Abstract
C4 photosynthesis characteristically features a cell-specific localization of enzymes involved in CO2 assimilation in bundle sheath cells (BSC) or mesophyll cells. Interestingly, enzymes of sulfur assimilation are also specifically present in BSC of maize and many other C4 species. This localization, however, could not be confirmed in C4 species of the genus Flaveria. It was, therefore, concluded that the bundle sheath localization of sulfate assimilation occurs only in C4 monocots. However, recently the sulfate assimilation pathway was found coordinately enriched in BSC of Arabidopsis, opening new questions about the significance of such cell-specific localization of the pathway. In addition, next generation sequencing revealed expression gradients of many genes from C3 to C4 species and mathematical modeling proposed a sequence of adaptations during the evolutionary path from C3 to C4. Indeed, such gradient, with higher expression of genes for sulfate reduction in C4 species, has been observed within the genus Flaveria. These new tools provide the basis for reexamining the intriguing question of compartmentalization of sulfur assimilation. Therefore, this review summarizes the findings on spatial separation of sulfur assimilation in C4 plants and Arabidopsis, assesses the information on sulfur assimilation provided by the recent transcriptomics data and discusses their possible impact on understanding this interesting feature of plant sulfur metabolism to find out whether changes in sulfate assimilation are part of a general evolutionary trajectory toward C4 photosynthesis.
Keywords: sulfate assimilation, C4 photosynthesis, bundle sheath cells, Flaveria, glutathione
INTRODUCTION
Sulfur is an important element in living organisms where it possesses a plethora of functions. As a component of the amino acids cysteine and methionine, sulfur is incorporated in peptides and proteins. The formation of disulfide bridges by cysteine residues of proteins is important for conformational and regulatory processes: however, sulfur in proteins possesses also catalytic and electrochemical functions and participates in electron transport in iron–sulfur clusters, is part of the catalytic sites of various enzymes and coenzymes. Furthermore, the sulfhydryl group of the tripeptide glutathione (GSH) is involved in redox reactions which protect the cell from oxidative stress (Leustek and Saito, 1999; Foyer and Noctor, 2009).
Plants have the ability to incorporate reduced sulfur components from the atmosphere in form of sulfur dioxide or hydrogen sulfide, but the majority of sulfur in higher plants derives from sulfate (Leustek et al., 2000; Saito, 2004). Sulfate (SO42–) is present in the soil as the most oxidized and accessible form of sulfur for plants, algae and many microorganisms. Sulfate uptake into the plant and its distribution within the cells, occurs via various sulfate transporters which differ in their properties and functions (Leustek and Saito, 1999; Buchner et al., 2004; Takahashi et al., 2011). Within plant cells, sulfate can either be stored in the vacuole or directly be incorporated in organic compounds. For synthesis of reduced sulfur compounds, sulfate needs to be reduced and assimilated to cysteine. The initial step of sulfur assimilation is the activation of sulfate, an inert and stable compound, by ATP sulfurylase (ATPS) to adenosine-5′-phosphosulfate (APS). APS can be reduced to sulfite by APS reductase (APR) or phosphorylated to the common sulfo-group donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) by APS kinase. APR uses two molecules of reduced GSH for the reduction of APS. Sulfite reductase (SIR) then reduces sulfite to sulfide by transferring electrons from the iron–sulfur protein ferredoxin. Finally, sulfide can be incorporated in the amino acid backbone of O-acetylserine (OAS) by OAS (thiol)lyase (OASTL). OAS is derived from serine by acetylation mediated by serine acetyltransferase (SAT; Leustek et al., 2000; Saito, 2004; Kopriva, 2006). Serine biosynthesis is closely linked to carbon and nitrogen assimilation, the synthesis of cysteine thus merges the three assimilatory pathways. Accordingly, light, carbon, and nitrogen compounds regulate sulfate assimilation (Kopriva et al., 1999, 2002; Koprivova et al., 2000). Using thiol- and stress-dependent regulation of APR expression as a tool, the reduction of activated sulfate was found to be a key step in sulfur assimilation (Vauclare et al., 2002; Scheerer et al., 2010).
Reduced sulfur is preferentially stored and transported in the form of GSH, the most abundant low molecular weight thiol in plants with a number of different functions. It is substantially involved in the detoxification of reactive oxygen species (ROS) and heavy metals, serves as sulfur donor in catalytic processes and is involved in redox signaling (Foyer and Noctor, 2009; Takahashi et al., 2011). Two enzymes catalyze the ATP-dependent reactions to form GSH from its constituent amino acids. γ-glutamylcysteine synthetase (γ-ECS) catalyzes the reaction of cysteine and glutamate to form γ-glutamylcysteine (γ-EC). Subsequently, glycine is added to γ-EC by GSH synthetase to form GSH.
The traditional view on the whole plant sulfur metabolism suggests that sulfate reduction predominantly takes place in leaves and reduced sulfur compounds are subsequently distributed to sink tissues (Herschbach and Rennenberg, 2001). However, although GSH is widely used to store and transport reduced sulfur, most plant tissues are capable of sulfate reduction and thus are able to cover their needs of reduced sulfate. Indeed, according to available microarray data in the Genevestigator database, APR and SIR transcripts are present in all Arabidopsis tissues, including reproductive organs (Zimmermann et al., 2004). The presence of sulfate reduction could also be detected in roots and developing seeds (Brunold and Suter, 1989; Kopriva et al., 2001; Tabe and Droux, 2002).
There are, however, exceptions and some plant tissues seem to have lost the capacity to reduce sulfate. Most notable among these tissues are the mesophyll cells (MC) in C4 plants, as in these plants the pathway is confined to a specific tissue, the bundle sheath cells (BSC; Gerwick et al., 1980; Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1984; Burgener et al., 1998). The intercellular distribution of sulfate (and nitrate) assimilation in C4 plants is one of the enigmatic open questions of sulfur metabolism, not just in terms of mechanisms but mainly of its biological significance. This question has been very recently revived by Aubry et al. (2014) who showed that genes involved in the pathway are mainly expressed in the BSC of Arabidopsis. Therefore, here we review what is known about the localization of sulfate assimilation in C4 plants, and discuss how the current progress in studies of C4 photosynthesis may contribute to understanding this interesting feature of plant sulfur metabolism.
