Abstract
Homology search and DNA strand–exchange reactions are central to homologous recombination in meiosis. During meiosis, these processes are regulated such that the probability of choosing a homolog chromatid as recombination partner is enhanced relative to that of choosing a sister chromatid. This regulatory process occurs as homologous chromosomes pair in preparation for assembly of the synaptonemal complex. Two strand–exchange proteins, Rad51 and Dmc1, cooperate in regulated homology search and strand exchange in most organisms. Here, we summarize studies on the properties of these two proteins and their accessory factors. In addition, we review current models for the assembly of meiotic strand–exchange complexes and the possible mechanisms through which the interhomolog bias of recombination partner choice is achieved.
In most eukaryotes, Rad51 and Dmc1 carry out the search for a recombination partner and perform strand exchange during meiosis. These proteins and their accessory factors promote pairing with a homologous chromatid, rather than the sister.
Meiotic recombination creates genetic diversity and forms physical connections between homologous chromosomes, called chiasmata, which allow proper bipolar spindle attachment and thereby promote reductional chromosome segregation. Like recombinational repair in mitotic cells, the core of the meiotic recombination pathway is the homology search for a recombination partner that results in the formation of heteroduplex DNA segments in which bases from the Watson strand of one chromatid are paired with bases from the Crick strand of the other. Meiotic recombination follows premeiotic S phase such that the maternal and paternal chromosomes are both present as a pair of two identical sister chromatids. For recombination to execute its meiotic functions (pairing and crossing over), a region of DNA from one chromatid must form heteroduplex with the corresponding allelic DNA sequence on one of the two homolog chromatids, rather than the corresponding sequence on the sister chromatid.
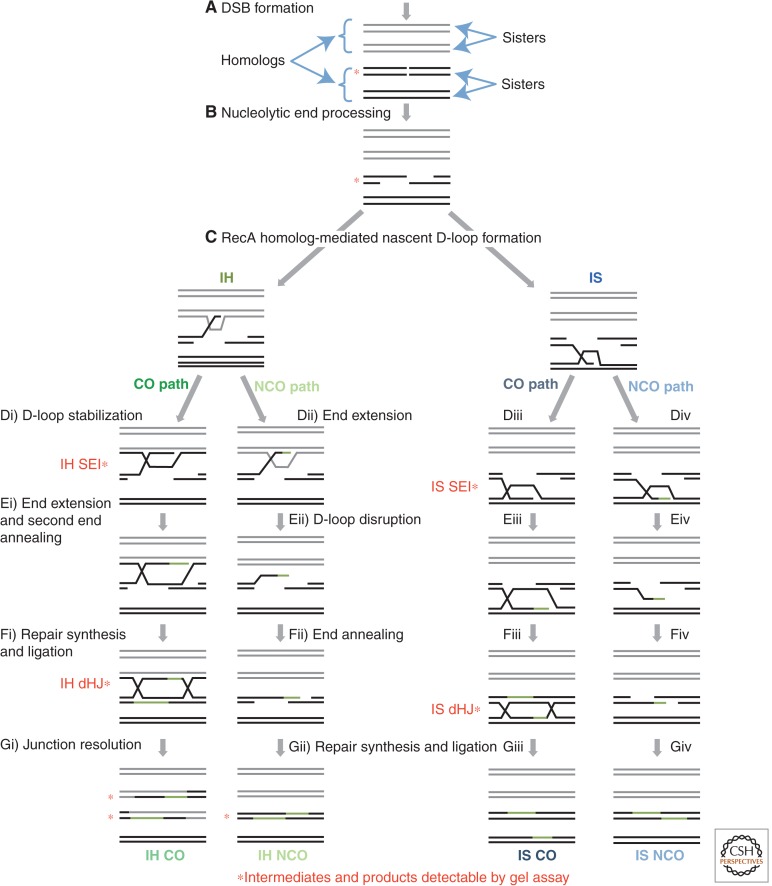
Meiotic recombination is initiated by double-strand breaks (DSBs) formed by the Spo11 transesterase and its many cofactors (Fig. 1) (see Hunter 2007 and Daley et al. 2014 for overview of meiotic recombination). The pair of DNA ends formed by Spo11 are nucleolytically processed, yielding 3′ single-stranded tails. Recombinosomes assemble de novo by binding 3′ single-stranded tails and then carry out homology search and DNA strand exchange. Initial homology recognition and strand exchange forms a displacement loop (D-loop) that is inferred to be unstable in vivo. D-loops can form by invasion of a homolog chromatid or a sister; however, although invasion of the sister can lead to DSB repair, it cannot produce a chiasma—the physical connection between homologs required for reductional segregation. The nascent D-loop formed by strand exchange has one of two fates. First, after extension of the invading end by polymerase, a D-loop can be dismantled, leading to DSB repair by the synthesis-dependent strand-annealing mechanism (Fig. 1C, Dii–Gii, and Div–Giv). In this case, the product of the recombination event is a noncrossover (NCO) recombinant, for instance, a product in which the chromosome arms that flank the site of recombination retain their starting configuration. The second potential fate of a nascent D-loop is stabilization to form a long-lived intermediate called a single-end invasion (SEI). SEIs usually go on to engage or “capture” the opposite or “second” DNA end from the same DSB, either by single-strand annealing or an independent strand invasion (Fig. 1C, Di–Gi, and Diii–Giii). Capture of the opposite DSB end then leads to formation of another metastable intermediate, the double Holliday junction (dHJ). Most dHJs are resolved to form reciprocal crossover (CO) products leading to formation of chiasmata.
Figure 1.
Meiotic recombination pathways. Four distinct recombination pathways can repair a meiotic DSB. Both strands of all four sister chromatids are shown with black and gray lines representing the two homologous chromosomes. Red asterisks indicate recombination intermediates and products that can be observed by Southern blots. (A) Meiotic recombination is initiated by the programmed introduction of DSBs. (B) 5′ to 3′ nucleolytic resection generates 3′ single-strand DNA (ssDNA) tails on both sides of the DSB. (C) RecA homologs locate an intact repair template on a homologous chromatid or the sister chromatid and catalyze strand exchange, generating a nascent D-loop intermediate. This D-loop structure is not stable enough to be observed in physical assays. (Di) Interhomolog (IH) crossover (CO) intermediates are formed when the nascent strand invasion of a homologous chromatid is stabilized. The resulting joint molecule (JM) is called an IH SEI. (Ei) Restorative DNA synthesis from the invading 3′ end (shown in green) extends the D-loop, allowing annealing of the second end of the DSB. (Fi) Further DNA synthesis and ligation of the remaining nicks generates an IH double Holliday junction (dHJ). (Gi) Resolution of the IH dHJ generates an IH CO in which the flanking DNA sequences are reciprocally exchanged. (Dii–Gii) IH NCOs are formed by a synthesis-dependent strand-annealing (SDSA) mechanism when an IH nascent JM is not stabilized. (Dii) DNA synthesis extends the 3′ end of a nascent D-loop. (Eii) The D-loop is disrupted. (Fii) This extended 3′ end anneals to the ssDNA tract on the opposite end. (Gii) Following further DNA synthesis and ligation, an IH NCO is formed. There is no reciprocal exchange of flanking regions in this recombination product. (Diii–Giii) Beside the choice of repair template, an IS CO is formed through the same biochemical steps as an IH CO. (Div–Giv) Similarly, an IS NCO forms through the same SDSA steps as an IH NCO. DSB, double-strand break; IS, intersister; NCO, noncrossover.
This review is focused on the activity of DNA strand–exchange proteins in forming D-loops and on factors that regulate that activity. Space limitations prevent us from discussing every significant finding. We, therefore, focus on studies in budding yeast for which in vivo analysis of recombination intermediates is most advanced, although a number of important findings from other organisms are discussed. We also recommend previous reviews on this subject by Zickler and Kleckner (1999) and Hunter (2007).
MEIOTIC STRAND–EXCHANGE PROTEINS
Most eukaryotic organisms possess two structural and functional homologs of the bacterial strand–exchange protein RecA, Rad51 and Dmc1, which cooperate during meiotic recombination. Although Rad51 is the only RecA-like strand-exchange protein that contributes to mitotic recombination, it also functions during meiotic recombination. Dmc1’s function is meiosis-specific. The RAD51 and DMC1 genes diverged following a gene duplication that occurred around the time of divergence of the pro- and eukaryotic kingdoms (Stassen et al. 1997; Ramesh et al. 2005). Most organisms that undergo meiosis fall into one of two categories with respect to Rad51 and Dmc1. The first category has orthologs of both Rad51 and Dmc1; in these organisms, both proteins are involved in meiotic recombination and required for the pairing and synapsis of homologs. The second category of organisms has Rad51, but lacks Dmc1. Organisms that possess both Rad51 and Dmc1 include budding and fission yeast, plants, and mammals. The “Rad51-only” group of organisms includes Drosophila melanogaster, Caenorhabditis elegans, and the fungus Sordaria macrospora. D. melanogaster and C. elegans are both capable of pairing and synapsing homologs in the absence of recombination (Dernburg et al. 1998; McKim et al. 1998; Villeneuve and Hillers 2001). S. macrospora is a counterexample in that it lacks Dmc1, but depends on Rad51 for homolog alignment and synapsis (Storlazzi et al. 2003). Phylogenetic studies show that “Rad51-only” organisms arose relatively recently by loss of the DMC1 gene rather than by divergence before the duplication from which it arose (Ramesh et al. 2005). Genes coding for key Dmc1 accessory factors, including the HOP2 and MND1 genes, are also absent in the lineages that have lost Dmc1, suggesting that the function of the three proteins is interdependent (Villeneuve and Hillers 2001; Ramesh et al. 2005). Keeping this history in mind may be useful in understanding the functional specialization of the two proteins and the mechanism(s) through which Dmc1-specific functions have been bypassed in “Rad51-only” species.
Rad51 and Dmc1 were first identified in budding yeast. The DMC1 gene was discovered on the basis of its meiosis-specific expression (Bishop et al. 1992). (Dmc1 officially stands for disrupted meiotic cDNA but the name was also intended as a homage to the rap group RUN DMC.) Sequencing of a DMC1 cDNA revealed 26% amino acid identity with RecA. Dmc1 is called Lim15 in Lilium longiflorum and Coprinus cinereus (Kobayashi et al. 1994; Hotta et al. 1995).
The RAD51 gene was discovered, along with the other members of the RAD50–57 series of genes, on the basis of its role in repairing DNA damage caused by ionizing radiation (Game and Mortimer 1974; Game et al. 1980). These genes were also shown to be required for production of viable spores. The structural similarity between Rad51 and RecA was discovered by cloning and sequencing members of the RAD52 epistasis group of recombination genes. This directed effort was based on the assumption that RecA’s structure and function would be conserved in evolution (Shinohara et al. 1992). The “core” domain of Rad51 was found to be 30% identical to RecA. Budding yeast Rad51 and Dmc1 are 45% identical to one another; however, the amino and carboxyl termini of the three proteins are unrelated. The name Rad51 is used to refer to orthologs in most species but the Drosophila protein is called SpnA and the Schizosaccharomyces pombe protein, Rhp51 (Jang et al. 1994; Staeva-Vieira et al. 2003).
Budding yeast dmc1 mutants are blocked at the strand-exchange step of recombination and accumulate processed DSBs (Bishop et al. 1992; Schwacha and Kleckner 1997). Homolog pairing and assembly of synaptonemal complexes are both partially defective. Most dmc1 mutant cells arrest in meiotic prophase owing to checkpoint activation. Budding yeast rad51 mutants also accumulate DSBs, although the amount of residual recombination is greater in rad51 than in dmc1 mutants and a subset of cells progress through meiosis and form spores (Bishop et al. 1992; Shinohara et al. 1992, 1997). However, the spores produced by rad51 mutants are dead because of inefficient DSB repair. If the checkpoint mechanism that causes prophase arrest in dmc1 mutants is inactivated, dmc1 mutant cells execute meiotic divisions in the presence of unrepaired DSBs and form dead spores (Lydall et al. 1996).
Either Rad51 or Dmc1 alone is capable of repairing meiotic DSBs; however, both RecA homologs are required to efficiently produce viable spores (Bishop et al. 1992; Shinohara et al. 1992; Tsubouchi and Roeder 2006). A recent study has clarified these roles by demonstrating that, although Rad51 is essential for efficient meiotic recombination, its strand-exchange activity is dispensable (Cloud et al. 2012). In accord with the dispensability of Rad51’s strand-exchange activity, this function is specifically inhibited during meiosis by direct binding of a protein called Hed1 (Tsubouchi and Roeder 2006; Busygina et al. 2008). Therefore, Dmc1 is the essential DNA strand–exchange factor for meiotic interhomolog recombination, whereas Rad51 performs a critical regulatory role (discussed below).
BIOCHEMICAL COMPARISON OF RAD51 AND DMC1 ACTIVITY
The biochemical properties of Rad51 are reviewed in detail in Morrical (2014); we will, therefore, limit this discussion to biochemical studies of Dmc1’s properties and compare them with those of Rad51.
