 2006; Bonfils et al. 2011; Miyoshi et al. 2012). Mde2 localizes preferentially to DSB hot spots and interacts with both SFT and DSBC (Fig. 3A) (Miyoshi et al. 2012). Through these physical interactions and Rec15 self-interaction, Mde2 at potential DSB sites (proposed to be on loop sequence) could connect with LinEs and recruit DSBC, thereby tethering DSB sites to LinEs and bringing DSB protein subcomplexes together. The model proposes that tethering of the loop to LinEs occurs before DSB formation. It is not known whether these proposed tethering interactions involve histone modifications analogous to S. cerevisiae, but S. pombe hot spots are enriched for H3K9 acetylation, and absence of this histone mark results in partially reduced DSB and Rec12-binding levels (Yamada et al. 2013). In contrast, hot spots are not associated with H3K4me3, although absence of Set1 results in more Rec12 binding, and reduced DSB and recombination activity at some hot spots (Yamada et al. 2013). It is possible that multiple chromatin-related factors influence DSB formation in S. pombe.
Smith and colleagues proposed an alternative scenario that does not invoke TLAC formation (Fowler et al. 2013; Martin-Castellanos et al. 2013). In this model, Rec12 binds both to hot spots and to DSB-cold regions, but only those binding events that occur in proximity to linear element proteins Rec25, Rec27, and Mug20 result in high-frequency DNA cleavage. Rec25, Rec27, and Mug20 are not absolutely required for DSBs, but they bind all hot spots with great specificity and are essential for DSBs at most hot spots (Martin-Castellanos et al. 2005; Davis et al. 2008; Fowler et al. 2013). Interestingly, Rec12 binding (as assessed in a catalytic-dead mutant) is higher within transcription start sites, but DSB frequency is higher between genes, suggesting that Rec12 binding to DNA is not enough to initiate DSB formation, but rather its activation is controlled separately, most likely mediated by Rec25, Rec27, and Mug20 (Fowler et al. 2013). According to Smith and colleagues, the high correlation between DSB hot spots and binding sites for a subset of LinE proteins, in addition to the absence of any significant anticorrelation between Rec8 binding and DSB hot spots, argues against a TLAC-based model (Fowler et al. 2013).
Further investigation should shed light on the mechanism for targeting DSBs in S. pombe—whether it involves a TLAC mechanism, or is determined by a set of hot spot determinants, or a combination of both models.
Targeting DSBs in Mouse and Human
PRDM9 has a PR/SET domain at the amino terminus with histone H3K4 trimethyltransferase activity, and multiple C2H2-type zinc-finger DNA-binding motifs at the carboxyl terminus (Hayashi et al. 2005). The zinc-finger array evolves rapidly, such that numerous alleles with distinct DNA-binding specificities are present in populations of humans, mice, and other mammalian species (Oliver et al. 2009; Thomas et al. 2009; Baudat et al. 2010; Berg et al. 2010; Parvanov et al. 2010). At least 73% of hot spots in mouse contain a consensus motif that matches the predicted binding site of PRDM9 (Smagulova et al. 2011), but this is likely an underestimate of PRDM9-binding sites in vivo because of current limitations on deducing PRDM9 binding from DNA sequence alone. Mouse strains with different Prdm9 alleles exhibit widely different hot spot distributions with only 1.1% overlap in DSB hot spot locations (Brick et al. 2012). In humans, a partially degenerate 13-bp motif is associated with at least 40% of human hot spots (Myers et al. 2008, 2010; Baudat et al. 2010), but this motif is specifically recognized by two PRDM9 variants frequently found in individuals of European descent, whereas variants common to other populations recognize different binding sites and, hence, are associated with different hot spots (Baudat et al. 2010; Berg et al. 2010; Kong et al. 2010; Hinch et al. 2011). These findings show that PRDM9 is responsible for defining most DSB hot spot locations in mouse and human.