C4 PHOTOSYNTHESIS
During photosynthesis, carbon fixation takes place in the Calvin–Benson cycle and is mediated by the dual-specific enzyme Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). The carboxylation reaction yields 3-phosphoglycerate (3-PGA) which is reduced to carbohydrates. In contrast, the oxygenation reaction yields 2-phosphoglycolate (2-PG), a toxic compound that enters photorespiration to be metabolized. The photorespiratory pathway releases one molecule of previously fixed CO2 and regenerates the other into 3-PGA. Although Rubisco shows higher affinity toward CO2 than O2, one third of the assimilated carbon is lost during photorespiration, increasing so the energy costs of photosynthesis. An efficient mechanism to minimize the oxygenation reaction of Rubisco and photorespiration is C4 photosynthesis. C4 plants avoid such loss of assimilated CO2 by splitting the CO2 fixing reactions among two differentiated cell types, MC and BSC. MC and BSC differ from each other by their morphological properties, distribution within the plant tissue and cell-specific localization of many enzymes. BSC build up a radial pattern around the vascular tissue. They exhibit thickened cell-walls and many starch containing chloroplasts. BSC are surrounded by an adjacent layer of MC with small and randomly distributed chloroplasts. The radial arrangement of BSC and MC around the vascular tissue is known as Kranz anatomy (Laetsch, 1974).
The primary CO2 fixation takes place in MC. This reaction, catalyzed by phosphoenolpyruvate carboxylase (PEPCase), results in oxaloacetate, a C4 compound. Oxaloacetate is immediately converted to malate or aspartate, C4 acids that enter BSC by diffusion. Here, the C4 acids are decarboxylated, resulting in enriched CO2 concentrations in the BSC. C4 photosynthesis can be divided into three subtypes defined by the enzymes which catalyze the decarboxylation reaction: NADP-dependent malic enzyme, NAD-dependent malic enzyme or phosphoenolpyruvate carboxykinase. The released CO2 enters the Calvin–Benson cycle within the BSC (Edwards and Huber, 1981; Edwards and Walker, 1983; Hatch, 1987). Besides Rubisco, additional genes involved in the carbon cycle are solely expressed in the BSC (Sheen, 1999; Edwards et al., 2001). The separation of PEPCase and Rubisco, as well as the Kranz anatomy and a lack of photosystem II (PSII) in BSC of most C4 plants (Hatch and Osmond, 1976) support more effective carbon assimilation. The CO2 enrichment in the BSC minimizes the rate of the oxygenation reaction. Any CO2, lost during residual photorespiration, is separated from the aerial space by the thick cell walls of the BSC and by the adjacent layer of MC, where it is directly re-assimilated. In addition, the lack of PSII in BSC restricts the emergence of oxygen by water splitting to MC chloroplasts. Paired with a BSC-specific expression of glycine decarboxylase (GDC), these mechanisms generate a low photosynthetic CO2 compensation point in C4 plants. The CO2 compensation point describes the concentration at which the photosynthetic CO2 uptake equals the rate of respiration.
The last decade has seen a revival of interest in C4 photosynthesis, as a potential route to increase crop productivity (Hibberd et al., 2008). Particular interest has been given the evolutionary route from C3 to C4 plants as this could identify steps needed to engineer C4 photosynthesis in C3 plants (Heckmann et al., 2013; Mallmann et al., 2014). C4 photosynthesis evolved not less than 62 times independently from C3 ancestors (Sage et al., 2011). Genera containing C3 and C4 as well as C3–C4 intermediate species, such as Flaveria, are especially suited to give information about the evolution from C3 to C4 photosynthesis (Kopriva et al., 1996; Heckmann et al., 2013; Williams et al., 2013; Mallmann et al., 2014). C3–C4 species are characterized by a CO2 compensation point intermediate of C3 and C4 species (Apel and Maass, 1981; Bauwe et al., 1987). While there are many different ways to achieve such CO2 compensation points (Bassüner et al., 1984; Bauwe, 1984; Monson et al., 1986), the underlying photosynthetic mechanism is universally based on a photorespiratory CO2 pump. Shortly, GDC activity is lost in the MC, resulting in the transport of photorespiratory glycine to BSC for decarboxylation, which causes a moderate increase in BSC CO2 concentration and a boost of carboxylation over oxygenation reaction. Since the two-carbon compound glycine serves as C transporter, this process is known as C2 photosynthesis (Bauwe and Kolukisaoglu, 2003; Sage, 2004; Sage et al., 2012; Schulze et al., 2013). C2 photosynthesis was postulated to be a prerequisite to C4 metabolism (Sage et al., 2012). This conclusion was confirmed by analysis of transcriptomes of nine Flaveria species and by flux balance analysis (Mallmann et al., 2014). However, the translocation of glycine decarboxylation from MC to BSC results in the shift of carbon and nitrogen balance. This disbalance can be counterbalanced by a basal activity of the C4 cycle, acting as an efficient ammonium recirculation pathway. Thus, the C2 photosynthesis triggers development of a basic C4 cycle which further leads to evolution of a full C4 photosynthesis. In another mathematical approach, a model of fitness landscape describes the evolutionary trajectory from C3 to C4 photosynthesis as a process of 30 individual steps, each of them gradually yielding a gain of biochemical fitness (Heckmann et al., 2013). This model positions the known C3–C4 intermediates as real transitory states in the evolution from C3 to C4 photosynthesis and explains the ease with which C4 photosynthesis evolved independently, making it feasible to be recreated by genetic engineering.