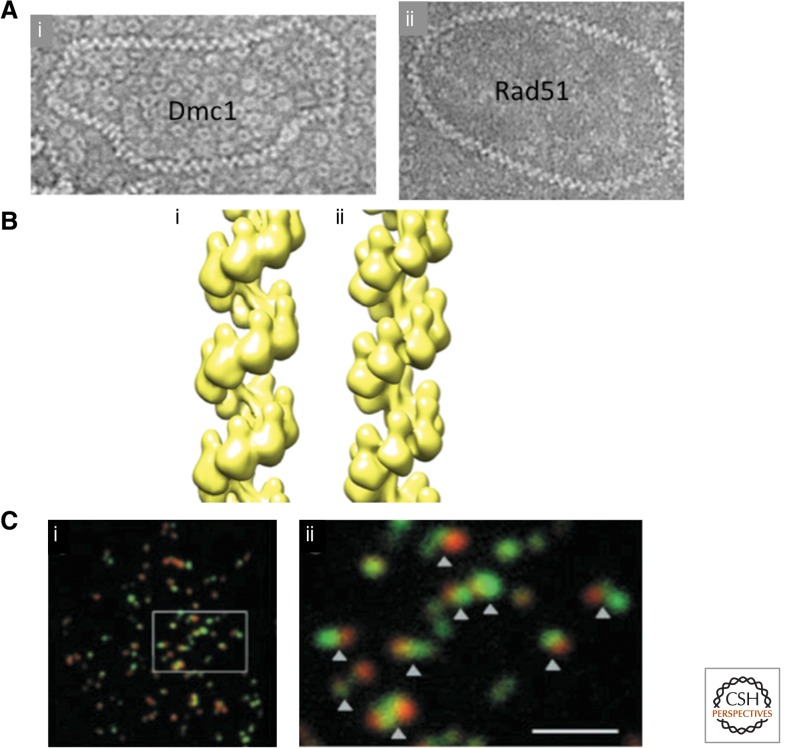
Biochemical studies of Dmc1 have, by and large, revealed striking similarities between its activity and that of Rad51 (Sung 1994; Baumann et al. 1996; Li et al. 1997; Hong et al. 2001). Both proteins have the ability to promote homology search and strand invasion/assimilation in the displacement loop (D-loop) assay. Both bind adenosine triphosphate (ATP) via a Walker box motif and are weak DNA-dependent ATPases, with kcat’s of ∼1.0 min−1 or less. Both proteins oligomerize on both single-strand DNA (ssDNA) and double-strand DNA (dsDNA) forming right-handed nucleoprotein filaments (Ogawa et al. 1993; Sehorn et al. 2004; Bugreev et al. 2005; Lee et al. 2005; Sauvageau et al. 2005; Sheridan et al. 2008). The DNA in these filaments is underwound and extended 1.5-fold, resulting in a conformation with 18 nt (or bp) and six protomers per helical turn. The filament has a helical pitch of 10 nm (Fig. 2A,B). It should be noted that two structural studies suggested Dmc1 might incorporate a different number of nucleotides per helical turn than Rad51 (Lee et al. 2005; Okorokov et al. 2010). However, more recent EM analysis provided definitive support for the canonical right-handed helical structure (Sheridan et al. 2008; Yu and Egelman 2010).
Figure 2.
Microscopic analysis of strand-exchange proteins. (A) Electron micrographs of human (i) Dmc1, and (ii) Rad51filaments coating a 1312 bp circular dsDNA plasmid. Note the high density of toroids in the background of the Dmc1 image (From Sheridan et al. 2008; reprinted, with permission, from Oxford University Press © 2008.) (B) Helical reconstructions of human (i) Dmc1, and (ii) Rad51 filaments (courtesy of E. Egelman). (C) Surface spread S. cerevisiae meiotic nuclei immunostained for Rad51 (green) and Dmc1 (red). (i) Low magnification view, and (ii) blow up of region indicated. Scale bar, 2 μm. (From Shinohara et al. 2000; reprinted, with permission, from the American Society of Plant Biologists © 1999.)
In addition to a high-affinity DNA-binding site (site I), which is responsible for binding of protomers to form nucleoprotein filaments, RecA has a second low-affinity-binding site (site II) that binds and extends dsDNA during the homology search (Mazin and Kowalczykowski 1998, 1996; Chen et al. 2008; Danilowicz et al. 2012). In RecA, site II also associates with the displaced ssDNA strand following strand exchange. Although studied in less detail than in RecA, site II appears to be conserved in Rad51 and Dmc1 (Cloud et al. 2012; Danilowicz et al. 2013).
Both Rad51 and Dmc1 promote D-loop formation by first assembling into nucleoprotein filaments on ssDNA and then searching for homology in duplex DNA. However, both proteins nucleate filaments on dsDNA more rapidly than does RecA, and this dsDNA-binding activity inhibits D-loop formation in vitro. As a result, D-loop reactions typically must be staged by allowing Dmc1-ssDNA filaments to form before addition of dsDNA substrates (Sung and Robberson 1995; Gupta et al. 1997; Hong et al. 2001).
Although the activity of these proteins in the D-loop reaction is quite limited in the presence of ATP and Mg2+ (0.5%–4%), substituting the nonhydrolyzable analogue AMP-PNP for ATP, or substituting Ca2+ for Mg2+ can stimulate D-loop formation ∼10-fold, to levels closer to those observed with RecA (∼50%) (Hong et al. 2001; Bugreev and Mazin 2004; Bugreev et al. 2005; Lee et al. 2005). Both of these conditions block ATP hydrolysis and stabilize the 100-nm pitch of the filament, which is active in homology search and strand exchange. These findings also show that, like the reaction promoted by RecA, the homology search and strand-exchange processes promoted by Rad51 and Dmc1 do not depend on the proteins’ ability to hydrolyze ATP. Strand exchange is driven by product stability (Menetski et al. 1990; Wittung et al. 1997; Peacock-Villada et al. 2012).
Importantly, addition of certain accessory factors dramatically enhances the strand-exchange activities of both Rad51 and Dmc1 under conditions that are thought to be physiologically relevant, involving Mg2+ and ATP as cofactors (Benson et al. 1998; New et al. 1998; Petukhova et al. 1998, 2005; Shinohara and Ogawa 1998). Although nonphysiologically high Ca2+ concentrations (low millimolar) have often been utilized to increase strand-exchange efficiency (see above), a recent study shows that micromolar Ca2+ promotes robust Dmc1 activity provided Mg2+ is also present at physiological concentrations (Chan et al. 2014). A crystal structure of an archael homolog of Dmc1 includes Ca2+ and Mg2+ bound to different sites in each protomer, and the same study showed Ca2+ stabilizes an active filament conformation (Qian et al. 2006). Thus, Ca2+ and Mg2+ are also likely to bind different sites in Dmc1.
One striking difference in the properties of Dmc1 and Rad51 is that Dmc1 has a much greater tendency to form octomeric ring structures in solution (see Fig. 2Ai) (Passy et al. 1999; Sehorn et al. 2004; Sheridan et al. 2008). The only crystal structure of Dmc1 at present is of the octomeric ring form, leading to the proposal that toroids might be the active form of the protein (Kinebuchi et al. 2004). However, the proposal is likely to be incorrect given that the same conditions that stimulate Dmc1 D-loop activity also stimulate the assembly of helical filaments on DNA (Sehorn et al. 2004; Lee et al. 2005). Thus, the functional significance of Dmc1’s strong tendency to form toroids, if any, remains to be determined.
Another set of studies comparing the activities of Dmc1 and Rad51 found that the two proteins were similar to one another, and to RecA, in that the strand-exchange reaction, represents a second, kinetically distinct phase that occurs after formation of a less stable homology-dependent ternary intermediate (Gupta et al. 2001). Furthermore, the efficiency of strand exchange displayed similar sensitivity to the guanine-cytosine (GC) content of substrates. These similarities suggest that the structure of the filament scaffold on which the core homologous recombination reaction occurs, and the general properties of the DNA–DNA interactions that occur on that scaffold are quite similar for Rad51 and Dmc1. This conclusion, along with the other similarities between Rad51 and Dmc1 described above, implies that interactions with specialized cofactors are likely to be important for functional specialization of the two proteins in vivo.
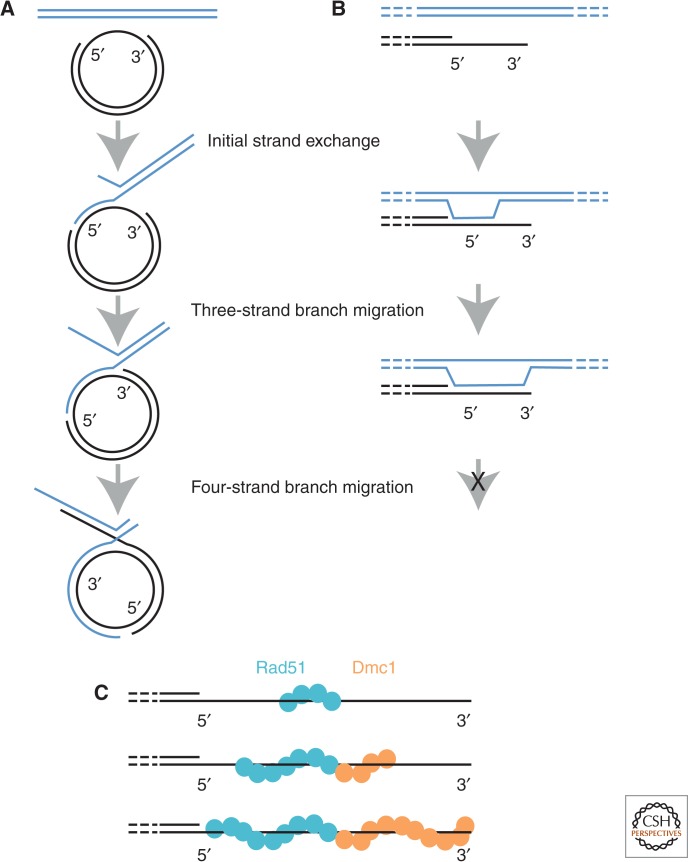
Although the activities of Rad51 and Dmc1 have been found to be similar in many respects, an intriguing difference has been reported for the fission yeast orthologs involving the ability of these proteins to promote reciprocal strand exchange in four-strand branch-migration reactions (Murayama et al. 2011). A circular plasmid containing a single-strand gap was tested for formation of joint molecules with a homologous linear duplex. One version of the linear duplex ended at the site of the 5′ end of the ssDNA gap, the second version at the 3′ end of the gap (Fig. 3A). The first of these substrates can only initiate a four-strand reaction in the 5′ to 3′ direction, the second only in the 3′ to 5′ direction. Rad51 and Dmc1 showed opposite substrate requirements for formation of four-strand joint molecules. Rad51 showed activity only for the 3′ to 5′ substrate and Dmc1 only for the 5′ to 3′ substrate. A key issue regarding interpretation of these results comes from the fact that meiotic recombination involves intermediates with 3′ single-stranded tails (Sun et al. 1991; Bishop et al. 1992). Dmc1-mediated exchange in the 5′ to 3′ direction could extend hybrid DNA by invasion of duplex and subsequent branch migration to a 3′ end. Once the 3′ end is reached, however, strand exchange is expected to cease; there is no obvious mechanism through which further branch migration could extend hybrid into a region of duplex such that a four-strand reaction would ensue (Fig. 3A). The same study that showed directional preferences in the four-strand reaction also used linear substrates to analyze the directionality of three-strand reactions and found no directional preference for Rad51 versus Dmc1 (Fig. 3B) (Murayama et al. 2011). Two possible roles for the four-strand reaction promoted by Dmc1 were proposed; removal of Spo11-linked oligonucleotides from 3′ ends, and extension of D-loops by assembly of Dmc1 on the displaced ssDNA strand. We note that it is also possible that the results of four-strand reactions reflect different directionalities of Rad51 and Dmc1 polymerization on DNA. This idea is suggested to us by analogy to RecA, which both polymerizes and promotes branch migration with a 5′ to 3′ bias (Register and Griffith 1985; Lindsley and Cox 1990; Roca et al. 1990). The possibility is interesting in light of our current model for Rad51-Dmc1 cooperation in which Dmc1 filaments are seeded at the end of Rad51 filaments (see below). If Dmc1 filaments elongate 5′ to 3′, they would tend to extend to the 3′ end of ssDNA tracts, whereas the Rad51 filaments would tend to extend toward ssDNA–dsDNA junctions (Fig. 3C). Furthermore, if Rad51 nucleates at a random position along an ssDNA tract, and polymerizes away from the 3′ end, the maximum length of a subsequent Dmc1 filament will be predetermined. Thus, depending on the position of Rad51 filament initiation, the relative lengths of Rad51 and Dmc1 filaments could differ from one event to the next (Fig. 2C).
Figure 3.
