Abstract
Yeast cell wall integrity (CWI) signaling serves as a model of the regulation of fungal cell wall synthesis and provides the basis for the development of antifungal drugs. A set of five membrane-spanning sensors (Wsc1 to Wsc3, Mid2, and Mtl1) detect cell surface stress and commence the signaling pathway upon perturbations of either the cell wall structure or the plasma membrane. We here summarize the latest advances in the structure/function relationship primarily of the Wsc1 sensor and critically review the evidence that it acts as a mechanosensor. The relevance and physiological significance of the information obtained for the function of the other CWI sensors, as well as expected future developments, are discussed.
INTRODUCTION
Fungal cell walls are essential structures that provide the engulfed cell with shape and mechanical stability (1). The enzymes and signal transduction systems that govern their synthesis are perfect targets for the development of antifungal drugs, which are urgently required in view of the increment of fungal diseases observed in clinics (2, 3). A second field of interest is the battle against plant fungal pathogens, which cause substantial damage in agriculture (4). Clearly, any agent that interferes with proper cell wall synthesis will result in cell lysis and death. In view of the diversity of the fungal kingdom, it is therefore of great interest to define common regulatory and biosynthetic pathways that can serve as targets for the development of broad-spectrum antifungals.
Carbohydrates in fungal cell walls are contributed by glucan chains (β-1,3 and β-1,6 linkages) and mannoproteins, which are intertwined with chitin chains to various degrees, depending on the fungal species (5, 6). This elastic cage is constantly remodeled and structurally supported by the incorporation of cell wall proteins (7). Activities of the latter are required for bud growth and cell division in exponentially growing cultures, as well as for shmoo formation and cell fusion in the mating process. Moreover, as a first barrier to the environment, fungal cell walls are also subject to all kinds of stresses, such as changing medium osmolarity, oxidative agents, microbial enzymes, or mechanical injuries. In the model yeast Saccharomyces cerevisiae and its close relative Kluyveromyces lactis, such environmental cues are detected by a conserved signal transduction cascade commonly referred to as the cell wall integrity (CWI) pathway (Fig. 1) (8–11). Although it has been established that the pathway is triggered by a set of five membrane-spanning sensors in S. cerevisiae, their exact mode of action has not yet been elucidated (12). This review summarizes the experimental evidence gathered in recent years that indicates that they act as mechanosensors whose signaling capacity, at least in some cases, is enhanced by clustering in specific microdomains within the plasma membrane. Finally, we discuss the use of single-molecule and superresolution techniques that could be applied to better understand exactly how the different sensors function.