However, one major aspect of C4 photosynthesis remains largely unexplained. Besides the cell-specific distribution of enzymes involved directly in photosynthesis and photorespiration, intercellular differences could be detected in the localization of enzymatic reactions involved in nitrogen and sulfur assimilation in C4 plants. The reduction of nitrate and nitrite is restricted to MC, whereas the incorporation of reduced nitrogen into the amino acids glutamate and glutamine takes place in the BSC or in MC and BSC (Rathnam and Edwards, 1976; Moore and Black, 1979; Becker et al., 2000) which accords with the translocation of GDC into BSC. C4 species also exhibit higher nitrogen use efficiency, presumably because of the concentration of Rubisco into BSC and so decreasing the amount of this protein per leaf area (Brown, 1978; Ghannoum et al., 2011). Whether the cell-specific localization of nitrate reduction contributes to the improved nitrogen nutrition is not clear yet. Furthermore, since spatial separation was reported for the assimilation of carbon and nitrogen, two nutrients that are taken up into the plant in their oxidized form (CO2 and NO3–) and since the translocation of GDC to the BSC restricts production of photorespiratory serine, the precursor of OAS and cysteine, to these cells, the question of intercellular distribution of sulfate assimilation in C4 plants has long been of major interest.
INTERCELLULAR COMPARTMENTATION OF SULFATE ASSIMILATION IN C4 PLANTS
The question of intercellular compartmentation of sulfate assimilation in C4 species was first addressed by Gerwick and Black (1979) in the crabgrass Digitaria sanguinalis. They focused on the initial step of sulfur assimilation, the activation of sulfate by ATPS, and showed that more than 90% of total leaf ATPS activity was restricted to BSC in crabgrass. Similar results could be obtained for 17 additional C4 monocot species, representing each of the three C4 subtypes (Gerwick et al., 1980; Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1984; Burgener et al., 1998). Analyses of the spatial separation of sulfate assimilation in maize leaves could additionally show restriction of APR activity to BSC (Figure 1; Schmutz and Brunold, 1984; Burgener et al., 1998), whereas activity of SIR and OASTL was equally detected in MC and BSC (Passera and Ghisi, 1982; Schmutz and Brunold, 1984, 1985).
FIGURE 1.
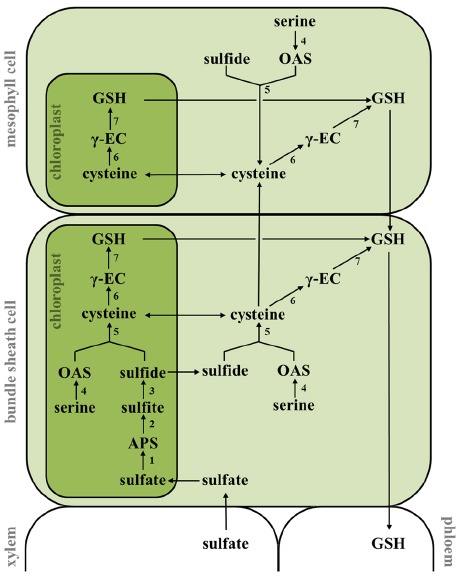
Intercellular compartmentation of sulfate assimilation and glutathione biosynthesis in maize. Sulfate is taken up from the soil and transported to the bundle sheath cells (BSC) through the xylem. The reduction of sulfate takes place exclusively in the plastids of BSC and is mediated by ATP sulfurylase (1), APS reductase (2), and sulfite reductase (3). Sulfide is further incorporated into the amino acid backbone of OAS by OAS (thiol)lyase (5) to form cysteine in chloroplasts, cytosol and mitochondria (not included) of BSC. OAS is derived from serine by serine acetyltransferase-mediated acetylation (4). Reduced sulfur is transported in form of cysteine to mesophyll cells where glutathione (GSH) synthesis is predominantly localized. GSH synthesis is driven by γ-EC synthetase (6) and GSH synthethase (7). APS, adenosine-5′-phosphosulfate; GSH, glutathione; OAS, O-acetylserine; γ-EC, γ-glutamylcysteine.
Corresponding to these findings, transcripts of ATPS and APR were found exclusively in the BSC and OASTL mRNA was present in MC and BSC in maize leaves (Kopriva et al., 2001). These findings were supported by recent analyses of the maize MC and BSC transcriptomes which showed a predominant expression of all four ATPS as well as two APR genes in the BSC and localization of OASTL transcripts in MC and BSC (Chang et al., 2012). The identical localization of APR, ATPS, and OASTL mRNA and their enzymes activity, indicates a transcriptional regulation of genes involved in sulfate assimilation. However, APR mRNA but not enzyme activity could also be detected in MC of maize leaves after exposure to chilling stress. This finding reveals participation of post-transcriptional processes in the compartmentalization of sulfate assimilation in maize (Kopriva et al., 2001). Interestingly, post-transcriptional mechanisms seem to be responsible also for the MC-specific localization of glutathione reductase (GR) in maize (Pastori et al., 2000).
Although SIR activity was originally detected in MC of maize leaves (Schmutz and Brunold, 1984), in further studies SIR transcripts were shown to accumulate only in BSC (Kopriva et al., 2001; Chang et al., 2012). The localization of SIR, particularly the discrepancy in protein and transcript, thus needs revisiting, although it is possible that in MC SIR fulfills a role independent from sulfate assimilation, e.g., protection against SO2 (Yarmolinsky et al., 2013). Burgener et al. (1998) were able to show that all steps of sulfate reduction take place in BSC. The restriction of sulfate reduction to BSC requires an efficient transport of reduced sulfate compounds to the adjacent MC. Feeding experiments with [35S]sulfate showed that cysteine is the transport metabolite in maize. It is transported from BSC to MC where GSH synthesis is predominantly localized (Burgener et al., 1998).
The question of evolutionary significance of the BSC-specific localization of sulfate assimilation was addressed in the genus Flaveria (Koprivova et al., 2001). Comparing the cell-specific localization of ATPS and APR mRNA by northern analysis and in situ hybridization in various Flaveria species, Koprivova et al. (2001) expected an accumulation of the transcripts in the BSC in C4 species and ubiquitous expression of ATPS and APR in all photosynthetic cells of C3 species. Remarkably, they found comparable transcript levels of both genes in each species, independent of the photosynthetic mechanism. Immunogold electron microscopy confirmed a similar distribution of APR protein in chloroplasts of BSC and MC in all species analyzed (Koprivova et al., 2001).