Directionality of Dmc1- versus Rad51-mediated branch migration. (A) Diagramatic representation of the four-strand reaction using substrates that can undergo conversion from three-strand branch-migration reactions to four-strand reactions. The left side of the linear duplex substrate (blue) is homologous to the ssDNA gap on the circular substrate. Strand exchange initiates in the ssDNA gap, branch migration extends the tract of heteroduplex to the ssDNA–dsDNA junction at the 3′ end of the ssDNA region. Then, further 5′-3′ branch migration results in reciprocal stand exchange via the four-strand reaction. (B) Inferred consequence of 5′-3′ branch migration on D-loops formed in vivo. Ends at DSB sites are processed to have 3′ overhanging ssDNA tails. Branch migration is expected to proceed to the 3′ end, but not be able to carry out a four-strand reaction because an end has been reached. (C) Model for strand-exchange filament elongation on 3′ ssDNA tails. Rad51 (blue) nucleates filament formation. Dmc1 filaments (orange) are seeded at the end of a Rad51 filament. The direction of filament elongation is proposed to be the same as the direction of branch migration for the four-strand reaction. Thus, the Rad51 filament is elongated in the 3′–5′ direction, the Dmc1 filament elongated in the 5′–3′ direction. This will tend to completely coat the entire ssDNA region, and perhaps lead to extension of the Rad51 filament into the flanking dsDNA.
Another study of human proteins examined the activity of the DNA translocase Rad54 on D-loops formed by Rad51 or Dmc1. Dmc1-associated D-loops were found to be more resistant to Rad54-mediated dissociation than were Rad51-associated D-loops (Bugreev et al. 2011). This result was interpreted to imply that the D-loops formed by Dmc1 are more stable than those formed by Rad51, and Dmc1-mediated stabilization of D-loops was interpreted to reflect a role for Dmc1 in channeling D-loop intermediates to the crossover rather than the noncrossover pathway. Although the experimental results of the study are convincing, the investigators’ interpretation of the results should be viewed with caution. Notably, this analysis did not take into account specificity of accessory factors for function with Rad51 versus Dmc1. Biochemical and genetic observations indicate that, despite a high degree of functional overlap, Tid1/Rdh54 displays a marked preference for Dmc1, whereas Rad54 functions more efficiently with Rad51 (Dresser et al. 1997; Shinohara et al. 2000; Nimonkar et al. 2012). Thus, Rad54 is not a “neutral” reagent for comparing the stability of Rad51- versus Dmc1-formed D-loops. It should also be noted that genetic studies indicate that Rad54 and Rdh54/Tid1 act to enhance the efficiency of the crossover pathway rather than channel intermediates away from it, as Mazin and colleagues suggested (as discussed in more detail below).
CYTOLOGICAL LOCALIZATION OF RAD51 AND DMC1
The functional cooperation of Rad51 and Dmc1 during meiotic recombination raises the question of how the two RecA homologs are arranged at a DSB. The first cytological characterization of Rad51 and Dmc1 in budding yeast showed that each formed a focal staining pattern in which the foci largely colocalized (Fig. 2Ci) (Bishop 1994). Focus formation depends on DSBs. This and other findings indicate that foci mark sites of ongoing recombination. Colocalization of Rad51 and Dmc1 foci, coupled with the functional cooperation of these RecA homologs in promoting the normal interhomolog bias of recombination, suggests that Rad51 and Dmc1 often reside at the same DSB site. However, Rad51 and Dmc1 foci do not precisely colocalize; rather, pairs of Rad51 and Dmc1 foci tend to be partially offset, lying side by side (Fig. 2Cii) (Shinohara et al. 2000). This separation is consistent with two-hybrid studies indicating that the two RecA homologs display strong homotypic interaction, and little heterotypic interaction (Dresser et al. 1997; Masson et al. 1999; Tarsounas et al. 1999). Cytological evidence for neighboring pairs of Rad51–Dmc1 homotypic filaments led to speculation that Rad51 might form a filament on one end of the DSB while Dmc1 forms a filament on the other (Shinohara et al. 2000). This speculation contributed to formal models in which the two ends of a DSB have distinct activities during recombination (Hunter and Kleckner 2001; Neale et al. 2005; Hunter 2007; Lao et al. 2008; Kim et al. 2010; Hong et al. 2013). Furthermore, a recent study of Rad51–Dmc1 staining patterns in Arabidopsis was interpreted as being consistent with the asymmetric loading model; Dmc1 foci were seen to lie adjacent to Rad51 foci (Kurzbauer et al. 2012). However, the asymmetric loading model does not easily accommodate our recent finding that Rad51 serves as an accessory factor for Dmc1-mediated D-loop formation both in vivo and in vitro (see below). Rad51 would need to stimulate Dmc1 filament assembly “in trans,” for instance, on the end opposite to that which it is bound. If, rather than loading on just one side, loading of Rad51 occurs on both sides of a DSB, the overlapping or side-by-side Rad51–Dmc1 foci would be explained if the end of a homofilament of Rad51 served as a seed for initiating polymerization of a homofilament of Dmc1 (Figs. 3C,4C). This model predicts that each DSB instigates assembly of a pair of Rad51 filaments, each of which stimulates assembly of a Dmc1 filament. Consistent with this idea, our unpublished studies in budding yeast provide evidence that side-by-side Rad51–Dmc1 foci come in pairs (MS Brown, J Grubb, and DK Bishop, unpubl.).
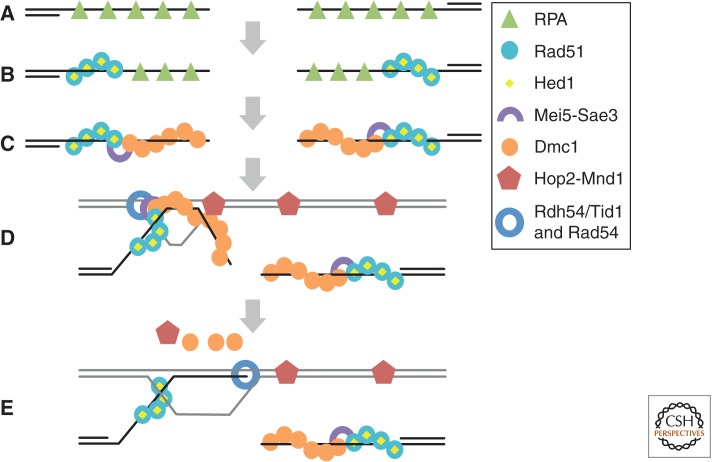
Figure 4.
Working model for assembly and function of Rad51–Dmc1 recombinosomes. (A) RPA binds to ssDNA regions formed by nucleolytic resection of DNA ends. (B) Rad51, with the aid of mediator proteins (not shown), displaces RPA. Rad51 is prevented from forming D-loops by the inhibitory protein Hed1. (C) Mei5–Sae3 promotes initiation of Dmc1 filaments at the end of a Rad51 filament. Once initiated, Dmc1 filaments elongate on DNA by homotypic protomer–protomer interactions. (D) Dmc1 carries out a homology search culminating in formation of a segment of heteroduplex DNA. Efficient formation of D-loops by Dmc1 requires interaction of the searching filament with a complex of Hop2–Mnd1 bound to the target dsDNA. (E) The Rdh54/Tid1 translocase, or the Rad54 translocase, binds the Rad51 filament and translocates along the heteroduplex, simultaneously displacing Dmc1 and extending the heteroduplex tract to the 3′ end. The end is, thus, rendered accessible for initiation of DNA synthesis.
It should also be noted that the longest dimension of the majority of Rad51 and Dmc1 structures visualized by conventional microscopy is usually shorter than the resolution limit of visible light (∼200 nm) (Bishop 1994). Thus, immunostaining foci are resolution-limited “airy disks” (Fig. 2C). Given that Rad51 and Dmc1 function by forming filaments on DNA, these filaments must usually be substantially shorter than 200 nm. If one assumes foci represent the helical nucleoprotein filaments described above, for instance, with one 10-nm helical turn containing 18 nt, the length of ssDNA within a diffraction limited focus is less than 400 nt. Thus, either filaments adopt a compressed configuration in cytological preparations or they contain DNA tracts of less than 400 bases. This is also true of the majority of Rad51 staining structures observed in association with mitotic DNA damage repair (Haaf et al. 1995; Bishop et al. 1998; Gasior et al. 2001).
ACCESSORY FACTORS FOR Dmc1-MEDIATED RECOMBINATION
Mei5–Sae3
Dmc1’s activity depends on the Mei5 and Sae3 proteins in budding yeast. Homologs of Mei5 and Sae3 are called Swi5 and Sfr1, respectively, in fission yeast and vertebrates. Mei5 and Sae3 bind directly to one another to form a complex important for recruitment of Dmc1 to recombinosomes in vivo (Hayase et al. 2004; Tsubouchi and Roeder 2004). Mei5 and Sae3 form immunonstaining foci that colocalize with each other and with foci formed by Dmc1. Formation of Mei5–Sae3 foci is not observed in dmc1 mutants indicating that normal association of Dmc1 and Mei5–Sae3 with chromatin is interdependent. Consistent with a role for Mei5–Sae3 in Dmc1 function, mei5 and sae3 mutants display clear phenotypic similarities to dmc1 mutants including failure to repair meiotic DSBs, accumulation of Rad51 foci, defective assembly of synaptonemal complexes, and uniform meiotic arrest in prophase (Hayase et al. 2004; Tsubouchi and Roeder 2004). In addition, Mei5–Sae3 foci do not form in a rad51 mutant in which weakly staining Dmc1 foci are assembled (Hayase et al. 2004). Together these findings suggest that Mei5–Sae3 promotes Dmc1 assembly downstream from Rad51 loading, a conclusion supported by biochemical observations described below.
The finding that Mei5–Sae3 is required for Dmc1 focus formation in vivo suggested that it might directly stimulate assembly of Dmc1 nucleoprotein filaments. A common characteristic of proteins that mediate nucleoprotein filament assembly is that they can overcome the inhibitory effects of RPA bound to ssDNA (Gasior et al. 2001; San Filippo et al. 2008). RPA is a highly expressed protein that binds specifically to ssDNA. RPA is essential for replication and also plays critical roles in recombination, as reviewed in detail in Zelensky et al. (2014). Factors that promote the assembly of strand-exchange proteins on RPA coated tracts of ssDNA are called recombination mediator proteins (see Zelensky et al. (2014) for a more complete discussion of mediators). The results of biochemical studies in budding yeast, fission yeast, and mouse indicate that Mei5–Sae3 and its homologs are capable of mediating Rad51 and Dmc1 filament formation.
Biochemical characterization of budding yeast Mei5 and Sae3 shows that the two proteins copurify as a single complex (Hayase et al. 2004; Ferrari et al. 2009), as is also the case for orthologs in fission yeast and mammals (Kokabu et al. 2011; Say et al. 2011; Yuan and Chen 2011). The complex binds both ssDNA and dsDNA via contacts with Mei5 (Say et al. 2011). Electrophoretic mobility shift assays show that binding of Mei5–Sae3 to ssDNA is preferred over dsDNA (Ferrari et al. 2009; Say et al. 2011). In addition, preferential binding to dsDNA–ssDNA forks has been observed and suggested to direct mammalian RAD51 or DMC1 filament assembly to the dsDNA–ssDNA junctions formed by nucleolytic 5′ end resection (Say et al. 2011). However, the in vivo relevance of this fork-binding activity remains to be tested. It should also be noted that the intrinsic DNA-binding activity of budding yeast Mei5–Sae3 is not sufficient for its stable association with recombination sites during meiosis because, as previously mentioned, dmc1 mutants are severely defective for formation of Mei5–Sae3 foci (Hayase et al. 2004; Tsubouchi and Roeder 2004). Similarly, Swi5–Sfr1 focus formation in mitotically cycling S. pombe cells requires Rhp51, the pombe Rad51 homolog (Akamatsu et al. 2007). In addition to binding DNA via Mei5, the Mei5–Sae3 complex has been shown to bind directly to three proteins: Rad51, Dmc1 (both via the amino terminus of Mei5), and RPA (Hayase et al. 2004; Ferrari et al. 2009; Say et al. 2011).
Budding yeast Mei5–Sae3 has recombination mediator activity; it suppresses the inhibitory effect of adding RPA to D-loop reactions before addition of Dmc1 (Ferrari et al. 2009). Although budding yeast Mei5–Sae3 only stimulates Dmc1 activity (Ferrari et al. 2009; Say et al. 2011), the Swi5–Sfr1 heterodimer from fission yeast can enhance the activity of both Rad51 and Dmc1 in vitro (Haruta et al. 2006; Kurokawa et al. 2008). Similarly, S. pombe and human Mei5–Sae3 orthologs stimulate Rad51 in vivo (Akamatsu et al. 2007; Yuan and Chen 2011). Thus, the specificity of Mei5–Sae3 for stimulating Dmc1 seen for budding yeast in vivo does not appear to be a conserved feature.