FIG 1.
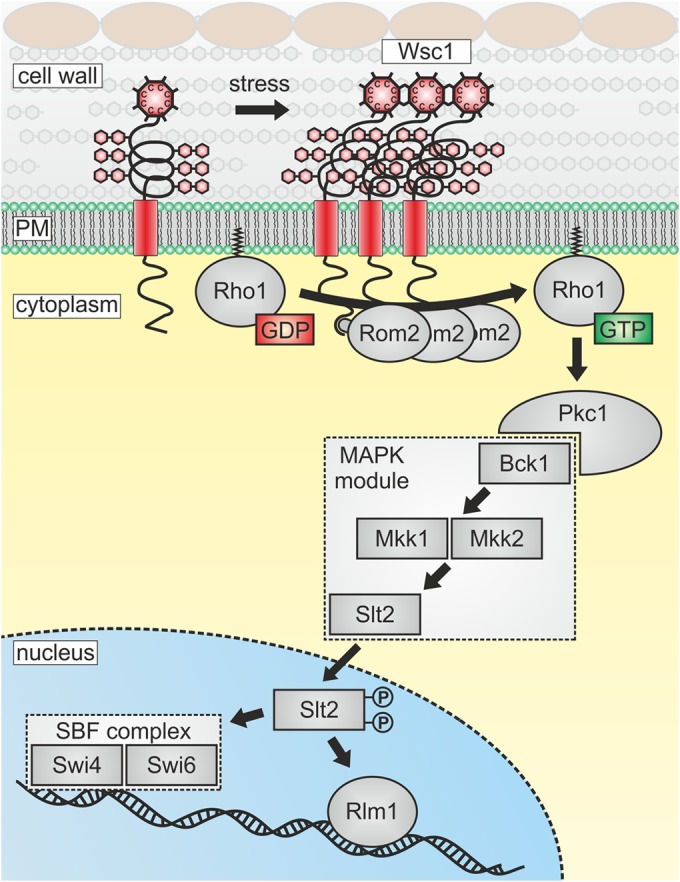
Schematic representation of the CWI pathway and its activation by sensor clustering. Wsc1 is depicted as a plasma membrane-spanning sensor that clusters upon the application of cell surface stress. Its cytoplasmic tail interacts with the GDP/GTP exchange factor Rom2, which activates the small GTPase Rho1 by catalyzing nucleotide exchange. Rho1-GTP then interacts with the sole yeast protein kinase C (Pkc1), which activates a conserved mitogen-activated protein kinase (MAPK) cascade comprising the MAPKKK Bck1, a pair of MAPKKs (Mkk1, Mkk2), and the MAPK Slt2. Target transcription factors are the SBF complex, which regulates cell cycle progression, and Rlm1, which activates genes whose products are required for cell wall synthesis, composition, and remodeling.
SENSOR STRUCTURES AND MECHANICAL PROPERTIES
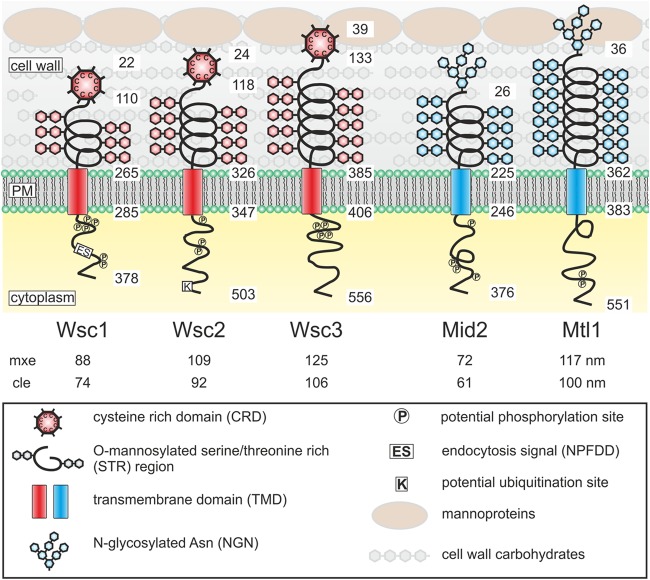
The five CWI sensors of S. cerevisiae can be divided into two small protein families, namely, (i) Wsc1 to Wsc3 and (ii) Mid2 and Mtl1 (Fig. 2). It should be noted that a similar protein, Wsc4, does not reside at the plasma membrane and thus is not a CWI sensor. The two families differ in the amino-terminal head group, which is presumed to physically connect with cell wall polysaccharides and/or proteins (13). In Wsc-type sensors, the head group is composed of a region comprising eight cysteine residues (CRD for cysteine-rich domain, also referred to as a WSC domain) that has features reminiscent of a lectin binding domain and is presumed to be in contact with the cell wall glucans (14, 15). The two Mid-type sensors each carry an N-glycosylated asparagine residue in a similar position, which is required for sensor function (16), and can be imagined to be intertwined with the glucan network or the mannoproteins in the outer layer (Fig. 2). The rest of the overall structures of all five sensors appear similar, in that a serine/threonine-rich (STR) region forms a spring-like structure that extends into the cell wall and a single transmembrane domain provides a second anchor for the sensors besides the head groups described above. Cytoplasmic tails of various lengths provide the link to the downstream components of the CWI signaling pathway, first by interacting with the GDP/GTP exchange factor Rom2 (17, 18). The cytoplasmic tails also determine the primary distribution of the sensors within the plasma membrane, as shown by fluorescence microscopy of hybrid Wsc1/Mid2 sensors (19).
FIG 2.