These findings contradicted the previously postulated compartmentation of sulfate assimilation in C4 plants (Gerwick et al., 1980; Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1984; Burgener et al., 1998). Admittedly, earlier studies were conducted in maize and 17 other C4 species, all belonging to the group of monocotelydons. F. trinervia and F. australasica were the first C4 dicots analyzed for the intercellular separation of sulfate assimilation (Koprivova et al., 2001). However, BSC-specific localization of sulfate assimilation is not a monocot-specific trait. In wheat, a C3 monocot, ATPS and APR activities were found at equivalent levels in MC and BSC (Schmutz and Brunold, 1984). In addition, recently, Aubry et al. (2014) described the preferential expression of genes associated with sulfate assimilation in the BSC of the C3 dicot Arabidopsis thaliana. Thus, BSC-specific localization of sulfate assimilation might no longer be seen as a C4-associated trait but possibly as species-specific characteristic. Therefore it is even more imperative to understand the biological reasons and/or consequences for this localization.
Indeed, the physiological significance of spatial separation in sulfate assimilation in maize has been widely discussed, unfortunately without a clear conclusion. The BSC of maize lack PSII and with it the water-splitting complex (Sheen and Bogorad, 1988; Pfundel et al., 1996). Burgener et al. (1998) considered that the consequential reduction in O2 concentration might be the reason for BSC-specific expression of sulfate assimilation in maize. The low levels of O2 might prevent the oxidation of sulfite and sulfide, the intermediates of sulfate assimilation, and so increase the efficiency of the pathway. However, such oxidative events, except the enzymatic sulfite oxidation by sulfite oxidase, have not been reported for C3 plants which possess PSII in the chloroplasts of all photosynthetic cells. As a precursor of cysteine, serine is necessary for sulfate reduction. In C3 plants, photorespiration is the main source of serine production (Douce et al., 2001; Ros et al., 2014). GDC and serine hydroxymethyltransferase, key enzymes in photorespiration and serine biosynthesis, are localized in BSC of C4 plants (Ohnishi and Kanai, 1983; Gardeström et al., 1985; Becker et al., 1993). Therefore, Burgener et al. (1998) hypothesized that the localization of sulfate reduction in maize coincides with the site of photorespiration because of serine availability. Several physiological aspects, however, challenge this hypothesis. C4 species of the genus Flaveria do not show BSC-specific localization of sulfur reduction, although GDC activity is restricted to this cell type. Moreover, serine would need to be transported from BSC to MC to participate, e.g., in protein biosynthesis. Indeed, an alternative pathway of serine biosynthesis exists and is vital for plants (Benstein et al., 2013), so that photorespiratory serine cannot be the link. The BSC localization of sulfate assimilation in Arabidopsis (Aubry et al., 2014) also contradicts the link to serine synthesis. The Arabidopsis data are derived from comparison of BSC expression vs. whole leaf expression, therefore the spatial separation may not be complete as it is in C4 monocots. The BSC expression of sulfate assimilation is linked with BSC expression of genes for synthesis of glucosinolates, sulfur-containing secondary metabolites. As these compounds are important in the vasculature for defense against insects (Shroff et al., 2008) the need for glucosinolate synthesis might drive the BSC specific expression of sulfate assimilation genes. This hypothesis, even though it has to be tested yet, seems to strengthen the view that the spatial distribution of sulfate assimilation may be a species-specific adaptation to diverse environmental conditions. Obviously, more research is needed to understand the biological reason(s) and consequences for plant fitness of such intercellular separation of sulfate assimilation, if there are any.
SULFUR ASSIMILATION PATHWAY FROM C3 TO C4 PLANTS
The analysis of sulfate assimilation in different Flaveria species revealed another interesting result. The activity of APR as well as cysteine and GSH levels were significantly higher in leaves of C4 and C4-like species compared to C3 and C3–C4 species (Koprivova et al., 2001). Enzyme activity, cysteine, and GSH content correlated with the degree of expression of C4 characteristics, considering the photosynthetic CO2 compensation point as a quantitative measure of C4 photosynthetic traits (Kopriva and Koprivova, 2005). The intriguing results of analyses of sulfate assimilation in Flaveria were unfortunately not followed by further studies. The question of intercellular localization of sulfur assimilation, and nitrate assimilation at the same time, seemed to be forgotten for more than a decade. However, with the increased interest in C4 photosynthesis as a means to improve plant productivity and with the more frequent use of next generation sequencing in plant science, these questions can be addressed from a different angle.
Indeed, the transcriptomes of nine Flaveria species were analyzed recently to assess the influence of photorespiration on the evolution of C4 photosynthesis (Mallmann et al., 2014). To obtain the transcriptomic data, the authors performed four independent cultivations during different seasons of two C3, four C3–C4, one C4-like, and two C4 species of the genus Flaveria (Figure 2). The analysis showed clear gradients in expression of many genes across the Flaveria species, the photorespiratory enzymes being expressed to lower degree with increasing degree of C4 photosynthesis, whereas the genes of C4 pathway being expressed more. Data mining of this transcriptome dataset focusing on genes associated with sulfate assimilation provides an interesting insight into the expression of ATPS, APR and SIR, depending on the photosynthetic mechanism and seasonal changes (Figure 2A). APS1 and APS2 are two isoforms of ATPS in Arabidopsis thaliana, with transcript sequences matching Flaveria ATPS mRNA. Whereas APS1 is exclusively localized in chloroplasts, APS2 can also be found in the cytosol. The expression of the ATPS genes does not show a recognizable correlation with the photosynthetic mechanism, but seems to follow seasonal changes. Direct comparison of APS1 and APS2 signals in each species indicate preferential expression of one of the isoforms for at least the C3 species, F. pringlei and F. robusta, and the C4-like species F. brownii (Figure 2B). Overall, the highest expression of ATPS genes can be detected in Flaveria C3 species.
FIGURE 2.