The requirement for both Rad51 and Mei5–Sae3 in Dmc1 focus formation suggested that Rad51 might function with Mei5–Sae3 to stimulate Dmc1 filament assembly (Bishop 1994; Hayase et al. 2004; Tsubouchi and Roeder 2004). This possibility is supported by the observation of robust simulation of Dmc1’s D-loop activity by a combination of Rad51 and Mei5–Sae3 (Cloud et al. 2012). This synergistic effect of combining Rad51 and Mei5–Sae3 was observed under more physiological conditions than those used to detect mediator activity in the absence of Rad51 (high-Mg2+ and low-Ca2+ concentration as opposed to high Ca2+ only). The role of Rad51 in stimulating Dmc1 assembly may normally occur via nucleation of a Dmc1 homopolymer on the end of a growing Rad51 filament, a scenario that would explain the side-by-side localization pattern of Rad51 and Dmc1 foci (Fig. 2C). Furthermore, genetic observations suggest that Rad51 can contribute to Dmc1 focus formation in a rad52 mutant background, in which cytologically visible Rad51 foci do not form (Bishop 1994; Gasior et al. 1998; Gasior 1999). This suggests that Rad51 can stimulate Dmc1 assembly without itself forming an extensive polymeric complex on DNA. Perhaps a minimal core complex composed of Rad51, Mei5, and Sae3 is sufficient to stimulate Dmc1 filament nucleation on ssDNA, or to stabilize such filaments once they form. Biochemical studies designed to determine the influence of Rad51, Mei5, and Sae3 on Dmc1 filament assembly are needed.
Overall, the biochemical properties of Mei5–Sae3 and its orthologs suggest that the protein has three related functions: assembly/stabilization of active Rad51 filaments, assembly/stabilization of Dmc1 filaments, and promoting the ability of Rad51 to enhance Dmc1 filament assembly/stabilization. All three of these activities are likely to be mechanistically related. Iwasaki and colleagues proposed that S. pombe Swi5/Sfr1 stabilizes filaments by binding to the helical groove, in which its elongated form could bridge adjacent protomers of Rad51 (Kokabu et al. 2011). Structural analysis of S. pombe Swi5/Sfr1 revealed it to be an elongated and kinked α-helical coiled-coil structure (Kuwabara et al. 2010, 2012; Kokabu et al. 2011) that could fit in the groove of a Rad51 or Dmc1 filament. If the protein does bind in the groove, it will be of interest to determine how such binding can occur without disruption of Rad51- (or Dmc1-) mediated homology search and strand-exchange activities, which occur within the groove (Haruta et al. 2008; Kokabu et al. 2011; Kuwabara et al. 2012). Following Iwasaki’s suggestion, an attractive possibility to explain the role of Mei5–Sae3’s role in promoting Rad51-dependent Dmc1 activity is that it stabilizes protein–protein contacts between adjacent Rad51 and Dmc1 protomers by forming a bridge at the junction between end-to-end Rad51 and Dmc1 homofilaments on the same piece of ssDNA (Fig. 4C).
The meiotic function of Mei5–Sae3 homologs in other organisms might not be entirely conserved with S. cerevisiae. At the time of writing, there is no information on the meiotic functions of the mammalian complex (called Swi5–Sfr1 or Swi5–Mei5). Mutant analysis of meiotic recombination in S. pombe indicates that Swi5–Sfr1 is specifically required for interhomolog joint molecule (JM) formation (but not for intersister [IS] JM formation) (Hyppa and Smith 2010). However, the role of this putative interhomolog-specific Swi5–Sfr1 function appears to be distinct from that of Mei5–Sae3 in Saccharomyces cerevisiae meiosis, because it seems to act through Rad51 and not Dmc1 at DSB hot spots (Hyppa and Smith 2010). Yet, JM formation at DSB cold spots requires Rad51, Swi5–Sfr1, and Dmc1 (Hyppa and Smith 2010); and Swi5–Sfr1 acts as a mediator of Dmc1 function in vitro (Haruta et al. 2006). These observations leave open the possibility that S. pombe Swi5–Sfr1 acts analogously to S. cerevisiae Mei5–Sae3 in wild-type S. pombe.
Hop2–Mnd1
A second critical accessory factor in meiotic strand invasion and exchange is the Hop2–Mnd1 heterodimer. The HOP2 gene was found in a screen for mutants defective in meiotic gene conversion (Leu et al. 1998). The MND1 gene was identified among genes that are specifically expressed in meiosis as being required for DSB repair, JM formation, and chromosome segregation (Rabitsch et al. 2001; Gerton and DeRisi 2002). Interaction between Hop2 and Mnd1 proteins was discovered following the demonstration that high copy numbers of MND1 can suppress phenotypes associated with a non-null allele of HOP2 (Tsubouchi and Roeder 2002). Hop2 has been shown to bind Mnd1 in crude extract pull down experiments (Tsubouchi and Roeder 2002) as well as by copurification (Chen et al. 2004; Petukhova et al. 2005; Pezza et al. 2006, 2007). Furthermore, molecular genetic and biophysical studies have established that the two proteins interact via coiled-coil motifs to form a tightly bound heterodimeric complex with a stoichiometry of 1:1 (Tsubouchi and Roeder 2002; Pezza et al. 2006). Small angle X-ray scattering analysis suggests that the heterodimer has a kinked, elongated V shape with the amino termini of both Hop2 and Mnd1 at the termini of the V (Zhao et al. 2013). Correspondingly, hop2 and mnd1 single mutant phenotypes are very similar to hop2 mnd1 double-mutant phenotypes. All show uniform prophase arrest in budding yeast with unrepaired DSBs and nonhomologous pairing/synapsis (Rabitsch et al. 2001; Gerton and DeRisi 2002; Tsubouchi and Roeder 2002; Chen et al. 2004; Zierhut et al. 2004; Henry et al. 2006). Defects in meiotic recombination have also been reported for single mutants in mouse, Arabidopsis, and fission yeast (Nabeshima et al. 2001; Petukhova et al. 2003; Schommer et al. 2003; Saito 2004; Domenichini et al. 2006; Kerzendorfer et al. 2006; Panoli et al. 2006; Vignard et al. 2007). Together, these findings suggest that the major functional form of Hop2 and Mnd1 is the heterodimeric form, although certain results involving the mouse homologs raise the possibility that Hop2 can contribute recombinogenic activity in the absence of Mnd1 both in vitro and in vivo (Pezza et al. 2006, 2013).
Additional findings led to the view that Hop2–Mnd1 functions to promote Dmc1-mediated homology search and strand exchange in budding yeast. First, there are strong phenotypic similarities between dmc1, hop2, and mnd1 single mutants, and dmc1 mnd1 or dmc1 hop2 double mutants resemble dmc1 single mutants (Tsubouchi and Roeder 2002; Zierhut et al. 2004; Henry et al. 2006). Second, overexpression of RAD51 was shown to bypass the arrest conferred by mnd1 or hop2 mutants, as was previously shown to be the case for dmc1 mutants (Tsubouchi and Roeder 2002; Zierhut et al. 2004) (see discussion below). Third, other mutations previously identified as being required for the uniform arrest observed in dmc1 mutants were shown to also be required for arrest conferred by hop2 or mnd1 mutants (Tsubouchi and Roeder 2002; Zierhut et al. 2004; Henry et al. 2006). Mutations that bypass arrest include those that block checkpoint signaling and axial element assembly (as described below in the section Regulation of Recombination Template Choice during Meiotic Recombination). Although no significant colocalization between Hop2–Mnd1 and Dmc1 has been observed (as discussed further below), Dmc1 foci and DSBs persist indefinitely in hop2 and mnd1 mutants suggesting Dmc1 is unable to promote JM formation in the absence of Hop2–Mnd1 in vivo (Leu et al. 1998; Gerton and DeRisi 2002; Tsubouchi and Roeder 2002; Zierhut et al. 2004; Lao et al. 2013). These findings support the hypothesis that Hop2–Mnd1 plays a specific role in promoting interhomolog recombination by functioning as a Dmc1 cofactor. The fact that Dmc1 foci form normally in the absence of Hop2–Mnd1 suggests that Hop2–Mnd1 acts on Dmc1 after the Mei5–Sae3 complex exerts its influence (Tsubouchi and Roeder 2002; Hayase et al. 2004; Zierhut et al. 2004; Vignard et al. 2007).
Initial biochemical studies of budding yeast Hop2–Mnd1 showed that the protein stimulates Dmc1 D-loop activity only threefold (Chen et al. 2004). However, this study was flawed because the Hop2 protein used lacked its normal carboxyl terminus, which is coded by a third exon in the HOP2 gene (Chan et al. 2014). The third exon had not been discovered at the time the expression construct used in the earlier experiments was built. Importantly, wild-type Hop2–Mnd1 stimulates Dmc1 activity 30-fold or more. This dramatic stimulation was also found for the mouse and fission yeast proteins (Petukhova et al. 2005; Enomoto et al. 2006; Ploquin et al. 2007). Thus, Hop2–Mnd1 can be viewed as an essential cofactor for Dmc1’s homology search and strand-exchange functions in vivo and in vitro.
A number of different biochemical interactions have been proposed to play a role in the stimulation of Dmc1’s strand-invasion activity by Hop2–Mnd1. Direct interaction of Hop2–Mnd1 with Dmc1 has been observed by surface plasmon resonance (Petukhova et al. 2005). This direct interaction is likely to be responsible for the Hop2–Mnd1-mediated stabilization of ssDNA–Dmc1 filaments that have been observed in vitro (Pezza et al. 2007; Zhao et al. 2013). However, Hop2–Mnd1 binds DNA directly, preferring dsDNA to ssDNA (Chen et al. 2004; Enomoto et al. 2006). Furthermore, the finding that preincubation of duplex DNA with Hop2–Mnd1 provides optimal stimulation of yeast Dmc1 suggested that either nonspecific dsDNA capture or alteration of the structure of duplex target might be important for the mechanism of stimulation (Chen et al. 2004; Chan et al. 2014). Indeed, affinity capture and cosedimentation experiments using the mouse and human orthologs of Hop2–Mnd1 and Dmc1, respectively, have provided evidence for homology-independent capture of dsDNA by ssDNA–Dmc1 filaments (Pezza et al. 2007). A similar activity of Hop2–Mnd1 has been seen with ssDNA–Rad51 filaments (Chi et al. 2007). In addition, Hop2–Mnd1 has been shown to dramatically alter the structure of duplex DNA via single molecule analysis, which revealed that Hop2–Mnd1 can promote extensive condensation of duplex DNA (Pezza et al. 2010). These findings led to the hypothesis that Hop2–Mnd1 stimulates Dmc1-mediated homology searching by promoting nonspecific binding of duplex by ssDNA–Dmc1 filaments followed by condensation of dsDNA in a manner that increases the rate of homology search (Fig. 6E) (Pezza et al. 2010).
Figure 6.
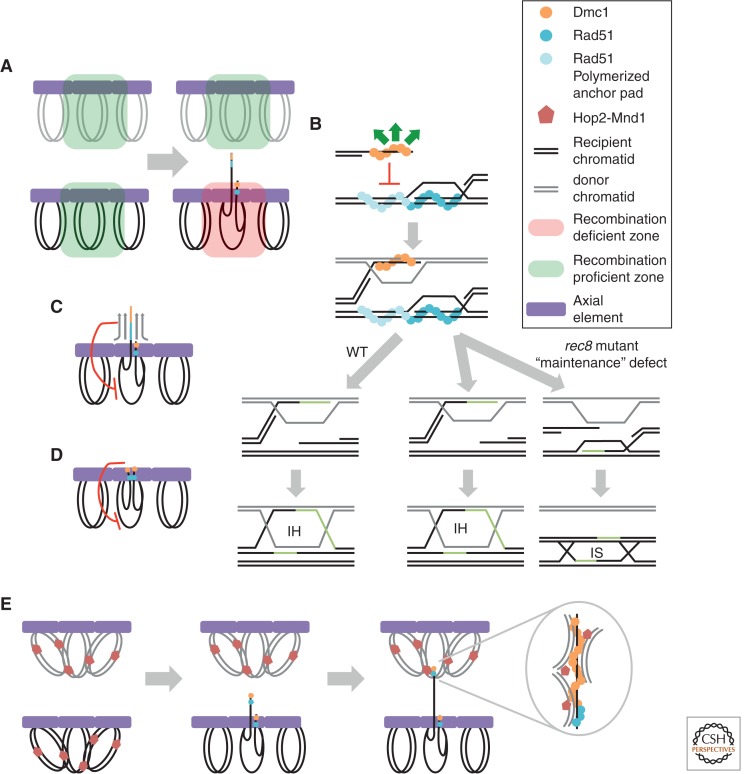
IH bias models. (A) Barrier to sister chromatid recombination (BSCR). In response to DSB formation, a local recombination deficient zone (red) is created on the recipient homolog. Because there are no DSBs at the allelic position on the homolog, the donor homolog remains a recombination proficient zone (green). (B) Anchor pad model. The Rad51 filament (blue) end of the DSB invades the sister chromatid. Further Rad51 polymerization beyond the D-loop (light blue) creates an “anchor pad” that blocks invasion of the sister chromatid by the Dmc1 end of the DSB. Left with no other choice, the Dmc1 end invades a homologous chromatid. In WT cells, this IH nascent interaction proceeds to an IH dHJ. In rec8 cells, the nascent IH interaction has an equal probability of forming an IH or an IS dHJ. Note that the version of the model shown has Rad51 and Dmc1 loaded on opposite DSB ends (Hong et al. 2013). However, the model could be modified to account for loading of the two RecA homologs on both ends, (C) steric hindrance model 1. A scaffold structure, dependent on Rad51, projects the Dmc1 filament away from the recipient homolog axis. The scaffold precludes invasion of the sister chromatid, (D) steric hindrance model 2. The DSB ends are held in close proximity to the recipient homolog axis, whereas the sister chromatid loop is distant from the axis. (E) Hop2–Mnd1 creates recombination proficient zones distant from DSBs. Hop2–Mnd1 associates with chromatin nonspecifically, locally clustering DNA. A DSB on the recipient homolog results in local depletion of Hop2–Mnd1. Aided by Hop2–Mnd1-mediated clustering of chromatin, the Dmc1 filament is able to simultaneously sample discontinuous regions of chromatin for homology, but precluded from searching in the vicinity of the DSB owing to the lack of Hop2–Mnd1. Note that, except for the cartoon of the anchor pad model in B, Rad51 and Dmc1 are both shown on both ends of a DSB. However, the originally proposed version of the anchor pad model (Hong et al. 2013) could be modified to account for symmetric RecA homolog loading.