Structures and proposed functional domains of CWI sensors. The five CWI sensors are depicted with the protein domains indicated. Numbers on each sensor refer to the amino acid positions of the proteins deduced from the coding sequences from the amino terminus to the carboxy terminus (note that the signal peptides are cleaved off during secretion). Numbers below the sensor designations refer to the calculated lengths of the sensor regions extending from the plasma membrane. mxe, maximal possible extension calculated from a peptide bond length of 0.36 nm; cle, computed live-cell extension. The latter value was calculated by multiplication of the mxe by a factor of 0.85. This factor was obtained as the ratio of the cell wall thickness determined by transmission electron microscopy (102 nm) (46) to the mxe of 120 nm of a sensor that just reaches the cell surface, among a set of Wsc1 rulers employed for live-cell measurements of cell wall thickness by AFM (22). Since the STR region of the sensors is assumed to have spring properties and the CRD may have a globular rather than a linear conformation, the factor thus corrects for the difference between a fully extended (unnatural) sensor and its three-dimensional structure in live yeast cells.
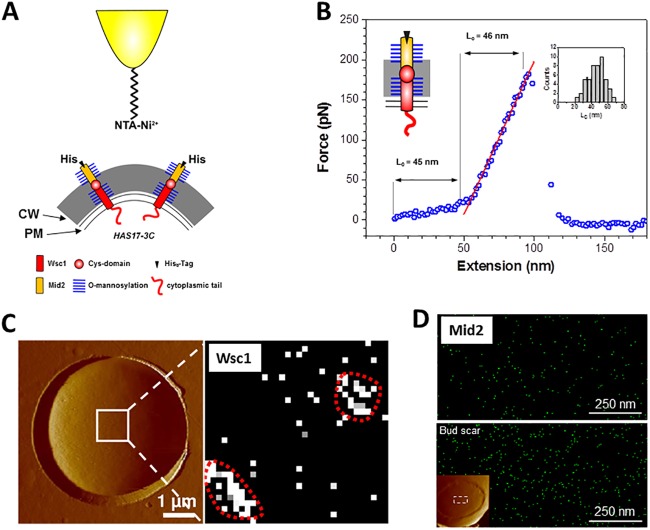
A major breakthrough in the determination of sensor functions came with the investigation of single Wsc1 sensors by atomic-force microscopy (AFM) (20), a technique that is increasingly used in microbiology (21). In brief, the CWI sensors are too short to reach the cell surface to be detected by AFM (22). Therefore, the Wsc1 sensor was elongated by adding the STR sequence of Mid2, as well as an 8×His tag, which could be probed with a Ni2+-nitrilotriacetic acid (NTA)-modified AFM tip (Fig. 3A) (23). Besides the detection of single sensor molecules, this also allowed the probing of their mechanical properties by pulling on them. These experiments showed that the Wsc1 sensor behaves like a Hookean spring (Fig. 3B), not only in its modified, elongated version, but also in a more native form carrying just an 8×His tag in the region of yeast bud scars, where the cell wall is considerably thinner (20). In agreement with previous suggestions (24), this led to a model in which Wsc1 acts as a mechanosensor that is anchored both in the cell wall and in the plasma membrane. The STR region in between would then act as a nanospring that stretches if either cell wall polysaccharides or membrane lipids are dislocated by external stress (or, for instance, by normal bud growth). This notion is consistent with the fact that compounds acting either on the cell wall or on the plasma membrane trigger CWI signaling (25). Conformational changes then transmitted to the cytoplasmic tail enable it to interact with the downstream signaling components and activate the CWI pathway. Although such a scenario seems logical, it has yet to be supported by direct experimental evidence of mechanoreception.
FIG 3.