Transcript abundance of genes involved in sulfate assimilation in leaves of C3, C4, and C3–C4 intermediate plants. Based on the transcriptomic data of nine Flaveria species by Mallmann et al. (2014) obtained in four independent cultivations harvested at different seasons (1, September 2009; 2, June 2010; 3, October 2010; 4, April 2011), the transcript levels of the individual cultivations (A) and mean values (B) were normalized and plotted in heat maps. C3: F. pringlei, F. robusta; C3–C4: F. chloraefolia, F. pubescens, F. anomala, F. ramosissima; C4-like: F. brownii; C4: F. bidentis, F. trinervia. (C) The relative transcript abundances of genes participating in sulfate assimilation in the Cleome species C. gynandra (C4) and C. spinosa (C3) are based on the Brautigam et al. (2011). Transcripts were annotated by allocation to their Arabidopsis homologues. APR1 and APR2, APS reductase; APS1 and APS2, ATP sulfurylase; SIR, sulfite reductase.
The transcript level of SIR was least variable among the different species and time points of harvest, with a tendency to higher levels in the C3 species (Figures 2A,B). This agrees with the low level of transcriptional regulation of this gene in the model species Arabidopsis (Takahashi et al., 2011). On the other hand, the differences in mRNA levels of genes similar to the Arabidopsis APR1 and APR2 sequences were striking (Figures 2A,B). Both isoforms show similar expression patterns, which indicates either a coordinate expression or a presence of a single APR isoform in the Flaveria species. Strongest expression of APR can be detected in the Flaveria C4 species F. bidentis and F. trinervia. The C3–C4 species F. pubescens constitutes an exception with rather high APR mRNA levels. The increased transcript levels of APR in C4 species correspond with the elevated APR enzyme activity in C4 and C4-like Flaveria, reported by Koprivova et al. (2001), and indicate transcriptional regulation of APR activity depending on the photosynthetic mechanism. To test whether this trait can be extended to other C4 species, we analyzed the comparative adult leaf transcriptomes of closely related C3 and C4 Cleomaceae species, Cleome gynandra (C4) and Cleome spinosa (C3; Brautigam et al., 2011). The ratio of ATPS and APR mRNA levels between the two species were comparable to transcript level relations in Flaveria C3 and C4 species (Figure 2C). The relative abundance of ATPS transcripts is higher in C. spinosa, whereas C. gynandra exhibits increased abundance of APR mRNA.
Adenosine-5′-phosphosulfate reductase has been reported to be the key enzyme of sulfate assimilation with strong regulation by many environmental factors (Kopriva et al., 2001; Vauclare et al., 2002; Scheerer et al., 2010). Among others, sulfur starvation and increased need for cysteine, required for GSH synthesis, induce APR transcription (Vauclare et al., 2002). Hence, increased transcript levels of APR in two independent C4 species compared to their close C3 relatives might indicate the need of enhanced levels of reduced sulfur in C4 plants. Indeed, C4 plants are particularly adapted to dry and warm habitats, environmental conditions which cause oxidative stress by formation of ROS. To protect the cells from damage, ROS is detoxified in the GSH–ascorbate cycle in which GSH serves as reductant of dehydroascorbate (Noctor and Foyer, 1998). Thus, exposure to oxidative stress increases the demand for GSH. Cysteine, required for GSH synthesis, is provided by sulfate reduction. Adaptation to exceptional habitats could explain the high levels of GSH and cysteine in C4 Flaveria described by Koprivova et al. (2001) as well as the increased APR activity and transcript levels. The C4 monocot maize is sensitive to chilling, another oxidative stress causing condition. Here as well exposure to chilling stress increases the mRNA levels and activity of APR and ATPS (Brunner et al., 1995; Kopriva et al., 2001). Remarkably, the species Zea mays also contains chilling-tolerant genotypes which show higher levels of GSH and cysteine as well as increased activity of APR and GR, the enzyme that reduces oxidized GSH (Kocsy et al., 1996, 1997). Kocsy et al. (2000) demonstrated a direct link between GSH synthesis and chilling tolerance by inhibiting γ-ECS using buthionine sulfoximine (BSO). BSO-treated maize plants lost their chilling tolerance which could be restored by supplementation with GSH or γ-EC.
Overall, these findings in maize fit the general hypothesis of demand-driven regulation of sulfate assimilation formulated from studies with C3 Brassicaceae species (Lappartient and Touraine, 1996). Sulfur starvation leads to increased transcript levels of ATPS, APR, and sulfate transporters in maize (Bolchi et al., 1999; Hopkins et al., 2004), whereas supplementation of reduced sulfur compounds results in repression of ATPS expression (Bolchi et al., 1999). Besides oxidative stress, cadmium has been reported to enhance ATPS and APR activity (Nussbaum et al., 1988). The detoxification of heavy metals requires the synthesis of phytochelatins which bind to the metals and enable their relocation to the vacuole. As GSH is indispensable for the formation of phytochelatins (Cobbett and Goldsbrough, 2002), exposure to heavy metals increases the demand for reduced sulfur. In Brassicacea species GSH was identified as the molecular regulator of demand-driven sulfate assimilation (Lappartient and Touraine, 1996; Vauclare et al., 2002). In contrast, cysteine directly regulates sulfate reduction in maize (Bolchi et al., 1999). Kopriva and Koprivova (2005) discussed a possible molecular background to the findings of Bolchi et al. (1999). They assumed that cysteine as regulatory molecule is a consequence of the BSC-specific localization of sulfate assimilation in maize. Whereas GSH can be synthesized in maize BSC and MC, cysteine synthesis is restricted to BSC and cysteine was shown to be transported from BSC protoplasts (Burgener et al., 1998; Gomez et al., 2004). This led to the conclusion, that the cysteine pools of MC and BSC are permanently connected and enable a rapid reaction to changes in cysteine concentration (Kopriva and Koprivova, 2005).
Regulation of sulfate assimilation in different Flaveria species has not been addressed yet. The elucidation of such regulation might give an insight view on the molecular mechanism causing a constitutionally higher activity of the sulfate assimilatory pathway in C4 species compared to C3 plants.