Interestingly, PRDM9 is not required for DSB formation, but instead targets the DSB machinery. DSB hot spots occur in different locations in the Prdm9−/− mouse frequently, but not exclusively, located at H3K4me3-enriched sites associated with promoters, more reminiscent of the distribution in wild-type yeast, which has no PRDM9 ortholog (Brick et al. 2012). It is not known how PRDM9 recruits SPO11 to generate breaks, for example, whether its methyltransferase activity is necessary or sufficient for DSB targeting. Many genomic regions, such as promoters, are enriched for H3K4me3 that is placed by non-PRDM9-dependent mechanisms (Ruthenburg et al. 2007; Shilatifard 2008). SPO11 rarely cuts in these regions in wild-type mice, but does so frequently when PRDM9 is missing (Brick et al. 2012). The preferential targeting of SPO11 activity to PRDM9-binding sites despite presence of many other sources of H3K4me3 suggests that this histone modification by itself is not sufficient for targeting. Perhaps PRDM9 recruits the DSB machinery directly through physical interaction with a DSB protein in addition to providing H3K4me3.
It is also not yet clear whether DSB locations in mammals also involves coordination of local (hot-spot-level) features with higher-order chromosome structure, but several lines of evidence suggest that it does. The HORMAD1 structural component of axes (ortholog of yeast Hop1) is required for normal DSB levels, reminiscent of Hop1 requirement for normal DSB levels in S. cerevisiae (Mao-Draayer et al. 1996; Woltering et al. 2000; Peciña et al. 2002; Niu et al. 2005; Carballo et al. 2008; Shin et al. 2010; Daniel et al. 2011). In addition, DSB proteins REC114 and MEI4 localize to chromosome axes in mouse (Kumar et al. 2010), so it is possible that breaks form in the context of axial structures (as opposed to broken segments being recruited to axes after DSB formation), as has been proposed for S. cerevisiae and S. pombe (see above). Last, in mouse, DNA in the pseudoautosomal region (PAR, the only region of homology shared between the X and Y chromosomes) is organized on a longer axis with shorter chromatin loops compared to autosomes (Kauppi et al. 2011). DNA organized as short loops means more loops along the axis; thus, more potential DSB sites, and consistent with the TLAC model, PARs exhibit higher DSB density (Kauppi et al. 2011).
REGULATION OF DSB TIMING
DSBs are tightly controlled so that they occur at the right time and place. Meiotic DSB formation appears to be universally restricted to a narrow window of time within prophase I (Padmore et al. 1991; Cervantes et al. 2000; Mahadevaiah et al. 2001; Colaiácovo et al. 2003; Jang et al. 2003; Mehrotra and McKim 2006). In yeasts, DSBs occur ∼1–1.5 h after premeiotic DNA replication (Borde et al. 2000; Cervantes et al. 2000; Murakami et al. 2003).
One way cells control when DSBs start to form is through gene expression, for instance, regulated meiosis-specific transcription of SPO11 and other genes required for meiotic DSBs, such as REC102, REC104, REC114, and MEI4 in S. cerevisiae, or by meiosis-specific splicing (e.g., MER2) (Keeney 2001, 2007). Another level of control is through coordination with premeiotic replication. In S. cerevisiae, DSB formation follows premeiotic replication through the dual roles of CDK-S and DDK in replication origin firing and DSB formation (Schild and Byers 1978; Sclafani 2000; Smith et al. 2001; Masai and Arai 2002; Benjamin et al. 2003), but premeiotic replication is not a prerequisite for DSB formation (Hochwagen et al. 2005; Blitzblau et al. 2012). The coordinate timing between replication and DSB formation is speculated to arise from competition for CDK-S and DDK kinase activities, whereby lower levels are sufficient for replication origin firing, but onset of DSB formation occurs only after increased levels of kinase activity are available (Murakami and Keeney 2008). Recent studies support this hypothesis (Murakami and Keeney 2014).