Application of AFM to study the distribution and mechanical properties of CWI sensors. (A) Schematic representation of the modifications of the Wsc1 sensor that enable it to traverse the cell wall and be detected by a Ni2+-NTA-loaded AFM tip. CW, cell wall; PM, plasma membrane. (B) A representative force-extension curve obtained for stretching a Wsc1 molecule on live yeast cells, demonstrating that it behaves like a Hookean spring. (C) AFM surface scan of a yeast cell trapped in a polycarbonate grid (left) and detection of single Wsc1 molecules in the contact mode with the modified AFM tip (right), demonstrating clustering on the cell surface. (D) High-resolution AFM detection of a modified Mid2 sensor. The sensor was modified basically as shown in panel A for Wsc1; i.e., the native Mid2 sensor was elongated by introduction of the STR region of Wsc1 and equipped with an 8×His tag. The upper image shows the network-like distribution of the sensor in the lateral cell wall, and the lower image depicts the accumulation in the bud scar (covering the area shown in the inset of the surface scan). Panels A and B are reproduced from reference 20 with permission. Panel C is reproduced from reference 26. Panel D is reproduced from reference 34 with permission of the publisher (copyright 2012 American Chemical Society).
SENSOR DISTRIBUTION AND FUNCTION
Another interesting feature deduced from the single-molecule AFM data was that the modified Wsc1 sensor was not distributed evenly at the yeast cell surface but rather was gathered in patches with an approximate diameter of 200 nm (Fig. 3C) (26). Both sensor density and patch sizes increased substantially upon heat and osmotic stress, indicating that sensor clustering may be related to CWI signaling capacity. Support for such a correlation between sensor abundance and cell wall synthesis also came from studies of mutants in which the rapid turnover of Wsc1 sensors by endocytosis was blocked. Thus, mutants with defects in clathrin-mediated endocytosis displayed a drastically increased Wsc1 concentration in the lateral plasma membrane, which led to a thicker cell wall, as judged from images generated by transmission electron microscopy (27). This was also confirmed by in vivo AFM studies of sensors lacking the endocytosis signal (22). The Wsc1 sensor accumulates at sites of polar cell growth, such as the bud tip and at the bud neck prior to cell division, which has been observed in fluorescence microscopy studies (19, 28). A similar distribution was also observed for the Wsc2 sensor, the lack of which, contrary to previous reports on its minor role in CWI signaling, reduced the fitness of yeast cells in growth competition experiments when they were mixed with wild-type cells (29). More importantly, green fluorescent protein (GFP)-tagged versions of the two Wsc-type sensors displayed a patchy distribution in the lateral plasma membrane in live-cell fluorescence microscopy, reminiscent of the patches observed in the AFM experiments described above. Although the size and stability of the Wsc1 patches are also reminiscent of yeast eisosomes (30), they do not colocalize (our unpublished results).
In the context of sensor clustering, single-molecule AFM studies on Wsc1 yielded another interesting observation. Mutants with cysteine-to-alanine changes in the CRD head group completely lost the ability to cluster at the cell surface (26). This phenotype could be copied by adding dithiothreitol as a reducing agent to cells carrying the modified wild-type sensor (31). Thus, the structure of the CRD, which presumably is conferred by disulfide bridges, is a prerequisite for its ability to mediate clustering. Regarding its in vivo performance, the cysteine-to-alanine mutants displayed growth phenotypes similar to those caused by complete wsc1 deletions, indicating that sensor function is lost concomitantly with the ability to form clusters. In fact, it was previously observed that truncated Wsc1 proteins lacking the entire CRD region also lacked their signaling capacity, which could be restored by overproduction of the truncated sensors (32). We suggest that this could be due to the increased sensor density in the membrane, which may mimic clustering.
As stated above, we also suggest that the double-anchored polypeptide chain of Wsc1 forms a crucial part of a sensor by which mechanical stress at the cell surface is detected by stretching of the nanospring. This leads to conformational changes in the CRD, stabilized by disulfide bridges, and triggers both sensor clustering and conformational changes in the cytoplasmic domain, which then interacts with the downstream CWI components. The assembly of sensors in plasma membrane microdomains that recruit intracellular signaling components thus leads to the formation of a Wsc1 sensosome and enhances CWI signaling capacity (33).