CONCLUSION AND OUTLOOK
The BSC localization of sulfate assimilation and the expression gradient from C3 to C4 species are interesting aspects of both sulfur metabolism and C4 photosynthesis. It is important to find out whether this localization and difference in expression give the plants an evolutionary advantage, which has to be considered in attempts to engineer C4 photosynthesis in C3 crops as well as to improve sulfur use efficiency of crop plants. The wealth of genomics and transcriptomics resources in C4 plants offers new ways to approach these questions, based not on a single model. The findings of preferential localization of sulfate assimilation in BSC in Arabidopsis could be the right trigger to assess the consequences of this localization for the general sulfur metabolism. All in all, while we cannot answer the question of importance of the changes in expression and localization of sulfate assimilation for evolution of C4 photosynthesis yet it seems that this enigmatic question of plant sulfur research might be answered in near future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENT
Research in Stanislav Kopriva’s lab is supported by Deutsche Forschungsgemeinschaft (EXC 1028).
REFERENCES
- Apel P., Maass L. (1981). Photosynthesis in species of Flaveria. CO2 compensation concentration, O2 influence on photosynthetic gas exchange and d13C values in species of Flaveria (Asteraceae). Biochem. Physiol. Pflanz. 176, 396–399 10.1016/S0015-3796(81)80052-2 [DOI] [Google Scholar]
- Aubry S., Smith-Unna R. D., Boursnell C. M., Kopriva S., Hibberd J. M. (2014). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulphur and glucosinolate metabolism. Plant J. 78, 659–673. 10.1111/tpj.12502 [DOI] [PubMed] [Google Scholar]
- Bassüner B., Keerberg O., Bauwe H., Pyarnik T., Keerberg H. (1984). Photosynthetic CO2 metabolism in C3–C4 intermediate and C4 species of Flaveria (Asteraceae). Biochem. Physiol. Pflanz. 179, 631–634 10.1016/S0015-3796(84)80039-6 [DOI] [Google Scholar]
- Bauwe H. (1984). Photosynthetic enzyme activities and immunofluorescence studies on the localization of ribulose-1, 5-bisphosphate carboxylase/oxygenase in leaves of C3, C4, and C3–C4 intermediate species of Flaveria (Asteraceae). Biochem. Physiol. Pflanz. 179, 253–268 10.1016/S0015-3796(84)80041-4 [DOI] [Google Scholar]
- Bauwe H., Keerberg O., Bassuner R., Parnik T., Bassuner B. (1987). Reassimilation of carbon dioxide by Flaveria (Asteraceae) species representing different types of photosynthesis. Planta 172, 214–218. 10.1007/BF00394590 [DOI] [PubMed] [Google Scholar]
- Bauwe H., Kolukisaoglu U. (2003). Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 54, 1523–1535. 10.1093/jxb/erg171 [DOI] [PubMed] [Google Scholar]
- Becker T. W., Carrayol E., Hirel B. (2000). Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211, 800–806. 10.1007/s004250000355 [DOI] [PubMed] [Google Scholar]
- Becker T. W., Perrot-Rechenmann C., Suzuki A., Hirel B. (1993). Subcellular and immunocytochemical localization of the enzymes involved in ammonia assimilation in mesophyll and bundle-sheath cells of maize leaves. Planta 191, 129–136 10.1007/BF00240904 [DOI] [Google Scholar]
- Benstein R. M., Ludewig K., Wulfert S., Wittek S., Gigolashvili T., Frerigmann H., et al. (2013). Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25, 5011–5029. 10.1105/tpc.113.118992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolchi A., Petrucco S., Tenca P. L., Foroni C., Ottonello S. (1999). Coordinate modulation of maize sulfate permease and ATP sulfurylase mRNAs in response to variations in sulfur nutritional status: stereospecific down-regulation by L-cysteine. Plant Mol. Biol. 39, 527–537. 10.1023/A:1006148815106 [DOI] [PubMed] [Google Scholar]
- Brautigam A., Kajala K., Wullenweber J., Sommer M., Gagneul D., Weber K. L., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155, 142–156. 10.1104/pp.110.159442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. H. (1978). A difference in N use efficiency in C3 and C4 plants and its implications in adaptation and evolution. Crop Sci. 18, 93–98. [Google Scholar]
- Brunner M., Kocsy G., Rftegsegger A., Schmutz D., Brunold C. (1995). Effect of chilling on assimilatory sulfate reduction and glutathione synthesis in maize. J. Plant Physiol. 146, 743–747 10.1016/S0176-1617(11)81942-5 [DOI] [Google Scholar]
- Brunold C., Suter M. (1989). Localization of enzymes of assimilatory sulfate reduction in pea roots. Planta 179, 228–234. 10.1007/BF00393693 [DOI] [PubMed] [Google Scholar]
- Buchner P., Takahashi H., Hawkesford M. J. (2004). Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55, 1765–1773. 10.1093/jxb/erh206 [DOI] [PubMed] [Google Scholar]
- Burgener M., Suter M., Jones S., Brunold C. (1998). Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol. 116, 1315–1322. 10.1104/pp.116.4.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell J. N. (1984). Sulfate assimilation in C4 plants: intercellular and intracellular location of ATP sulfurylase, cysteine synthase, and cystathionine β-lyase in maize leaves. Plant Physiol. 75, 873–875. 10.1104/pp.75.3.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. M., Liu W. Y., Shih A. C., Shen M. N., Lu C. H., Lu M. Y., et al. (2012). Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160, 165–177. 10.1104/pp.112.203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C., Goldsbrough P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. 10.1146/annurev.arplant.53.100301.135154 [DOI] [PubMed] [Google Scholar]
- Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001). The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6, 167–176 10.1016/S1360-1385(01)01892-1 [DOI] [PubMed] [Google Scholar]
- Edwards G. E., Franceschi V. R., Ku M. S., Voznesenskaya E. V., Pyankov V. I., Andreo C. S. (2001). Compartmentation of photosynthesis in cells and tissues of C4 plants. J. Exp. Bot. 52, 577–590. 10.1093/jexbot/52.356.577 [DOI] [PubMed] [Google Scholar]