Cells also regulate the termination of DSB activity, but how this is controlled is less clear (Padmore et al. 1991; Keeney 2001; Henderson et al. 2006). In S. cerevisiae, Spo11 and other DSB proteins persist on chromosomes past the time of DSB formation, and this is also observed for mouse SPO11 (Romanienko and Camerini-Otero 2000; Arora et al. 2004; Kee et al. 2004; Prieler et al. 2005; Henderson et al. 2006; Li et al. 2006; Maleki et al. 2007). This suggests DSB formation is not simply regulated by eliminating the participating proteins. Instead, restriction of DSB formation to a narrow window of time may be under more direct cell cycle control. S. cerevisiae mutants that arrest in pachytene (such as ndt80, cdc28, cdc36, and cdc39) exhibit increased recombination frequency and detectable DSBs at later meiotic time points (Shuster and Byers 1989; Xu et al. 1995; Allers and Lichten 2001), suggesting that DSBs continue to form in pachytene-arrested cells and further implying that progression past pachytene and/or prophase terminates the window of opportunity for break formation (Allers and Lichten 2001; Keeney 2001; Henderson et al. 2006).
REGULATION OF DSB NUMBERS VIA FEEDBACK CONTROL
Mechanisms for regulating DSB numbers have been described in different species. In S. cerevisiae, a DSB on one chromosome decreases the frequency of DSB formation on its homolog at the same and nearby positions (Xu and Kleckner 1995; Rocco and Nicolas 1996; Fukuda et al. 2008). This phenomenon, known as trans inhibition, is proposed to be dependent on the DNA damage signal transduction kinases Tel1 and Mec1, and tends to constrain DSBs to one per pair of homologs (Zhang et al. 2011). Inhibition in cis in which a strong DSB hot spot suppresses DSB formation on the same chromatid has also been reported, but the mechanism for this is not known (Wu and Lichten 1995; Xu and Kleckner 1995; Fan et al. 1997; Fukuda et al. 2008). Mice and flies appear to have a negative feedback loop, whereby DSBs catalyzed by SPO11 activate the Tel1 homolog ATM, which inhibits further DSB formation (Joyce et al. 2011; Lange et al. 2011). It is not clear how DSB inhibition is mediated in mice and flies. In S. cerevisiae, the negative feedback loop may involve phosphorylation of Rec114 by Tel1 and/or Mec1 (Carballo et al. 2013).
DSB numbers are also regulated through a feedback mechanism mediated by homolog engagement. In mouse, unsynapsed chromosome regions continue to form DSBs, suggesting existence of a mechanism by which DSB formation ceases once interhomolog interactions have been achieved, or by which unsynapsed regions are actively targeted for de novo DSB formation (Kauppi et al. 2013). A similar conclusion is suggested by the occurrence of elevated DSB numbers in S. cerevisiae mutants defective for engagement of homologous chromosomes (Thacker et al. 2014). A similar mechanism has been proposed in worms (Hayashi et al. 2010; Henzel et al. 2011), based on elevated levels and presence of RAD-51 foci at later stages in mutants with synapsis defects (him-3, him-8, rec-8, and syp-1), or with chromosomal translocations that prevent homologous synapsis (Alpi et al. 2003; Nabeshima et al. 2004; Carlton et al. 2006; Hayashi et al. 2007). CO precursors have also been proposed in worms to regulate the DSB-permissive state via chromatin association of DSB-1 and DSB-2, potentially either as a negative feedback loop, or an obligate CO checkpoint (Rosu et al. 2013; Stamper et al. 2013).
CONCLUDING REMARKS
Findings in recent years from studies of meiotic recombination initiation in different organisms are providing a better understanding of the mechanism of meiotic DSB formation by Spo11, particularly of the cast of players required for Spo11-mediated break formation, the spatial distribution of DSBs and how Spo11 activity is targeted to these sites, as well as regulation of DSB timing, and feedback control of DSB numbers. However, many questions remain unanswered. For example, DSB proteins appear to function in controlling Spo11 activity through various levels of regulation (coordination with replication, formation, and recruitment of DSB protein subcomplexes and coordination with higher-order chromosome structure) to ensure that DSBs occur in the proper spatial and temporal contexts. Nonetheless, the precise molecular functions of many of the DSB proteins, and their absolute requirement for break formation are still not completely understood. Further questions remain on the relationship between DSB proteins and hot spot determinants in S. pombe (Rec25, Rec27, and Mug20) and mammals (PRDM9), as well as the molecular details of mechanisms regulating the timing of Spo11 activity. It will be interesting to see what future research uncovers with respect to Spo11-mediated meiotic DSBs.