NOT ALL SENSORS ARE THE SAME
The similarity of the overall structures of the five CWI sensors described above suggests that the data obtained for Wsc1 also apply to the other sensors. Indeed, AFM revealed nanospring properties similar to those of Wsc1 in a modified Mid2 sensor (34). However, one major difference between Wsc- and Mid-type sensors is the nature of their head group, i.e., CRD in the former and an N-glycosylated asparagine in the latter. Indeed, using high-resolution AFM imaging, clustering could not be observed for an elongated, His-tagged Mid2 sensor, either under normal growth conditions or under heat or osmotic stress, indicating that, at least during vegetative growth, Mid2 does not form sensosomes (Fig. 3D) (34). This observation is consistent with live-cell fluorescence imaging, where Mid2-GFP shows a more uniform distribution in a dense, network-like structure, rather than forming patches (35). In high-resolution AFM images, Mid2 appeared to be three times as abundant in bud scars as on the lateral cell surface, probably reflecting a greater chance of detection because of the thinner cell wall in this region (34).
Because of the severity of the growth defects in different sensor deletion mutants, Wsc1 and Mid2 have been suggested to contribute to the bulk of CWI signaling, with partially overlapping functions (13, 14). However, the differential responses of wsc1- and mid2-null mutants to different stress agents in serial-dilution assays and in transcriptome analyses suggest that the sensors evolved to specialize in different responses (19, 36). This may not be closely correlated with their membrane distribution, since switching of the Wsc1 endocytosis signal to a Mid2 sensor resulted in its untypical accumulation at sites of polar growth (similar to that of Wsc1) but could not suppress the growth defects of a wsc1 deletion (29). Alternatively, with regard to the mechanosensor hypothesis, the differential responses of the sensors to different stress agents could be mediated by probing different points in the lateral cell wall. Thus, the STR domains of the five sensors vary in length, so that their head groups would be anchored at corresponding distances from the plasma membrane (Fig. 2) (12). Different stress agents could affect the different layers of the cell wall to various degrees and thereby act primarily on a specific sensor. This would then trigger the appropriate adaptive physiological responses. Whether the distribution of the sensors in specific microdomains within the plasma membrane (see above) also contributes to the differential response remains to be elucidated.
It should be mentioned that Mtl1 has been proposed to serve a function in the oxidative-stress response (18, 37). Recently, it has been suggested to work in conjunction with Wsc1 or Mid2 to regulate the nuclear/cytoplasmic distribution of cyclin C and thereby programmed cell death in yeast (38). If and how these functions are related to the presumed mechanosensing properties of the sensors remain to be determined. In this context, comparative studies with other yeast species may be of value. Thus, in K. lactis, a closely related yeast that has not undergone a whole-genome duplication (11), only three CWI sensors have been identified, excluding an Mtl1 homolog (39). Since CWI signaling functions seem to be similar in K. lactis and S. cerevisiae (8), it would be interesting to see which of the remaining sensors is involved in the oxidative-stress response of the milk yeast. In the more distantly related yeast Schizosaccharomyces pombe, Wsc1 and Mid2 homologs have also been characterized (with the latter designated SpMtl2), but they did not trigger the CWI pathway (40). It has also been suggested that Wsc1 has a role in biofilm formation in a certain S. cerevisiae strain independent from the downstream CWI signaling pathway (41). Thus, mechanosensing may trigger not-yet-anticipated cellular responses in addition to cell wall remodeling.
QUO VADIS?
Despite the progress made in recent years regarding the functional characterization of CWI sensors as summarized above, the most pertinent question to be addressed is if they indeed react to mechanical cues at the yeast cell surface. The AFM tools developed so far to pull on specific sensor molecules could be exploited to this end. In combination with live-cell fluorescence microscopy and superresolution imaging techniques (42), one can design the appropriate experimental setup. A broad AFM tip modified with Ni2+-NTA could be employed to probe the surface of a yeast cell carrying the elongated, His-tagged Wsc1 sensor. Pulling on several sensor molecules at a time and following the distribution of GFP-tagged downstream components of the CWI pathway would provide conclusive evidence of the formation of sensosomes. Alternatively, the AFM tip could be modified with an antibody and used in conjunction with a sensor tagged accordingly, in order to enhance the strength of the interaction.
It should also be noted that similar AFM approaches could be applied to study any cell surface proteins that can be elongated to penetrate the yeast cell wall (e.g., the pheromone receptors) and also for the investigation of basically any cell surface protein in other organisms (33).