- Edwards G. E., Huber S. C. (1981). “The C4 pathway,” in Biochemistry of Plants, eds Hatch M. D., Boardman N. K. (New York: Academic Press; ), 238–281. [Google Scholar]
- Edwards G. E., Walker D. A. (1983). C3, C4: Mechanisms and Cellular and Environmental Regulation of Photosynthesis. Oxford: Blackwell Scientific. [Google Scholar]
- Foyer C. H., Noctor G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905. 10.1089/ars.2008.2177 [DOI] [PubMed] [Google Scholar]
- Gardeström P., Edwards G. E., Henricson D., Ericson I. (1985). The localization of serine hydroxymethyltransferase in leaves of C3 and C4 species. Physiol. Plant. 64, 29–33 10.1111/j.1399-3054.1985.tb01208.x [DOI] [Google Scholar]
- Gerwick B. C., Black C. C. (1979). Sulfur assimilation in C4 plants. Plant Physiol. 64, 590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick B. C., Ku S. B., Black C. C. (1980). Initiation of sulfate activation: a variation in C4 photosynthesis plants. Science 209, 513–515. 10.1126/science.209.4455.513 [DOI] [PubMed] [Google Scholar]
- Ghannoum O., Evans J., von Caemmerer S. (2011). “Nitrogen and water use efficiency of C4 plants,” in Advances in Photosynthesis and Respiration, Vol. 32, C4 photosynthesis and related CO2 concentrating mechanisms, eds Raghavendra A. S., Sage R. F. (Dordrecht: Springer; ), 109–128. [Google Scholar]
- Gomez L. D., Vanacker H., Buchner P., Noctor G., Foyer C. H. (2004). Intercellular distribution of glutathione synthesis in maize leaves and its response to short-term chilling. Plant Physiol. 134, 1662–1671. 10.1104/pp.103.033027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M. D. (1987). C4 Photosynthesis: a unique blend of modified biochemistry, anatomy, and ultrastructure. Biochim. Biophys. Acta 895, 81–106 10.1016/S0304-4173(87)80009-5 [DOI] [Google Scholar]
- Hatch M. D., Osmond C. B. (1976). “Compartmentation and transport in C4 photosynthesis,” in Transport in Plants III, eds Stocking C. R., Heber U. (Berlin: Springer; ), 144–184. [Google Scholar]
- Heckmann D., Schulze S., Denton A., Gowik U., Westhoff P., Weber A. P., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. 10.1016/j.cell.2013.04.058 [DOI] [PubMed] [Google Scholar]
- Herschbach C., Rennenberg H. (2001). “Significance of phloem-translocated organic sulfur compounds for the regulation of sulfur nutrition,” in Progress in Botany, eds Esser K., Lüttge U., Kadereit J. W., Beyschlag W. (Berlin: Springer; ), 177–193. [Google Scholar]
- Hibberd J. M., Sheehy J. E., Langdale J. A. (2008). Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr. Opin. Plant Biol. 11, 228–231. 10.1016/j.pbi.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Hopkins L., Parmar S., Bouranis D. L., Howarth J. R., Hawkesford M. J. (2004). Coordinated expression of sulfate uptake and components of the sulfate assimilatory pathway in maize. Plant Biol. (Stuttg.) 6, 408–414. 10.1055/s-2004-820872 [DOI] [PubMed] [Google Scholar]
- Kocsy G., Brunner M., Rüegsegger A., Stamp P., Brunold C. (1996). Glutathione synthesis in maize genotypes with different sensitivities to chilling. Planta 198, 365–370 10.1007/BF00620052 [DOI] [Google Scholar]
- Kocsy G., Owttrim G., Brander K., Brunold C. (1997). Effect of chilling on the diurnal rhythm of enzymes involved in protection against oxidative stress in a chilling-tolerant and a chilling-sensitive maize genotype. Physiol. Plant. 99, 249–254 10.1111/j.1399-3054.1997.tb05409.x [DOI] [Google Scholar]
- Kocsy G., Von Ballmoos P., Suter M., Ruegsegger A., Galli U., Szalai G., et al. (2000). Inhibition of glutathione synthesis reduces chilling tolerance in maize. Planta 211, 528–536. 10.1007/s004250000308 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 97, 479–495. 10.1093/aob/mcl006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S., Chu C. C., Bauwe H. (1996). Molecular phylogeny of Flaveria as deduced from the analysis of nucleotide sequences encoding the H-protein of the glycine cleavage system. Plant Cell Environ. 19, 1028–1036 10.1111/j.1365-3040.1996.tb00209.x [DOI] [Google Scholar]
- Kopriva S., Jones S., Koprivova A., Suter M., Ballmoos P., Brander K., et al. (2001). Influence of chilling stress on the intercellular distribution of assimilatory sulfate reduction and thiols in Zea mays. Plant Biol. 3, 24–31 10.1055/s-2001-11745 [DOI] [Google Scholar]
- Kopriva S., Koprivova A. (2005). Sulfate assimilation and glutathione synthesis in C4 plants. Photosynth. Res. 86, 363–372. 10.1007/s11120-005-3482-z [DOI] [PubMed] [Google Scholar]
- Kopriva S., Muheim R., Koprivova A., Trachsel N., Catalano C., Suter M., et al. (1999). Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. Plant J. 20, 37–44. 10.1046/j.1365-313X.1999.00573.x [DOI] [PubMed] [Google Scholar]
- Kopriva S., Suter M., Von Ballmoos P., Hesse H., Krahenbuhl U., Rennenberg H., et al. (2002). Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiol. 130, 1406–1413. 10.1104/pp.007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Melzer M., Von Ballmoos P., Mandel T., Brunold C., Kopriva S. (2001). Assimilatory sulfate reduction in C3, C3–C4, and C4 species of Flaveria. Plant Physiol. 127, 543–550. 10.1104/pp.010144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Suter M., Den Camp R. O., Brunold C., Kopriva S. (2000). Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 122, 737–746. 10.1104/pp.122.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch W. M. (1974). The C4 syndrome: a structural analysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 25, 27–52 10.1146/annurev.pp.25.060174.000331 [DOI] [Google Scholar]