Although Hop2–Mnd1 can stimulate certain Rad51 activities in vitro, it is unclear at present whether this activity plays a significant role in vivo (Petukhova et al. 2005). One report suggested that Arabidopsis Mnd1 is capable of enhancing Rad51-mediated IS repair in a Dmc1−/− mutant (Vignard et al. 2007), but a more recent and extensive study came to the opposite conclusion (Uanschou et al. 2013). Similarly, there are conflicting reports concerning the radiation sensitivity of somatic cells in Mnd1−/− mutants (Domenichini et al. 2006; Kerzendorfer et al. 2006). In budding yeast, Hop2 and Mnd1 are not required for Rad51-mediated meiotic recombination in the dmc1 hed1 double-mutant background (Lao et al. 2013). Furthermore, neither budding yeast nor fission yeast Hop2–Mnd1 stimulates Rad51’s D-loop activity in vitro (Ploquin et al. 2007; Chan et al. 2014). Finally, no evidence of Hop2–Mnd1 stimulation of Rad51 activity has been reported for Hop2−/− or Mnd1−/− knockout mice (Petukhova et al. 2003). Thus, further evidence is required to determine whether Hop2–Mnd1 influences Rad51 activity in vivo. If it does, the function is likely to be a more recent evolutionary invention than the role of Hop2–Mnd1 in controlling Dmc1’s activity.
Cytological characterization of Hop2–Mnd1 indicates that the protein associates with chromosomes independent of other components of the meiotic recombinosomes. Hop2 and Mnd1 localize to chromosomes in a dense focal staining pattern (Leu et al. 1998; Tsubouchi and Roeder 2002; Zierhut et al. 2004). Surprisingly, Hop2–Mnd1 foci show key differences compared with those formed by other proteins that play direct roles in D-loop formation. First, Hop2–Mnd1 foci form independently of DSBs (Leu et al. 1998; Tsubouchi and Roeder 2002; Zierhut et al. 2004; Vignard et al. 2007). Second, in contrast to other accessory proteins, Hop2–Mnd1 foci do not show high levels of colocalization with Rad51 or Dmc1 (Tsubouchi and Roeder 2002; Zierhut et al. 2004; Vignard et al. 2007). These findings led to the suggestion that Hop2–Mnd1 might influence the activity of recombinosomes indirectly via control of chromosome pairing interactions. This idea was supported by the finding that mutation of the S. pombe homolog of Hop2 (Meu13) reduced chromosome pairing somewhat in a spo11 mutant background and that S. cerevisiae hop2 mutants build synaptonemal complexes between nonhomologous chromosomes (Nabeshima et al. 2001; Tsubouchi and Roeder 2003). However, biochemical evidence for direct interaction between Hop2–Mnd1 and strand-exchange proteins (Petukhova et al. 2005), as well as evidence showing that Hop2–Mnd1 can stimulate strand exchange in purified systems (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Ploquin et al. 2007; Chan et al. 2014), suggest Hop2–Mnd1 functions directly at sites of meiotic recombination in vivo. The simplest explanation for the absence of significant colocalization between Dmc1 and Hop2–Mnd1 is that the dsDNA-binding activity of Hop2–Mnd1 accounts for the presence of DSB-independent foci, and only a small fraction of Hop2–Mnd1 foci are associated with DNA recombination intermediates at any given moment; for instance, the preponderance of Hop2–Mnd1 complexes are not engaged with Dmc1 (and/or Rad51) and may obscure the relatively small subset of complexes that are.
Tid1/Rdh54 and Rad54
The DNA translocase enzymes Tid1/Rdh54 and Rad54 both play a role in meiotic recombination. They possess a potent dsDNA-dependent ATPase activity and function as molecular motors, translocating along dsDNA. Although multiple mechanisms of translocase action have been proposed, it is important to emphasize that both translocases positively regulate meiotic recombination including crossing over (Ceballos and Heyer 2011; also reviewed in Daley et al. 2014). A tid1 single mutant displays a substantial delay in forming JM crossover intermediates but high levels of CO products are eventually formed (Shinohara et al. 1997, 2000, 2003b). A rad54 mutant displays normal CO levels but reduced spore viability (Shinohara et al. 1997; Schmuckli-Maurer and Heyer 2000). Importantly, rad54 tid1 double mutants rarely produce spores, and those produced are mostly inviable (Shinohara et al. 1997). Furthermore, recombinant products fail to form, DSBs persist, and end hyperresection occurs demonstrating a substantially redundant role for Rad54 and Tid1 in promoting both crossover and noncrossover recombination (Shinohara et al. 1997). In addition to this role in DSB repair, Rad54 and Tid1 have another role in preventing the sequestration of Rad51 and Dmc1 in nonrecombinogenic complexes by displacing the proteins from dsDNA (Holzen et al. 2006; Shah et al. 2010).
The same dsDNA translocation activity can account for Rad54 and Tid1’s roles in both promoting productive recombination and preventing nonrecombinogenic complex formation. Biochemical studies have showed that Rad54 is capable of stripping Rad51 from the heteroduplex formed at sites of strand invasion (Kiianitsa et al. 2006; Wright and Heyer 2014). Given their partial redundancy and similar phenotypes in vivo, it seems likely that Tid1/Rdh54 and Rad54 function similarly. Thus, the translocases are probably recruited to the ends of Dmc1 and Rad51 filaments—whether recombinogenic or nonrecombinogenic—and mediate their disassembly. In the case of recombinogenic filament disassembly, homology between the donor and acceptor chromatids would allow the translocase to act as a “heteroduplex pump,” which mechanistically couples the extension of heteroduplex DNA to stabilize the nascent joint molecule with removal of RecA homolog protomers from the heteroduplex DNA product (Fig. 4D,E) (Li and Heyer 2008; Wright and Heyer 2014). Finally, the DNA translocases may also contribute to the efficiency of repair by virtue of their nucleosome remodeling activity, which could also act to stabilize D-loops (Alexeev et al. 2003; Zhang et al. 2007; Hicks et al. 2011).
MEIOB and SPATA22
Recently, genetic studies in mice identified two proteins, MEIOB and SPATA22, which are critical for fertility (La Salle et al. 2012; Ishishita et al. 2013; Luo et al. 2013; Souquet et al. 2013). MEIOB is of particular interest as it contains a so-called oligonucleotide/oligosaccharide binding (OB) fold, which is common to proteins that bind specifically to tracts of ssDNA, including RPA. Indeed, biochemical experiments show that MEIOB binds ssDNA (Luo et al. 2013; Souquet et al. 2013). A truncated form of the protein was also found to have ssDNA-specific 3′ exonuclease activity (Luo et al. 2013). Immunostaining studies show that both proteins display very high levels of interdependent colocalizing foci, which also colocalize with RPA. Meiob−/− single mutants display evidence of DNA damage accumulation and fail to form MLH1 foci (Luo et al. 2013; Souquet et al. 2013), which mark positions of crossovers in wild-type cells (Anderson et al. 1999). RAD51 and DMC1 foci form in these mutants, but two studies reported that the foci do not persist as long as in wild type (Ishishita et al. 2013; Souquet et al. 2013). These results implicate MEIOB and SPATA22 as components of the meiotic recombinosome that play a role during the RAD51/DMC1-dependent stage of recombination.
REGULATION OF RECOMBINATION TEMPLATE CHOICE DURING MEIOTIC RECOMBINATION
In diploids, each meiotic DSB could form a D-loop with any of three intact repair templates, one on the sister chromatid and two on the pair of homolog chromatids (Fig. 1). Yet, meiotic DSBs are preferentially repaired using a homolog chromatid; IH bias in budding yeast yields a 5:1 ratio of interhomolog to IS JM species, exceeding the 2:1 ratio expected by random choice between the three available intact chromatids. This is in stark contrast with mitotic DSB repair, which shows strong IS bias (Jackson and Fink 1985; Kadyk and Hartwell 1992; Schwacha and Kleckner 1994; Bzymek et al. 2010).
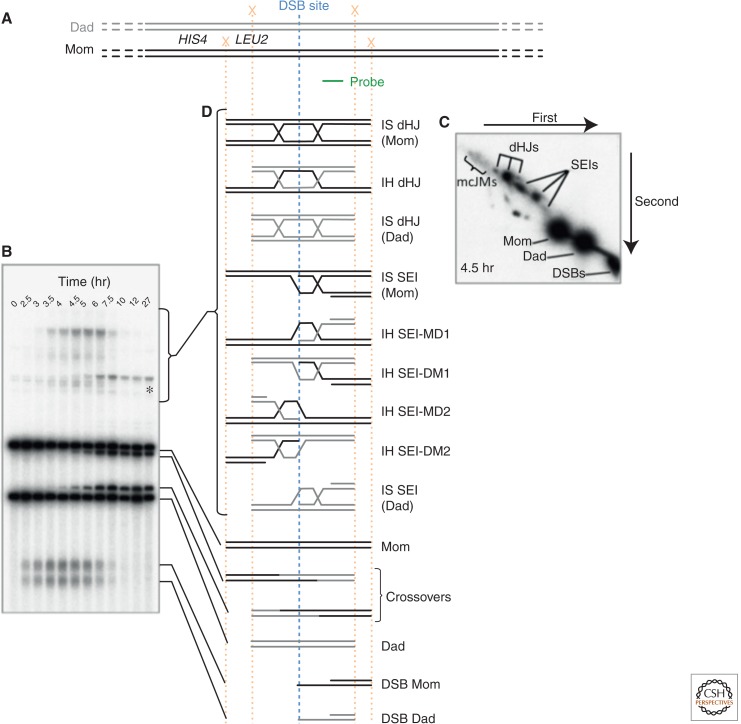
Southern blotting assays that allow detection of DNA intermediates and products of meiotic recombination have facilitated the study of IH bias in fungi (Fig. 5) (Cao et al. 1990; Schwacha and Kleckner 1994; Cromie et al. 2006). In budding yeast, Southern blots of two-dimensional (2D) gels revealed that two JM intermediates—the SEI and the dHJ—are biased toward the homolog chromatid rather than the sister chromatid (Schwacha and Kleckner 1995; Hunter and Kleckner 2001). In addition to these CO intermediates, one-dimensional (1D) gel electrophoresis variations can reveal both IH CO and NCO product levels. However, these physical assays cannot detect nascent D-loops—the last common intermediate between CO and NCO pathways—and are similarly unable to detect IS CO and NCO products. Additionally, assumptions are required to determine the lifetimes of intermediates. Despite these significant limitations, JM analysis via 2D gels has driven our current understanding of the mechanism of partner choice in meiosis.
Figure 5.
HIS4::LEU2 DSB hot spot. (A) Structure of the HIS4::LEU2 DSB hot spot. The paternal and maternal homologs are shown in gray and black, respectively. A single DSB hot spot is located downstream from the LEU2 gene, which was inserted adjacent to the HIS4 locus (Cao et al. 1990). XhoI restriction endonuclease cleavage sites (X) are engineered such that cleavage of the paternal and maternal homologs yields fragments of distinct sizes on Southern blots (probe location in green). (B) A representative 1D gel, showing the structure of chromatids at the HIS4::LEU2 DSB hot spot throughout a meiotic time course experiment. *A meiosis-specific band resulting from gene conversion of the XhoI site closest to the DSB site. (C) Single time point of a representative 2D gel showing branched molecules (SEIs, dHJs, and multichromatid joint molecules [mcJMs]) migrating off of the linear arc. (D) Schematic of the various species observable at the HIS4::LEU2 DSB hot spot. Note that although both products and intermediates of meiotic recombination are observable, crosslinking of the DNA by ultraviolet light before DNA purification is required to stabilize the branched JMs. SEI nomenclature is from Kim et al. 2010. (From data in Figure 2 in Oh et al. 2007; modified, with permission.)