Finally, apart from the interesting questions of exactly how the sensors function and if there is mechanosensing in yeast cells, there are also two applied aspects to this research. (i) The data available so far suggest that the nanospring properties of the sensors are essential for their signaling function. These mechanical features are conferred by the highly mannosylated STR region and probably also by the anchoring of the head group in the cell wall. Thus, drugs interfering with either protein mannosylation or specific inhibitors of the CRD interactions within the Wsc-type sensors would be expected to impair cellular integrity, leading to cell lysis. In fact, mutants defective in O-mannosylation have been shown to have cell lysis defects (43). Expressing CWI sensors of pathogenic fungi, preferably in a deletion mutant of S. cerevisiae lacking its endogenous sensor genes, should allow screening for such specific drugs. (ii) We already elongated the Wsc1 and Mid2 sensors to reach through the cell wall in order to be detectable by single-molecule AFM (23, 34). Instead of displaying a His tag, similar constructs could be used to display specific antigens on the yeast cell surface, e.g., to trigger the immune system to attack and destroy tumor cells, as already reported for other yeast surface display systems (44, 45). The use of the CWI sensors for surface display would have the advantage that the antigens remain tightly attached to the yeast cells since they form one polypeptide chain with the downstream transmembrane domain.
ACKNOWLEDGMENTS
Work at the Université Catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université Catholique de Louvain (Fonds Spéciaux de Recherche), the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté Française de Belgique (Concerted Research Action). Y.F.D. is a Research Director at the FNRS. Work in the laboratory of J.J.H. was supported by a grant from the Deutsche Forschungsgemeinschaft within the framework of SFB944.
Biographies

Christian Kock studied biology at the University of Osnabrück and received his M.S. in 2011. He is currently working on his doctoral thesis at the Department of Genetics at the University of Osnabrück. The main subject of his thesis is the investigation of CWI sensor functions and their distribution in the yeast plasma membrane.

Yves Dufrêne is a Research Director at the National Fund for Scientific Research at the Université Catholique de Louvain (UCL), Louvain, Belgium. He studied bioengineering at UCL and received his Ph.D. in 1996. He is currently heading a team interested in understanding the nanoscale properties of biomolecules, membranes, and living cells by using AFM techniques. The ultimate goal is to elucidate the molecular bases of crucial cellular events such as cell adhesion and mechanosensing and their biomedical implications like pathogen-host interactions.

Jürgen Heinisch heads the Genetics Department at the University of Osnabrück. He studied biology and received his doctoral degree in 1985 at the University of Darmstadt for his studies on yeast phosphofructokinase. From 1986 to 1988, he was a postdoctoral fellow at the University of Alberta, Edmonton, Canada, and returned from 1988 to 1999 as a research assistant to the University of Düsseldorf, where his work on yeast glycolysis was continued and the project on yeast cell wall integrity signaling was started around 1995. From 2000 to 2002, he was professor for fermentation technology at the University of Hohenheim, applying his experience in yeast glycolysis to the production of beverages and spirits. He resumed his basic research focusing on cell wall integrity sensors in 2003 after accepting the professorship in Osnabrück. Within the past 10 years, his research interests have been extended to yeast cytokinesis, and the milk yeast K. lactis was established as an alternative model organism.