ACKNOWLEDGMENTS
We thank Shintaro Yamada for comments on the manuscript. Work from the authors’ laboratory is supported in part by National Institutes of Health (NIH) R01 Grants GM058673 (S.K.), HD053855 (S.K. and M. Jasin), and GM105421 (M. Jasin and S.K.). I.L. is supported in part by NIH fellowship F31 GM097861. S.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint-André C, Nicolas A, Géli V 2012. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Agashe B, Prasad CK, Siddiqi I 2002. Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129: 3935–3943. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Offenberg HH, Verkuijlen WMHC, Heyting C 1997. RecA-like proteins are components of early meiotic nodules in lily. Proc Natl Acad Sci 94: 6868–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, Calabrese P, Tiemann-Boege I 2007. Mammalian meiotic recombination hot spots. Annu Rev Genet 41: 369–399. [DOI] [PubMed] [Google Scholar]
- Arora C, Kee K, Maleki S, Keeney S 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559. [DOI] [PubMed] [Google Scholar]
- Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC 1995. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104: 19–28. [DOI] [PubMed] [Google Scholar]
- Barlow AL, Benson FE, West SC, Hultén MA 1997. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J 16: 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Nicolas A 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B 2010. PRDM9 is a major determinant of meiotic recombination hot spots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrimont A, Penkner A, Woglar A, Mamnun YM, Hulek M, Struck C, Schnabel R, Loidl J, Jantsch V 2011. A new thermosensitive smc-3 allele reveals involvement of cohesin in homologous recombination in C. elegans. PLoS ONE 6: e24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP 2009. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res 19: 2245–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42: 859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JM, Fass D, Wang JC, Harrison SC 1998. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci 95: 7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P 1997. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF 2008. Prelude to a division. Annu Rev Cell Dev Biol 24: 397–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI 2004. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113: 197–203. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A 2007. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol 17: 2003–2012. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP 2012. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet 8: e1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng KA, Yang X, Dong F, Owen HA, Makaroff CA 2008. SWI1 is required for meiotic chromosome remodeling events. Mol Plant 1: 620–633. [DOI] [PubMed] [Google Scholar]
- Bonfils S, Rozalen AE, Smith GR, Moreno S, Martin-Castellanos C 2011. Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J Cell Sci 124: 1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V 2007. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res 15: 551–563. [DOI] [PubMed] [Google Scholar]
- Borde V, Wu T-C, Lichten M 1999. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol 19: 4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Goldman ASH, Lichten M 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell 13: 389–401. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broverman SA, Meneely PM 1994. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics 136: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthès P, Grey C, de Massy B 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28: 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM 2002. Homologous chromosome associations and nuclear order in meiotic and mitotically dividing cells of budding yeast. Adv Genet 46: 49–90. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Panizza S, Serrentino M-E, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS 2013. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 9: e1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM, Farruggio AP, Dernburg AF 2006. A link between meiotic prophase progression and crossover control. PLoS Genet 2: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 489–484. [DOI] [PubMed] [Google Scholar]
- Cherry SM, Adelman CA, Theunissen JW, Hassold TJ, Hunt PA, Petrini JH 2007. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr Biol 17: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin GM, Villeneuve AM 2001. C elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G2 DNA damage checkpoint. Genes Dev 15: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M 2010. Evolutionary conservation of meiotic DSB proteins: More than just Spo11. Genes Dev 24: 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Couteau F, Zetka M 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev 19: 2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Nabeshima K, Villeneuve A, Zetka M 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol 14: 585–592. [DOI] [PubMed] [Google Scholar]
- Cromie G, Smith GR 2007a. Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. In Recombination and meiosis (ed. Egel R, Lankenau DH), pp. 195–230 Springer, Berlin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Smith GR 2007b. Branching out: Meiotic recombination and its regulation. Trends Cell Biol 17: 448–455. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet 3: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, et al. 2011. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Rozalen AE, Moreno S, Smith GR, Martin-Castellanos C 2008. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr Biol 18: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Soriano I, Marin L, Serrano R, Quintales L, Antequera F 2012. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J 31: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P-M, Géli V 2006. The multiple faces of Set1. Biochem Cell Biol 84: 536–548. [DOI] [PubMed] [Google Scholar]
- Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, et al. 2006. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281: 35404–35412. [DOI] [PubMed] [Google Scholar]
- de Massy B, Rocco V, Nicolas A 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J 14: 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Vezon D, Gendrot G, Gallois JL, Stevens R, Grelon M 2007. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J 26: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, Laine-Choinard S, Pelletier G, Mercier R, Nogue F, et al. 2009. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Diaz RL, Alcid AD, Berger JM, Keeney S 2002. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol Cell Biol 22: 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M 2005. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci 102: 737–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlinger B, Schlögelhofer 2011. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J Exp Bot 62: 1545–1563. [DOI] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR 2005. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci 102: 10952–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estreicher A, Lorenz A, Loidl J 2012. Mug20, a novel protein associated with linear elements in fission yeast meiosis. Curr Genet 58: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DH, Li YF, Fox ME, Smith GR 1997. A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics 146: 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QQ, Xu F, White MA, Petes TD 1997. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics 145: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Gutierrez-Velasco S, Martin-Castellanos C, Smith GR 2013. Protein determinants of meiotic DNA break hot spots. Mol Cell 49: 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K 2008. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res 36: 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SEL, Gray S, Neale MJ 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner JM, Bullard SA, Chrome C, Malone RE 1997. Molecular and genetic analysis of REC103, an early meiotic recombination gene in yeast. Genetics 146: 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu H-G, Mistrot C, Unal E, Koshland DE, DeRisi J, Gerton JL 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Lichten M 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14: 263–274. [DOI] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K 2005. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol 15: 1663–1669. [DOI] [PubMed] [Google Scholar]
- Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 9: e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M, Conti E 2013. The yeast Ski complex: Crystal structure and RNA channeling to the exosome complex. Cell 154: 814–826. [DOI] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ 2006. Genetics of meiotic prophase I in plants. Annu Rev Plant Biol 57: 267–302. [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC 2010. Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM 2007. C elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet 3: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Mlynarczyk-Evans S, Villeneuve AM 2010. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S 2006. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzel JV, Nabeshima K, Schvarzstein M, Turner BE, Villeneuve AM, Hillers KJ 2011. An asymmetric chromosome pair undergoes synaptic adjustment and crossover redistribution during Caenorhabditis elegans meiosis: Implications for sex chromosome evolution. Genetics 187: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. 2011. The landscape of recombination in African Americans. Nature 476: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Steiner WW, Shibata T, Ohta K 2007. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot Cell 6: 2072–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Tham W-H, Brar GA, Amon A 2005. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell 122: 861–873. [DOI] [PubMed] [Google Scholar]
- Hunter N 2007. Meiotic recombination. In Molecular genetics of recombination (ed. Aguilera A, Rothstein R), pp. 381–442 Springer, Berlin. [Google Scholar]
- Hyppa RW, Smith GR 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JK, Sherizen DE, Bhagat R, Manheim EA, McKim KS 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Jolivet S, Vezon D, Froger N, Mercier R 2006. Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11: 615–622. [DOI] [PubMed] [Google Scholar]
- Joyce EF, Pedersen M, Tiong S, White-Brown SK, Paul A, Campbell SD, McKim KS 2011. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J Cell Biol 195: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan F, Davidson MK, Wahls WP 2010. Meiotic recombination protein Rec12: Functional conservation, crossover homeostasis and early crossover/non-crossover decision. Nucleic Acids Res 39: 1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S 2004. Where the crossovers are: Recombination distributions in mammals. Nat Rev Genet 5: 413–424. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K, Protacio RU, Arora C, Keeney S 2004. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J 23: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S 2001. Mechanisms and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney S 2007. Spo11 and the formation of DNA double-strand breaks in meiosis. In Recombination and meiosis (ed. Egel R, Lankenau DH), pp. 81–123 Springer, Berlin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Kleckner N 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci 92: 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N 1996. Meiosis: How could it work? Proc Natl Acad Sci 93: 8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N 2006. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci 94: 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, et al. 2009. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell 20: 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, de Massy B 2010. Initiation of meiotic recombination in mammals. Genes 1: 521–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bourbon HM, de Massy B 2010. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake CM, Nielsen RJ, Hawley RS 2011. The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet 7: e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S 2011. ATM controls meiotic double-strand-break formation. Nature 479: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J 2010. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol Cell Biol 30: 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS 2006. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O’Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC 2002. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 242: 174–187. [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC 2003. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci 100: 15706–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, de Massy B 2011. The impressionistic landscape of meiotic recombination. Cell 147: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goldman ASH 1995. Meiotic recombination hotspots. Annu Rev Genet 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: Evidence for a covalently linked DNA-protein intermediate. EMBO J 14: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jang JK, Kato N, McKim KS 2002. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J 2006. S. pombe linear elements: The modest cousins of synaptonemal complexes. Chromosoma 115: 260–271. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F, McFarlane RJ, Loidl J 2004. S pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci 117: 3343–3351. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J 2006. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115: 330–340. [DOI] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, et al. 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci 96: 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276. [DOI] [PubMed] [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S 2007. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 116: 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Pittman DL, Nau JJ 1997. Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol Gen Genet 255: 410–419. [DOI] [PubMed] [Google Scholar]
- Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE 1996. Analysis of meiotic recombination pathways in yeast Saccharomyces cerevisiae. Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, et al. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Fowler KR, Smith GR 2013. Making chromosomes hot for breakage. Cell Cycle 12: 1327–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19: 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Arai K 2002. Cdc7 kinase complex: A key regulator in the initiation of DNA replication. J Cell Physiol 190: 287–296. [DOI] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK 2007. Synthesis-dependent strand annealing in meiosis. PLoS Biol 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, McKim KS 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, McGovern OL, Heinis FI, Yanowitz JL 2012. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefevre F, Pelletier G, Horlow C 2001. SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15: 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FC 2003. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K 2012. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell 47: 722–733. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Ohta K 2013. Spatiotemporal regulation of meiotic recombination by Liaisonin. Bioarchitecture 3: 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Pearlman RE, Heng HHQ, Traut W 1998. Chromosome cores and chromatin at meiotic prophase. Curr Top Dev Biol 37: 241–262. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J 2001. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci 116: 1719–1731. [DOI] [PubMed] [Google Scholar]
- Munroe RJ, Bergstrom RA, Zheng QY, Libby B, Smith R, John SW, Schimenti KJ, Browning VL, Schimenti JC 2000. Mouse mutants from chemically mutagenized embryonic stem cells. Nat Genet 24: 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S 2008. Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev 22: 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S 2014. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell 158: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Borde V, Shibata T, Lichten M, Ohta K 2003. Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res 31: 4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murton BL, Chin WL, Ponting CP, Itzhaki LS 2010. Characterising the binding specificities of the subunits associated with the KMT2/Set1 histone lysine methyltransferase. J Mol Biol 398: 481–488. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet 40: 1124–1129. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Villeneuve AM, Hillers KJ 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM 1999. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J 18: 6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N 2004. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Masai H 2006. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication. Mei4-dependent transcription as a potential target of meiotic checkpoint. J Biol Chem 281: 1338–1344. [DOI] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J 13: 5754–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Page SL, Nielsen RJ, Teeter K, Lake CM, Ong S, Wright KR, Dean KL, Agne D, Gilliland WD, Hawley RS 2007. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly 1: 172–181. [DOI] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Timofejeva L, Meeley RB, Sheridan WF, Cande WZ 2004. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303: 89–92. [DOI] [PubMed] [Google Scholar]
- Peciña A, Smith KN, Mezard C, Murakami H, Ohta K, Nicolas A 2002. Targeted stimulation of meiotic recombination. Cell 111: 173–184. [DOI] [PubMed] [Google Scholar]