Using the 2D gel assay, a number of proteins have been implicated by genetic analysis as being required for the fivefold homolog bias observed in budding yeast in vivo (Hunter 2007). Positive regulators of homolog bias include the RecA homologs Rad51 and Dmc1 (Schwacha and Kleckner 1997), as well as DNA damage checkpoint signaling proteins (Grushcow et al. 1999; Thompson and Stahl 1999; J Grubb and DK Bishop, unpubl.), including components of the checkpoint clamp, the clamp loader, and the ataxia telangiectasia- and RAD3-related (ATR/Mec1) checkpoint kinase. In budding yeast, the checkpoint clamp is composed of Rad17, Mec3, and Ddc1; the clamp loader is a derivative of the replication factor C (RFC) complex that contains the checkpoint specific subunit Rad24 (Hochwagen and Amon 2006). Also critical for IH bias is a set of proteins that are associated with the axial/lateral elements of the synaptonemal complex. These proteins include Red1 and Hop1, two abundant proteins that are thought to be structural components of axial/lateral elements of the synaptonemal complex (SC), as well as Mek1, a key meiotic kinase whose activity depends on Red1 and Hop1. Importantly, Red1, Hop1, and Mek1 are downstream components of a DSB-dependent checkpoint signal in which Mec1- and Tel1-mediated phosphorylation of Hop1 is translated into a DSB-proximal Mek1 signal (Wan et al. 2004; Niu et al. 2005; Carballo et al. 2008). Finally, cohesin has been proposed to be a negative regulator of meiotic IH bias (Kim et al. 2010).
The identification of these players led to a number of molecular models of IH bias, although the critical aspects of the mechanism remain unclear. A key point of discussion has been whether homolog bias results from negative regulation of IS recombination or positive regulation of IH recombination (Callender and Hollingsworth 2010; Hong et al. 2013). However, the two possibilities are not mutually exclusive and a number of results suggest that both modes of regulation are involved.
Rad51 and Dmc1 Must Cooperate to Impose Interhomolog Bias of Template Choice
Although mutation of Dmc1 was shown to result in a strong block to meiotic recombination, it was later shown that this strong block requires Hed1, a protein capable of inhibiting the activity of Rad51 (Tsubouchi and Roeder 2006). If Hed1 is eliminated, Rad51 is capable of promoting DSB repair. This Dmc1-independent activity of Rad51 results in eventual completion of the meiotic program for most cells and generation of tetrads in which ∼65% of spores are viable. Among these spores, the frequency of IH recombination is only modestly reduced relative to that observed in wild type. These and earlier observations suggested that Rad51 is capable of substituting for Dmc1 or that Dmc1 play a relatively minor role in supporting Rad51’s strand-exchange activity, with Hed1 acting to prevent Rad51 from acting until Dmc1 is incorporated into recombinosomes. However, analysis of recombination intermediates showed that interhomolog bias is drastically diminished when Rad51 is activated to take the place of Dmc1 (Hong et al. 2013; Lao et al. 2013). This defect in interhomolog interactions is manifested by defects in the formation of interhomolog joint molecules, chromosome pairing and synapsis, and exit from prophase (Lao et al. 2013). Given that nearly normal levels of crossovers do eventually form in dmc1 hed1 despite an extreme deficiency in interhomolog bias, it is clear that homeostatic mechanisms compensate, albeit incompletely, for the deficiency. Two mechanisms explain this compensation (Lao et al. 2013). First, a larger fraction of IH interactions results in formation of a CO as opposed to an NCO recombinant (Martini et al. 2006; Lao et al. 2013). Second, additional DSBs are inferred to form when homolog pairing/synapsis is inefficient and prophase is extended (Argunhan et al. 2013; Gray et al. 2013; Lao et al. 2013; Thacker et al. 2014). As a consequence of these two compensatory processes, normal or near-normal levels of COs eventually form in hed1 dmc1 mutants. Thus, as in organisms that lack a Dmc1 ortholog, budding yeast Rad51 can promote high levels of IH recombination in budding yeast, albeit with greatly reduced efficiency. However, homolog bias requires Dmc1.
Additional evidence that Dmc1 and Rad51 cooperate in meiosis comes from analysis of the rad51 mutant, in which Dmc1 can still form immunostaining foci and promote strand exchange, albeit inefficiently (Schwacha and Kleckner 1997; Shinohara et al. 2003a). The Dmc1 foci formed in rad51 mutants are threefold dimmer on average than those formed in wild type suggesting that they reflect incomplete filament elongation (Bishop 1994; Shinohara et al. 2003a). However, as seen when Rad51 catalyzes recombination in the absence of Dmc1, interhomolog bias is lost when Dmc1 catalyzes recombination in the absence of Rad51 (Schwacha and Kleckner 1997).
Additionally, the formation of axial associations, which connect aligned but unsynapsed homologs in budding yeast zip1 mutants, requires both Rad51 and Dmc1 (Rockmill et al. 1995). Yet, despite the ability of either Rad51 or Dmc1 to catalyze strand invasion independently, the strand-invasion activity of Rad51 is dispensable during meiosis (Cloud et al. 2012); a separation of function mutant that retains the ability to form helical filaments, but is deficient in D-loop formation, has normal IH bias and forms normal levels of recombination products. Thus, the RecA homologs normally cooperate during meiosis. Dmc1 catalyzes strand invasion and Rad51 directs this invasion toward a homolog chromatid.
Rad51 and Dmc1 appear to similarly cooperate in plants (Couteau et al. 1999). Like S. cerevisiae, Dmc1 appears to be the catalytic recombinase in Arabidopsis thaliana meiosis, because a Rad51–GFP fusion supports normal meiosis despite being deficient in mitotic DNA repair (Cloud et al. 2012; Da Ines et al. 2013). Also similar to budding yeast, formation of wild-type levels of Dmc1 foci requires Rad51 (Vignard et al. 2007; Kurzbauer et al. 2012). Interestingly, the Rad51-independent assembly of Dmc1 foci is suppressed by ATR kinase in A. thaliana (Kurzbauer et al. 2012). It is currently unclear whether a similar mode of regulation is present in other organisms. rad51, dmc1, and rad51 dmc1 A. thaliana mutants all display aberrant homolog alignment and synapsis (Couteau et al. 1999; Li et al. 2004; Vignard et al. 2007; Kurzbauer et al. 2012). However, in contrast to the arrest of budding yeast dmc1 mutants, plant dmc1 mutants progress and repair all meiotic DSBs via Rad51-mediated IS repair (Couteau et al. 1999; Vignard et al. 2007). This finding is consistent with the apparent absence of a Hed1 homolog in plants. It is possible that local assembly of Dmc1 inhibits Rad51’s IS repair activity in plants, as appears to be the case in budding yeast (Hong et al. 2013; Lao et al. 2013; Cole 2014; Liu et al. 2014).
Despite the overwhelming evidence of cooperation between Rad51 and Dmc1, there are examples in which the two RecA homologs appear to contribute different functions based on the location of a DSB. In A. thaliana, Dmc1-mediated recombination can promote centromere pairing independently of Rad51, whereas both Rad51 and Dmc1 are required for IH recombination and pairing of chromosome arms (Da Ines et al. 2012). In S. pombe, IH recombination at DSB hot spots appears to be independent of Dmc1; however, at DSB coldspots, both Rad51 and Dmc1 are required for IH JM formation (Hyppa and Smith 2010). It should be noted that, in contrast to S. cerevisiae, there appears to be no IH bias at hot spots in S. pombe (Cromie et al. 2006; Hyppa and Smith 2010); however, it is unclear whether and how the lack of IH bias is connected to differential use of Dmc1 at DSB hot spots and coldspots. In both of these cases, at least one mode of IH recombination still requires both Rad51 and Dmc1.
Evidence for Negative Regulation of IS Recombination
A number of observations indicate that cells can negatively regulate IS recombination, leading to the proposal that homolog bias results from a kinetic impediment or barrier to sister chromatid recombination (BSCR; Fig. 6A). Key observations have utilized dmc1 mutants, which assemble Rad51 complexes but fail to repair DSBs (Bishop et al. 1992; Bishop 1994). In the absence of RED1 or MEK1 activity, dmc1 mutants efficiently repair DSBs almost exclusively using the sister chromatid through a Rad51/Rad54-dependent pathway, suggesting the relaxation of a Red1–Mek1-imposed constraint on IS recombination (Schwacha and Kleckner 1997; Niu et al. 2005, 2009). As mentioned above, a similar result pertains when a direct inhibitor of Rad51’s activity, Hed1, is mutated in the absence of Dmc1: homolog bias is reduced 25-fold relative to that in wild-type cells (Hong et al. 2013; Lao et al. 2013). Notably, Dmc1 itself also blocks Rad51’s ability to promote IS recombination given that a hed1 single mutant has only a modest reduction in homolog bias relative to a hed1 dmc1 double mutant (Hong et al. 2013; Lao et al. 2013). Importantly, red1, mek1, and hed1 single mutants also display defects in homolog bias, indicating that they play important roles during wild-type meiosis (Rockmill and Roeder 1991, 1990; Kim et al. 2010; Hong et al. 2013; Lao et al. 2013). Thus, Red1, Mek1, and Hed1 are all required to block Rad51-mediated IS recombination in dmc1 mutant cells, whereas Red1, Mek1, and Dmc1 all act to limit Rad51’s IS activity in the absence of Hed1.
Importantly, the BSCR is not specific to Rad51-mediated events. In addition to blocking Rad51-mediated events in a dmc1 mutant, Mek1 appears to partially inhibit Dmc1-mediated IS DSB repair during haploid meiosis in a mutant in which Rad51 activity is diminished and no homologous chromatid is available (Callender and Hollingsworth 2010). Although the nature of this BSCR is still uncertain, Red1 clearly controls Mek1 kinase activity and specific inhibition of the Mek1 kinase, using an ATP analog sensitive mek1-as allele, strongly suggests that target phosphorylation is responsible for Mek1’s role in homolog bias (Wan et al. 2004).
Three substrates of Mek1 kinase have been reported to date. First, Mek1 self-activates its kinase activity by autophosphorylation on threonine 327 (Niu et al. 2007). Second, Mek1 dependent phosphorylation occurs at threonine 11 of histone H3 (Govin et al. 2010). Although subsequent work showed that H3 is a direct substrate of Mek1, no overt recombination phenotypes have been found to be associated with a mutant form of H3 that cannot be phosphorylated (S Keeney, pers. comm.). Finally, Mek1 phosporylates Rad54 on threonine 132. This modification of Rad54 can inhibit its activity in vitro and in vivo (Niu et al. 2005, 2009; Busygina et al. 2008). Nonetheless, it is currently unclear whether phosphorylation of Rad54 by Mek1 contributes to IH bias in wild-type cells. This is because single mutant strains harboring the nonphosphorylatable allele of the RAD54 gene, rad54-T132A, produce IH recombinants with normal efficiency, and also form viable meiotic products. Thus, phosphorylation of Rad54 is unlikely to be a primary mechanism for promoting homolog bias. Two-dimensional gel analysis of IH bias in the rad54-T132A single mutant would address this outstanding question.
It is possible to infer that Mek1 phosphorylation of substrates other than Rad54 contributes to IH bias (Niu et al. 2009). One key observation is that the Rad51-repressive activity of Hed1 is largely, although not completely, Mek1 dependent (Niu et al. 2007; Lao et al. 2013). However, there were no reports implicating Hed1 as a substrate of Mek1 at the time this review was prepared.
Mek1-mediated repression of Rad51’s strand-exchange activity via Hed1 may be lifted late in meiosis, during a “clean-up” phase in which a mitotic-like IS repair pathway ensures that all DSBs are repaired (Sheridan and Bishop 2006). Consistent with this late idea, in C. elegans, IH recombination is shut down late in meiosis and IS recombination predominates (Rosu et al. 2011). In addition to contributing to the BSCR, Mek1 activity is also inferred to be a positive regulator of IH recombination (discussed below), further emphasizing the fact that additional Mek1 substrates remain to be discovered.
Evidence for Positive Regulation of IH Recombination
Because Dmc1 directly catalyzes homology search and strand invasion, the incorrect choice of recombination partner in rad51 mutants implies that IH bias is positively imposed during normal meiosis, overruling the default propensity for IS recombination (Schwacha and Kleckner 1997; Cloud et al. 2012). Furthermore, mutant analysis suggests that Mek1 kinase activity may confer two distinct regulatory effects on Dmc1’s IH strand-exchange activity. First, completely eliminating Red1 or Mek1 function using null mutations blocks Dmc1-dependent JM formation (Hong et al. 2013). Second, low levels of Red1 function allow Dmc1-mediated joint molecule formation, but do not support IH bias (Schwacha and Kleckner 1997). Thus, in addition to negatively regulating Rad51’s activity via Hed1, Mek1 acts as a positive regulator of Dmc1’s JM forming activity.