REFERENCES
- 1.Klis FM, Mol P, Hellingwerf K, Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 2.Heinisch JJ. 2005. Baker's yeast as a tool for the development of antifungal kinase inhibitors—targeting protein kinase C and the cell integrity pathway. Biochim Biophys Acta 1754:171–182. doi: 10.1016/j.bbapap.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 3.Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol 11:272–279. doi: 10.1016/S0966-842X(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 4.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Free SJ. 2013. Fungal cell wall organization and biosynthesis. Adv Genet 81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 6.Orlean P. 2012. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192:775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klis FM, Brul S, De Groot PW. 2010. Covalently linked wall proteins in ascomycetous fungi. Yeast 27:489–493. doi: 10.1002/yea.1747. [DOI] [PubMed] [Google Scholar]
- 8.Backhaus K, Buchwald U, Heppeler N, Schmitz HP, Rodicio R, Heinisch JJ. 2011. Milk and sugar: regulation of cell wall synthesis in the milk yeast Kluyveromyces lactis. Eur J Cell Biol 90:745–750. doi: 10.1016/j.ejcb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol 32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- 10.Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodicio R, Heinisch JJ. 2013. Yeast on the milky way: genetics, physiology and biotechnology of Kluyveromyces lactis. Yeast 30:165–177. doi: 10.1002/yea.2954. [DOI] [PubMed] [Google Scholar]
- 12.Rodicio R, Heinisch JJ. 2010. Together we are strong—cell wall integrity sensors in yeasts. Yeast 27:531–540. doi: 10.1002/yea.1785. [DOI] [PubMed] [Google Scholar]
- 13.Jendretzki A, Wittland J, Wilk S, Straede A, Heinisch JJ. 2011. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur J Cell Biol 90:740–744. doi: 10.1016/j.ejcb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponting CP, Hofmann K, Bork P. 1999. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr Biol 9:R585–R588. doi: 10.1016/S0960-9822(99)80379-0. [DOI] [PubMed] [Google Scholar]
- 16.Hutzler F, Gerstl R, Lommel M, Strahl S. 2008. Protein N-glycosylation determines functionality of the Saccharomyces cerevisiae cell wall integrity sensor Mid2p. Mol Microbiol 68:1438–1449. doi: 10.1111/j.1365-2958.2008.06243.x. [DOI] [PubMed] [Google Scholar]
- 17.Vay HA, Philip B, Levin DE. 2004. Mutational analysis of the cytoplasmic domain of the Wsc1 cell wall stress sensor. Microbiology 150:3281–3288. doi: 10.1099/mic.0.27264-0. [DOI] [PubMed] [Google Scholar]
- 18.Petkova MI, Pujol-Carrion N, de la Torre-Ruiz MA. 2012. Mtl1 O-mannosylation mediated by both Pmt1 and Pmt2 is important for cell survival under oxidative conditions and TOR blockade. Fungal Genet Biol 49:903–914. doi: 10.1016/j.fgb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Straede A, Heinisch JJ. 2007. Functional analyses of the extra- and intracellular domains of the yeast cell wall integrity sensors Mid2 and Wsc1. FEBS Lett 581:4495–4500. doi: 10.1016/j.febslet.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Dupres V, Alsteens D, Wilk S, Hansen B, Heinisch JJ, Dufrene YF. 2009. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat Chem Biol 5:857–862. doi: 10.1038/nchembio.220. [DOI] [PubMed] [Google Scholar]
- 21.Alsteens D, Beaussart A, El-Kirat-Chatel S, Sullan RM, Dufrene YF. 2013. Atomic force microscopy: a new look at pathogens. PLoS Pathog 9:e1003516. doi: 10.1371/journal.ppat.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupres V, Dufrene YF, Heinisch JJ. 2010. Measuring cell wall thickness in living yeast cells using single molecular rulers. ACS Nano 4:5498–5504. doi: 10.1021/nn101598v. [DOI] [PubMed] [Google Scholar]
- 23.Heinisch JJ, Dupres V, Alsteens D, Dufrene YF. 2010. Measurement of the mechanical behavior of yeast membrane sensors using single-molecule atomic force microscopy. Nat Protoc 5:670–677. doi: 10.1038/nprot.2010.19. [DOI] [PubMed] [Google Scholar]
- 24.Philip B, Levin DE. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol Cell Biol 21:271–280. doi: 10.1128/MCB.21.1.271-280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straede A, Corran A, Bundy J, Heinisch JJ. 2007. The effect of tea tree oil and antifungal agents on a reporter for yeast cell integrity signalling. Yeast 24:321–334. doi: 10.1002/yea.1478. [DOI] [PubMed] [Google Scholar]