- Petes TD 2001. Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Prieler S, Penkner A, Borde V, Klein F 2005. The control of Spo11’s interaction with meiotic recombination hotspots. Genes Dev 19: 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K 2004. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Villeneuve AM 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Reinholdt LG, Schimenti JC 2005. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114: 127–134. [DOI] [PubMed] [Google Scholar]
- Richard G-F, Kerrest A, Lafontaine I, Dujon B 2005. Comparative genomics of hemiascomycete yeasts: Genes involved in DNA replication, repair, and recombination. Mol Biol Evol 22: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Rocco V, Nicolas A 1996. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells 1: 645–661. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, Roeder GS 2006. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J 2007. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, Franklin FC 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S 2010. Genome destabilization by homologous recombination in the germ line. Nature 11: 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Murakami H, Fukuda T, Shibata T, Nicolas A, Ohta K 2007. Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic Acids Res 35: 1119–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, Kawasaki Y, Shibata T, Masai H, Ohta K 2008. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev 22: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Byers B 1978. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma 70: 109–130. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Sclafani RA 2000. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci 113: 2111–2117. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, Chin GM, Deneen B, Force SJ, Hari KL, et al. 1999. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino M-E, Borde V 2012. The spatial regulation of meiotic recombination hotspots: Are all DSB hotspots crossover hotspots? Exp Cell Res 318: 1347–1352. [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YH, Choi Y, Erdin SU, Yatsenko SA, Kloc M, Yang F, Wang PJ, Meistrich ML, Rajkovic A 2010. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet 6: e1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu Y, Mikawa T, Onuma M, Hirayama T, Shibata T 2010. A DNA-binding surface of SPO11–1, an Arabidopsis SPO11 orthologue required for normal meiosis. FEBS J 277: 2360–2374. [DOI] [PubMed] [Google Scholar]
- Shuster EO, Byers B 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136: 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Penkner A, Ohta K, Klein F, Nicolas A 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr Biol 11: 88–97. [DOI] [PubMed] [Google Scholar]
- Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, de La Roche Saint-André C 2004. Set1 in required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J 23: 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Béneut C, Chaplais E, Serrentino M-E, Borde V 2013. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Spirek M, Estreicher A, Csaszar E, Wells J, McFarlane RJ, Watts FZ, Loidl J 2010. SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe. Chromosoma 119: 59–72. [DOI] [PubMed] [Google Scholar]
- Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in C. elegans, illuminates a checkpoint that promotes crossover assurance. PLoS Genet 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Kohli J, Ludin K 2010. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr Genet 56: 237–249. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Székvölgyi L, Nicolas A 2010. From meiosis to postmeiotic events: Homologous recombination is obligatory but flexible. FEBS J 277: 571–589. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Morita T, Pearlman RE, Moens PB 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147: 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Xuan Z, Keeney S 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO, Shendure J 2009. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS ONE 4: e8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield SE, Keeney S 2012. Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Smith GR 1994. A heteromeric protein that binds to a meiotic homologous recombination hot spot: Correlation of binding and hot spot activity. Genes Dev 8: 1693–1702. [DOI] [PubMed] [Google Scholar]
- Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, Hollingsworth NM 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev 22: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JO, Reinholdt LG, Hartford SA, Wilson LA, Munroe RJ, Schimenti KJ, Libby BJ, O'Brien M, Pendola JK, Eppig J, et al. 2003. Toward the genetics of mammalian reproduction: Induction and mapping of gametogenesis mutants in mice. Biol Reprod 69: 1615–1625. [DOI] [PubMed] [Google Scholar]
- Woltering D, Baumgartner B, Bagchi S, Larkin B, Loidl J, De los Santos T, Hollingsworth NM 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol 20: 6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TC, Lichten M 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M 1995. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Gao J, Sharif WD, Davidson MK, Wahls WP 2004. Purification, folding and characterization of Rec12 (Spo11) meiotic recombinase of fission yeast. Protein Expr Purif 38: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res 25: 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kleckner N 1995. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J 14: 5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Ohta K, Yamada T 2013. Acetylated histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res 41: 3504–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY-H, Boiteux S, Hunter N 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol Cell 40: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kim KP, Kleckner NE, Storlazzi A 2011. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci 108: 20036–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Song Y, Cheng ZH, Wang YX, Zhu J, Ma H, Xu L, Yang ZN 2012. The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J 72: 271–281. [DOI] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11: 105–109. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]