In addition to the positive regulation of Dmc1’s activity by Rad51 and Mek1 signaling, a recent study concluded that local regulation of the meiosis-specific cohesin component Rec8 positively regulates IH recombination (Kim et al. 2010; Hong et al. 2013). Specifically, cohesin is inferred to direct recombination between sisters and the Rad51–Mek1 ensemble removes this constraint to IH recombination. This study distinguished two classes of homolog bias mutants, a highly defective class with IH:IS dHJ ratios of 1:5 or less (wild-type bias is 5:1) and a second class, that results in the less severe IH:IS dHJ ratio of exactly 1:1. Mutants lacking a RecA homolog, Red1, or Mek1 represent the first class, whereas mutants lacking Rec8 constitute the second class. Importantly, rec8 red1, rec8 rad51, and rec8 hed1 dmc1 mutants all display the less severe 1:1 IH:IS defect of the rec8 single mutant rather than the more severe defects of red1, rad51, or hed1 dmc1 mutants (Kim et al. 2010; Hong et al. 2013). These results support the inference that Rec8 negatively regulates IH recombination and Red1–Mek1 signaling overcomes this negative effect to enhance the efficiency of IH recombination.
Recombination between Sisters Is Not Completely Blocked in Dmc1+ Meiosis
Although IS repair of DSBs is completely blocked in dmc1 mutants, IS recombination does occur during normal meiotic recombination. DSBs formed at hemizgyous loci—in which repair is only possible via IS recombination—are efficiently repaired during otherwise wild-type meiosis (Goldfarb and Lichten 2010). Interestingly, the timing of DSB repair was equivalent whether the DSB occurred in a homozygous or hemizygous region; however, the rate of this IS repair at both homozygous and hemizygous loci is increased in mek1 mutants. To account for these findings, a kinetic model of IH bias was proposed in which the relative rates of IS and IH strand exchange are skewed to favor IH recombination (Goldfarb and Lichten 2010). Specifically, Mek1 signaling imposes a kinetic brake on IS recombination and the degree of this negative regulation determines the relative rates at which IH and IS repair proceed. Similarly, in haploid yeast engineered to enter meiosis—in which every DSB lacks a homologous chromatid repair template—there is a Mek1-dependent delay in IS recombination (De Massy et al. 1994; Callender and Hollingsworth 2010). Thus, regardless of the presence of a nonsister chromatid, Mek1 limits the rate of IS recombination. In addition to controlling the relative rates of IH and IS recombination, Mek1 signaling modulates CO versus NCO repair choice (Goldfarb and Lichten 2010). The increased rate of IS DSB repair at hemizygous sites in a mek1 mutant is not associated with a corresponding increase in IS JMs, suggesting increased use of an NCO pathway. This emphasizes the need for caution in relying solely on IH:IS JM ratios to characterize partner choice defects, because these measures only detect CO intermediates.
Both DNA Ends Often Form D-Loops Independently of One Another
Hunter and colleagues reported that mcJM intermediates, comprising three and four interconnected chromatids, occur during normal meiotic recombination and that these structures accumulate to high levels in mutants that lack the JM disrupting helicase Sgs1 (Oh et al. 2007). Invasion of both homolog and sister chromatids within the same mcJM is consistent with the inference that IS recombination occurs frequently during normal meiosis (above), and that a given DNA end may engage more than one chromatid before a DSB is eventually repaired (McMahill et al. 2007). The existence of mcJMs further suggests that both ends created by a DSB are capable of promoting homology search and strand exchange. This possibility is consistent with the loading of Dmc1 on both DNA ends (as discussed above) rather than Rad51 being the sole strand-exchange protein located on one of the two ends. Another implication of these findings is that IH bias may involve processes that preferentially stabilize nascent IH interactions, possibly dependent on chromosome pairing and/or synapsis (Börner et al. 2004; Lao and Hunter 2010).
Partner Choice in Organisms with DSB-Independent Synapsis
Although most organisms require recombination to properly align and synapse homologs, D. melanogaster and C. elegans synapse homologs independent of DSBs (Dernburg et al. 1998; McKim et al. 1998). In these organisms—which lack Dmc1, Hop2, and Mnd1— the stable, colinear juxtaposition of homologs in the context of the synaptonemal complex could constitute a distinct mechanism of IH bias in which the spatial proximity of homolog chromatids to a DSB increases the likelihood of IH recombination, perhaps approaching the 2:1 IH:IS ratio predicted from random invasion of one of the three available chromatids. Consistent with this idea, IH CO recombination in these organisms is absolutely dependent on synapsis (Page and Hawley 2001; MacQueen et al. 2002). Furthermore, DSBs—manifested as cytologically visible γ-H2AX or Rad51 foci—appear to form after full synapsis in these organisms (Jang et al. 1994; Colaiacovo et al. 2003).
MODELS FOR IH BIAS
The Anchor Pad Model
The analysis of Rec8 function described above led to the development of a model for the mechanism of IH bias (Fig. 6B) (Kim et al. 2010; Hong et al. 2013). This model breaks IH bias into two distinct phases: “establishment” and “maintenance.” Establishment is the process through which a homologous chromatid is chosen as the initial recombination partner. Technically exquisite 2D gel analysis, in which both IS and IH SEIs were distinguished, showed that rad51, hed1 dmc1, red1, and mek1 mutants are defective for establishment of IH bias, forming high levels of IS SEIs rather than IH SEIs (Kim et al. 2010; Hong et al. 2013). The maintenance phase is proposed to occur at the SEI-to-dHJ transition. rec8 single mutants are inferred to properly establish IH bias at the SEI stage, but subsequently manifest a precise 1:1 ratio of IH:IS dHJs (Kim et al. 2010; Hong et al. 2013). This 1:1 dHJ ratio was argued to reflect defective processing of an “ends-apart” recombination intermediate in which one DSB end forms an IH SEI, and the opposite end forms a homology-dependent complex with the sister chromatid. This putative homology-dependent IS complex was presumed to be undetectable by the current 2D gel method. The 1:1 dHJ ratio seen in rec8 mutants was thus explained by disruption of the ends-apart intermediate as the SEI-to-dHJ transition occurs. If maintenance is normal, the IS complex is preferentially lost and the IH SEI is converted to an IH dHJ. In the rec8 mutant, the mechanism that selectively disrupts the IS complex is defective such that the IH SEI and the IS complex are lost with equal probability, leading to subsequent formation of an IS or an IH dHJ, respectively. The putative IS connection is given the name “anchor pad.” During normal (REC8+) meiosis, the anchor pad blocks the opposite (partner) DSB end from a homology-dependent interaction with the sister by seeding polymerization of the strand-exchange nucleoprotein filament beyond the 3′ end of the IS complex (which we presume would be a nascent D-loop) into the adjacent duplex DNA region. This elongated filament could block invasion of the sister by the opposite DSB end, leaving the homolog chromatids as the only available partners for that end. This model has yet to be supported by evidence for the existence of anchor pads.
Homology-Independent Steric Hindrance Models
Other models to explain IH bias propose that recombinosomes are subject to steric hindrance that reduces access to the target DNA sequence on the sister chromatid. One proposal suggested that the homology search activity of Dmc1 filaments is confined by an extended structure or scaffold that projects the active portion of the filament away from the axial element on which the scaffold is assembled (Fig. 6C) (Sheridan and Bishop 2006). The integrity of this structure was proposed to require local assembly of Rad51 as well as phosphorylation of a nearby Mek1 substrate. Ultrastructural studies in Allium, showing recombinosomes associated with proteinacious bridges between coaligned axial elements, are consistent with this idea (reviewed by Zickler and Kleckner 1999). However, it is not known whether these extended structures form before the homology search or not. Another model involving steric hindrance proposes that recombinosomes are locally tethered to the axial/lateral element (Fig. 6D) (Blat et al. 2002; Hunter 2007). Such confinement could preclude interaction with corresponding sequences on a sister chromatid that are physically separated from the axis in loop regions. The model rests on the assumption that both DSB ends remain associated with the axis during the homology search.
Activation of Strand Exchange in DSB-Distal Regions
Klein and colleagues proposed a model in which Mnd1 (and by extension, Hop2) mediates IH bias by creating recombination-proficient donor templates (Fig. 6E) (Zierhut et al. 2004). This model was based on the fact that Mnd1 forms immunostaining foci independently of, and spatially separated from, DSBs (as discussed above). Mnd1 was proposed to bind to chromatin nonspecifically, which is now understood to reflect the dsDNA-binding activity of the Hop2–Mnd1 heterodimer, and then to be locally displaced (or inactivated) from DSB proximal regions, which would include the target sequence on the sister chromatid. Consequently, Mnd1-dependent activation of strand exchange is only possible with homolog templates, in which Mnd1 has not been displaced. The biochemical results described above led us to propose an updated version of this model in which homolog bias results from local activation of Dmc1’s D-loop activity by Hop2–Mnd1, together with rapid displacement of Hop2–Mnd1 from DSB-proximal regions during the time between DSB formation and the completion of recombinosome assembly (Chan et al. 2014).
It should be noted that the models presented above are compatible with one another and IH bias could involve aspects of more than one of these hypothetical mechanisms.
CONCLUDING REMARKS
In conclusion, recent progress has identified many, perhaps all, of the proteins that carry out the search for DNA homology and catalyze DNA strand exchange during meiotic recombination. Many of these proteins appear to modulate mitotic-like DNA repair pathways, contributing meiosis-specific functions that promote the invasion of a homolog rather than the sister chromatid. These studies have generated a number of hypotheses to explain the mechanism of IH bias. However, critical evidence for any of the models is still lacking. Of particular importance will be the improvement of methods to detect all joint molecule species, especially those that go on to form NCO recombinants. Other key questions include: Does local regulation of Rec8 occur at DSB sites, and, if so, how does this contribute to IH bias? Does an IS anchor pad form as a required intermediate on the pathway to IH–JM formation? More generally, how are the activities of the two ends of a given DSB differentiated? Is activation of Mek1 kinase normally limited to DSB proximal regions? Does inhibitory phosphorylation of Rad54 by Mek1 normally play any role in normal IH bias? What are all the Mek1 substrates that play a role in IH bias and how does phosphorylation contribute to those roles? Does local, DSB-dependent redistribution or inactivation of Hop2–Mnd1 dictate template choice? What is the 3D structure of the recombinosome? How does that structure contribute to regulation of recombinosome function? By what mechanisms do organisms that lack Dmc1 promote IH recombination? We expect answers to many of these questions in the near future.