- Lappartient A. G., Touraine B. (1996). Demand-driven control of root ATP sulfurylase activity and SO42– uptake in intact canola (the role of phloem-translocated glutathione). Plant Physiol. 111, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M. N., Bick J. A., Davies J. P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 141–165. 10.1146/annurev.arplant.51.1.141 [DOI] [PubMed] [Google Scholar]
- Leustek T., Saito K. (1999). Sulfate transport and assimilation in plants. Plant Physiol. 120, 637–644. 10.1104/pp.120.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallmann J., Heckmann D., Brautigam A., Lercher M. J., Weber A. P., Westhoff P., et al. (2014). The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3, e02478. 10.7554/eLife.02478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson R. K., Moore B. D., Ku M. S. B., Edwards G. E. (1986). Co-function of C3-and C4-photosynthetic pathways in C3, C4 and C3–C4 intermediate Flaveria species. Planta 168, 493–502. 10.1007/BF00392268 [DOI] [PubMed] [Google Scholar]
- Moore R., Black C. C. (1979). Nitrogen assimilation pathways in leaf mesophyll and bundle sheath cells of C4 photosynthesis plants formulated from comparative studies with Digitaria sanguinalis (L.) Scop. Plant Physiol. 64, 309–313. 10.1104/pp.64.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C. H. (1998). ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. 10.1146/annurev.arplant.49.1.249 [DOI] [PubMed] [Google Scholar]
- Nussbaum S., Schmutz D., Brunold C. (1988). Regulation of assimilatory sulfate reduction by cadmium in Zea mays L. Plant Physiol. 88, 1407–1410. 10.1104/pp.88.4.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi J.-I., Kanai R. (1983). Differentiation of photorespiratory activity between mesophyll and bundle sheath cells of C4 plants I. Glycine oxidation by mitochondria. Plant Cell Physiol. 24, 1411–1420. [Google Scholar]
- Passera C., Ghisi R. (1982). ATP sulphurylase and O-acetylserine sulphydrylase in isolated mesophyll protoplasts and bundle sheath strands of S-deprived maize leaves. J. Exp. Bot. 33, 432–438 10.1093/jxb/33.3.432 [DOI] [Google Scholar]
- Pastori G. M., Mullineaux P. M., Foyer C. H. (2000). Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiol. 122, 667–675. 10.1104/pp.122.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfundel E., Nagel E., Meister A. (1996). Analyzing the light energy distribution in the photosynthetic apparatus of C4 plants using highly purified mesophyll and bundle-sheath thylakoids. Plant Physiol. 112, 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnam C. K., Edwards G. E. (1976). Distribution of nitrate-assimilating enzymes between mesophyll protoplasts and bundle sheath cells in leaves of three groups of C4 plants. Plant Physiol. 57, 881–885. 10.1104/pp.57.6.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R., Munoz-Bertomeu J., Krueger S. (2014). Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci. 19, 564–569. 10.1016/j.tplants.2014.06.003 [DOI] [PubMed] [Google Scholar]
- Sage R. F. (2004). The evolution of C4 photosynthesis. New phytol. 161, 341–370 10.1111/j.1469-8137.2004.00974.x [DOI] [PubMed] [Google Scholar]
- Sage R. F., Christin P. A., Edwards E. J. (2011). The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169. 10.1093/jxb/err048 [DOI] [PubMed] [Google Scholar]
- Sage R. F., Sage T. L., Kocacinar F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. 10.1146/annurev-arplant-042811-105511 [DOI] [PubMed] [Google Scholar]
- Saito K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136, 2443–2450. 10.1104/pp.104.046755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer U., Haensch R., Mendel R. R., Kopriva S., Rennenberg H., Herschbach C. (2010). Sulphur flux through the sulphate assimilation pathway is differently controlled by adenosine 5′-phosphosulphate reductase under stress and in transgenic poplar plants overexpressing γ-ECS, SO, or APR. J. Exp. Bot. 61, 609–622. 10.1093/jxb/erp327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz D., Brunold C. (1984). Intercellular localization of assimilatory sulfate reduction in leaves of Zea mays and Triticum aestivum. Plant Physiol. 74, 866–870. 10.1104/pp.74.4.866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz D., Brunold C. (1985). Localization of nitrite and sulfite reductase in bundle sheath and mesophyll cells of maize leaves. Physiol. Plant. 64, 523–528 10.1111/j.1399-3054.1985.tb08533.x [DOI] [Google Scholar]
- Schulze S., Mallmann J., Burscheidt J., Koczor M., Streubel M., Bauwe H., et al. (2013). Evolution of C4 photosynthesis in the genus Flaveria: establishment of a photorespiratory CO2 pump. Plant Cell 25, 2522–2535. 10.1105/tpc.113.114520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1999). C4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 187–217. 10.1146/annurev.arplant.50.1.187 [DOI] [PubMed] [Google Scholar]
- Sheen J. Y., Bogorad L. (1988). Differential expression in bundle sheath and mesophyll cells of maize of genes for photosystem II components encoded by the plastid genome. Plant Physiol. 86, 1020–1026. 10.1104/pp.86.4.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. U.S.A. 105, 6196–6201. 10.1073/pnas.0711730105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L. M., Droux M. (2002). Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol. 128, 1137–1148. 10.1104/pp.010935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Vauclare P., Kopriva S., Fell D., Suter M., Sticher L., Von Ballmoos P., et al. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31, 729–740. 10.1046/j.1365-313X.2002.01391.x [DOI] [PubMed] [Google Scholar]
- Williams B. P., Johnston I. G., Covshoff S., Hibberd J. M. (2013). Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. Elife 2, e00961. 10.7554/eLife.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky D., Brychkova G., Fluhr R., Sagi M. (2013). Sulfite reductase protects plants against sulfite toxicity. Plant Physiol. 161, 725–743. 10.1104/pp.112.207712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]