ACKNOWLEDGMENTS
We thank Edward Egelman for contributing to Figure 2 as well as Diedre Reitz and Yuen-Ling Chan for comments on the manuscript.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Akamatsu Y, Tsutsui Y, Morishita T, Siddique MSP, Kurokawa Y, Ikeguchi M, Yamao F, Arcangioli B, Iwasaki H 2007. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J 26: 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeev A, Mazin A, Kowalczykowski SC 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51–ssDNA nucleoprotein filament. Nat Struct Biol 10: 182–186. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argunhan B, Farmer S, Leung W-K, Terentyev Y, Humphryes N, Tsubouchi T, Toyoizumi H, Tsubouchi H 2013. Direct and indirect control of the initiation of meiotic recombination by DNA damage checkpoint mechanisms in budding yeast. PLoS ONE 8: e65875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC 1996. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87: 757–766. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391: 401–404. [DOI] [PubMed] [Google Scholar]
- Bishop DK 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N 1992. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A 1998. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273: 21482–21488. [DOI] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV 2004. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci 101: 9988–9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV 2005. Activation of human meiosis-specific recombinase Dmc1 by Ca2+. J Biol Chem 280: 26886–26895. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Pezza RJ, Mazina OM, Voloshin ON, Camerini-Otero RD, Mazin AV 2011. The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat Struct Mol Biol 18: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev 22: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender TL, Hollingsworth NM 2010. Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770. [DOI] [PubMed] [Google Scholar]
- Ceballos SJ, Heyer W-D 2011. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta 1809: 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Brown MS, Qin D, Handa N, Bishop DK 2014. The 3rd exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J Biol Chem 27: 18076–18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, Ti SC, Lo YH, Wang AHJ, Chang CS, et al. 2004. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc Natl Acad Sci 101: 10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 489–494. [DOI] [PubMed] [Google Scholar]
- Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P 2007. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev 21: 1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, Bishop DK 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Cole F 2014. The sense and sensibility of strand exchange in recombination homeostasis. PLoS Genet 10: e1004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP 1999. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O, Abe K, Goubely C, Gallego ME, White CI 2012. Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet 8: e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI 2013. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet 9: e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Daley JM, Gaines WA, Kwon Y, Sung P 2014. Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Feinstein E, Conover A, Coljee VW, Vlassakis J, Chan Y-L, Bishop DK, Prentiss M 2012. RecA homology search is promoted by mechanical stress along the scanned duplex DNA. Nucleic Acids Res 40: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Peacock-Villada A, Vlassakis J, Facon A, Feinstein E, Kleckner N, Prentiss M 2013. The differential extension in dsDNA bound to Rad51 filaments may play important roles in homology recognition and strand exchange. Nucleic Acids Res 42: 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Massy B, Baudat F, Nicolas A 1994. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci 91: 11929–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Domenichini S, Raynaud C, Ni D-A, Henry Y, Bergounioux C 2006. Atmnd1-Δ1 is sensitive to γ-irradiation and defective in meiotic DNA repair. DNA Repair (Amst) 5: 455–464. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto R, Kinebuchi T, Sato M, Yagi H, Kurumizaka H, Yokoyama S 2006. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J Biol Chem 281: 5575–5581. [DOI] [PubMed] [Google Scholar]
- Ferrari SR, Grubb J, Bishop DK 2009. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem 284: 11766–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Mortimer RK 1974. A genetic study of X-ray sensitive mutants in yeast. Mutat Res 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Game JC, Zamb TJ, Braun RJ, Resnick M, Roth RM 1980. The role of radiation (rad) genes in meiotic recombination in yeast. Genetics 94: 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S 1999. “Assembly of recombination complexes in Saccharomyces cerevisiae.” Dissertation, University of Chicago, Chicago. [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK 1998. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev 12: 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK 2001. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci 98: 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi JL 2002. Mnd1p: An evolutionarily conserved protein required for meiotic recombination. Proc Natl Acad Sci 99: 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Lichten M 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8: e1000520–e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger S 2010. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev 2: 1772–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Allison RM, Garcia V, Goldman ASH, Neale MJ 2013. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1 (ATR). Open Biol 3: 130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC, Bazemore LR, Golub EI, Radding CM 1997. Activities of human recombination protein Rad51. Proc Natl Acad Sci 94: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC, Golub E, Bi B, Radding CM 2001. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc Natl Acad Sci 98: 8433–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Golub EI, Reddy G, Radding CM, Ward DC 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci 92: 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H 2006. The Swi5–Sfr1 complex stimulates Rhp51/Rad51–and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol 13: 823–830. [DOI] [PubMed] [Google Scholar]
- Haruta N, Akamatsu Y, Tsutsui Y, Kurokawa Y, Murayama Y, Arcangioli B, Iwasaki H 2008. Fission yeast Swi5 protein, a novel DNA recombination mediator. DNA Repair (Amst) 7: 1–9. [DOI] [PubMed] [Google Scholar]
- Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A 2004. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119: 927–940. [DOI] [PubMed] [Google Scholar]
- Henry JM, Camahort R, Rice DA, Florens L, Swanson SK, Washburn MP, Gerton JL 2006. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol Cell Biol 26: 2913–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Yamaguchi M, Haber JE 2011. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci 108: 3108–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Amon A 2006. Checking your breaks: Surveillance mechanisms of meiotic recombination. Curr Biol 16: R217–R228. [DOI] [PubMed] [Google Scholar]
- Holzen TM, Shah PP, Olivares HA, Bishop DK 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev 20: 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, Shinohara A, Bishop DK 2001. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem 276: 41906–41912. [DOI] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP 2013. The logic and mechanism of homologous recombination partner choice. Mol Cell 51: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Furukawa K, Tabata S 1995. Meiosis specific transcription and functional proteins. Adv Biophys 31: 101–115. [DOI] [PubMed] [Google Scholar]
- Hunter N 2007. Meiotic recombination. In Molecular genetics of recombination, Vol. 17 (ed. Aguilera A, Rothstein R), pp. 381–442 Springer, Berlin. [Google Scholar]
- Hunter N, Kleckner N 2001. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishishita S, Inui T, Matsuda Y, Serikawa T, Kitada K 2013. Infertility associated with meiotic failure in the tremor rat (tm/tm) is caused by the deletion of Spermatogenesis Associated 22. Exp Anim 62: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JA, Fink GR 1985. Meiotic recombination between duplicated genetic elements in Saccharomyces cerevisiae. Genetics 109: 303–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YK, Jin YH, Kim EM, Fabre F, Hong SH, Park SD 1994. Cloning and sequence analysis of rhp51+, a Schizosaccharomyces pombe homolog of the Saccharomyces cerevisiae RAD51 gene. Gene 142: 207–211. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C, Vignard J, Pedrosa-Harand A, Siwiec T, Akimcheva S, Jolivet S, Sablowski R, Armstrong S, Schweizer D, Mercier R, et al. 2006. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J Cell Sci 119: 2486–2496. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer W-D 2006. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc Natl Acad Sci 103: 9767–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinebuchi TT, Kagawa WW, Enomoto RR, Tanaka KK, Miyagawa KK, Shibata TT, Kurumizaka HH, Yokoyama SS 2004. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell 14: 363–374. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kobayashi E, Sato S, Hotta Y, Miyajima N, Tanaka A, Tabata S 1994. Characterization of cDNAs induced in meiotic prophase in lily microsporocytes. DNA Res 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Kokabu Y, Murayama Y, Kuwabara N, Oroguchi T, Hashimoto H, Tsutsui Y, Nozaki N, Akashi S, Unzai S, Shimizu T, et al. 2011. Fission yeast Swi5-Sfr1 protein complex, an activator of Rad51 recombinase, forms an extremely elongated dogleg-shaped structure. J Biol Chem 286: 43569–43576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H 2008. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol 6: e88–e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbauer M-T, Uanschou C, Chen D, Schlögelhofer P 2012. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24: 2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N, Hashimoto H, Yamada N, Unzai S, Ikeguchi M, Sato M, Murayama Y, Iwasaki H, Shimizu T 2010. Expression, purification and crystallization of Swi5 and the Swi5–Sfr1 complex from fission yeast. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1124–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N, Murayama Y, Hashimoto H, Kokabu Y, Ikeguchi M, Sato M, Mayanagi K, Tsutsui Y, Iwasaki H, Shimizu T 2012. Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex. Structure 20: 440–449. [DOI] [PubMed] [Google Scholar]
- Lao JP, Hunter N 2010. Trying to avoid your sister. PLoS Biol 8: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N 2008. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang C-C, Grubb J, Thacker D, Lee C-Y, Dresser ME, Hunter N, Bishop DK 2013. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet 9: e1003978–e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Palmer K, O'Brien M, Schimenti JC, Eppig J, Handel MA 2012. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol Reprod 86: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-H, Chang Y-C, Hong EL, Grubb J, Chang C-S, Bishop DK, Wang T-F 2005. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem 280: 40980–40984. [DOI] [PubMed] [Google Scholar]
- Leu JY, Chua PR, Roeder GS 1998. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94: 375–386. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD 2008. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Golub EI, Gupta R, Radding CM 1997. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci 94: 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B 2004. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci 101: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley JE, Cox MM 1990. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J Biol Chem 265: 9043–9054. [PubMed] [Google Scholar]
- Liu Y, Gaines WA, Callender T, Busygina V, Oke A, Sung P, Fung JC, Hollingsworth NM 2014. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet 10: e1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ 2013. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun 4: 2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J-Y, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC 1999. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J 18: 6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Kowalczykowski SC 1996. The specificity of the secondary DNA-binding site of RecA protein defines its role in DNA strand exchange. Proc Natl Acad Sci 93: 10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Kowalczykowski SC 1998. The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J 17: 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK 2007. Synthesis-dependent strand annealing in meiosis. PLoS Biol 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Bear DG, Kowalczykowski SC 1990. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci 87: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrical SW 2014. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Tsutsui Y, Iwasaki H 2011. The fission yeast meiosis-specific Dmc1 recombinase mediates formation and branch migration of Holliday junctions by preferentially promoting strand exchange in a direction opposite to that of Rad51. Genes Dev 25: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Kakihara Y, Hiraoka Y, Nojima H 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J 20: 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, Kowalczykowski SC 2012. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem 287: 28727–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM 2007. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol 27: 5456–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259: 1896–1899. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY-H, Taylor AF, Smith GR, Hunter N 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorokov AL, Chaban YL, Bugreev DV, Hodgkinson J, Mazin AV, Orlova EV 2010. Structure of the hDmc1-ssDNA filament reveals the principles of its architecture. PLoS ONE 5: e8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoli AP, Ravi M, Sebastian J, Nishal B, Reddy TV, Marimuthu MPA, Subbiah V, Vijaybhaskar V, Siddiqi I 2006. AtMND1 is required for homologous pairing during meiosis in Arabidopsis. BMC Mol Biol 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Masson JY, West SC, Egelman EH 1999. Human Dmc1 protein binds DNA as an octameric ring. Proc Natl Acad Sci 96: 10684–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock-Villada A, Yang D, Danilowicz C, Feinstein E, Pollock N, McShan S, Coljee V, Prentiss M 2012. Complementary strand relocation may play vital roles in RecA-based homology recognition. Nucleic Acids Res 40: 10441–10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393: 91–94. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Romanienko PJ, Camerini-Otero RD 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell 5: 927–936. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson J-Y, Camerini-Otero RD 2005. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol 12: 449–453. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD 2006. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J Biol Chem 281: 18426–18434. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD 2007. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev 21: 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Camerini-Otero RD, Bianco PR 2010. Hop2-Mnd1 condenses DNA to stimulate the synapsis phase of DNA strand exchange. Biophys J 99: 3763–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Volodin AA, Boateng KA, Bellani MA, Mazin AV, Camerini-Otero RD 2013. The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res 42: 2346–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin M, Petukhova GV, Morneau D, Déry U, Bransi A, Stasiak A, Camerini-Otero RD, Masson J-Y 2007. Stimulation of fission yeast and mouse Hop2-Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res 35: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, He Y, Ma X, Fodje MN, Grochulski P, Luo Y 2006. Calcium stiffens archaeal Rad51 recombinase from Methanococcus voltae for homologous recombination. J Biol Chem 281: 39380–39387. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Tóth A, Gálová M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol 11: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM Jr 2005. A phylogenomic inventory of meiotic genes. Curr Biol 15: 185–191. [DOI] [PubMed] [Google Scholar]
- Register JC, Griffith J 1985. The direction of RecA protein assembly onto single-strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem 260: 12308–12312. [PubMed] [Google Scholar]
- Roca AI, Cox MM, Brenner SL 1990. The RecA protein: Structure and function. Crit Rev Biochem Mol Biol 25: 415–456. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS 1991. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev 5: 2392–2404. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS 1995. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev 9: 2684–2695. [DOI] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT 2004. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res 32: 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson J-Y 2005. Fission yeast Rad51 and Dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol Cell Biol 25: 4377–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Say AF, Ledford LL, Sharma D, Singh AK, Leung W-K, Sehorn HA, Tsubouchi H, Sung P, Sehorn MG 2011. The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst) 10: 586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuckli-Maurer J, Heyer W-D 2000. Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma 109: 86–93. [DOI] [PubMed] [Google Scholar]
- Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R 2003. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J 36: 1–11. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N 1997. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P 2004. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429: 433–437. [DOI] [PubMed] [Google Scholar]
- Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL 2010. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell 39: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S, Bishop DK 2006. Red-Hed regulation: Recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev 20: 1685–1691. [DOI] [PubMed] [Google Scholar]
- Sheridan SD, Yu X, Roth R, Heuser JE, Sehorn MG, Sung P, Egelman EH, Bishop DK 2008. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res 36: 4057–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Gasior SL, Bishop DK, Shinohara A 2000. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc Natl Acad Sci 97: 10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK 2003a. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Shinohara A, Bishop DK 2003b. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54-and DMC1-dependent pathway. Genetics 163: 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquet B, Abby E, Hervé R, Finsterbusch F, Tourpin S, Le Bouffant R, Duquenne C, Messiaen S, Martini E, Bernardino-Sgherri J, et al. 2013. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet 9: e1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira E, Yoo S, Lehmann R 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen NY, Logsdon JM Jr, Vora GJ, Offenberg HH, Palmer JD, Zolan ME 1997. Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum, and phylogenetic analysis of eukaryotic recA homologs. Curr Genet 31: 144–157. [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Sung P 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82: 453–461. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Morita T, Pearlman RE, Moens PB 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147: 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Stahl FW 1999. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153: 621–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS 2002. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol 22: 3078–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS 2003. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell 5: 915–925. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS 2004. The budding yeast Mei5 and Sae3 proteins act together with Dmc1 during meiotic recombination. Genetics 168: 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev 20: 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou C, Ronceret A, Harder Von M, De Muyt A, Vezon D, Pereira L, Chelysheva L, Kobayashi W, Kurumizaka H, Schlögelhofer P, et al. 2013. Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell 25: 4924–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, Armstrong SJ, Schlögelhofer P, Mercier R 2007. The interplay of RecA-related proteins and the MND1–HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet 3: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ 2001. Whence meiosis? Cell 106: 647–650. [DOI] [PubMed] [Google Scholar]
- Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell 15: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittung P, Ellouze C, Maraboeuf F, Takahashi M, Nordén B 1997. Thermochemical and kinetic evidence for nucleotide-sequence-dependent RECA-DNA interactions. Eur J Biochem 245: 715–719. [DOI] [PubMed] [Google Scholar]
- Wright WD, Heyer W-D 2014. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol Cell 53: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Egelman EH 2010. Helical filaments of human dmc1 protein on single-stranded DNA: A cautionary tale. J Mol Biol 401: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J 2011. The role of the human SWI5-MEI5 complex in homologous recombination repair. J Biol Chem 286: 9888–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zelensky A, Kanaar R, Wyman C 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fan H-Y, Goldman JA, Kingston RE 2007. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol 14: 397–405. [DOI] [PubMed] [Google Scholar]
- Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, Williams GJ, Chi P, Lu L, Pezza RJ, et al. 2013. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res 42: 906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F 2004. Mnd1 is required for meiotic interhomolog repair. Curr Biol 14: 752–762. [DOI] [PubMed] [Google Scholar]