- 26.Heinisch JJ, Dupres V, Wilk S, Jendretzki A, Dufrene YF. 2010. Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS One 5:e11104. doi: 10.1371/journal.pone.0011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piao HL, Machado IM, Payne GS. 2007. NPFXD-mediated endocytosis is required for polarity and function of a yeast cell wall stress sensor. Mol Biol Cell 18:57–65. doi: 10.1091/mbc.E06-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verna J, Lodder A, Lee K, Vagts A, Ballester R. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilk S, Wittland J, Thywissen A, Schmitz HP, Heinisch JJ. 2010. A block of endocytosis of the yeast cell wall integrity sensors Wsc1 and Wsc2 results in reduced fitness in vivo. Mol Genet Genomics 284:217–229. doi: 10.1007/s00438-010-0563-2. [DOI] [PubMed] [Google Scholar]
- 30.Merzendorfer H, Heinisch JJ. 2013. Microcompartments within the yeast plasma membrane. Biol Chem 394:189–202. doi: 10.1515/hsz-2012-0241. [DOI] [PubMed] [Google Scholar]
- 31.Dupres V, Heinisch JJ, Dufrene YF. 2011. Atomic force microscopy demonstrates that disulfide bridges are required for clustering of the yeast cell wall integrity sensor Wsc1. Langmuir 27:15129–15134. doi: 10.1021/la203679s. [DOI] [PubMed] [Google Scholar]
- 32.Lodder AL, Lee TK, Ballester R. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinisch JJ, Dufrene YF. 2010. Is there anyone out there? Single-molecule atomic force microscopy meets yeast genetics to study sensor functions. Integr Biol (Camb) 2:408–415. doi: 10.1039/c0ib00012d. [DOI] [PubMed] [Google Scholar]
- 34.Alsteens D, Dupres V, Yunus S, Latge JP, Heinisch JJ, Dufrene YF. 2012. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir 28:16738–16744. doi: 10.1021/la303891j. [DOI] [PubMed] [Google Scholar]
- 35.Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Söldner R. 2012. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 36.Bermejo C, Garcia R, Straede A, Rodriguez-Pena JM, Nombela C, Heinisch JJ, Arroyo J. 2010. Characterization of sensor-specific stress response by transcriptional profiling of wsc1 and mid2 deletion strains and chimeric sensors in Saccharomyces cerevisiae. OMICS 14:679–688. doi: 10.1089/omi.2010.0060. [DOI] [PubMed] [Google Scholar]
- 37.Petkova MI, Pujol-Carrion N, de la Torre-Ruiz MA. 2010. Signal flow between CWI/TOR and CWI/RAS in budding yeast under conditions of oxidative stress and glucose starvation. Commun Integr Biol 3:555–557. doi: 10.4161/cib.3.6.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin C, Parshin AV, Daly I, Strich R, Cooper KF. 2013. The cell wall sensors Mtl1, Wsc1, and Mid2 are required for stress-induced nuclear to cytoplasmic translocation of cyclin C and programmed cell death in yeast. Oxid Med Cell Longev 2013:320823. doi: 10.1155/2013/320823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodicio R, Buchwald U, Schmitz HP, Heinisch JJ. 2008. Dissecting sensor functions in cell wall integrity signaling in Kluyveromyces lactis. Fungal Genet Biol 45:422–435. doi: 10.1016/j.fgb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Cruz S, Munoz S, Manjon E, Garcia P, Sanchez Y. 2013. The fission yeast cell wall stress sensor-like proteins Mtl2 and Wsc1 act by turning on the GTPase Rho1p but act independently of the cell wall integrity pathway. Microbiologyopen 2:778–794. doi: 10.1002/mbo3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarode N, Davis SE, Tams RN, Reynolds TB. 2014. The Wsc1p cell wall signaling protein controls biofilm (Mat) formation independently of Flo11p in Saccharomyces cerevisiae. G3 (Bethesda) 4:199–207. doi: 10.1534/g3.113.006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gahlmann A, Moerner WE. 2014. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat Rev Microbiol 12:9–22. doi: 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loibl M, Strahl S. 2013. Protein O-mannosylation: what we have learned from baker's yeast. Biochim Biophys Acta 1833:2438–2446. doi: 10.1016/j.bbamcr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Griffin MD, Holman PO, Tang Q, Ashourian N, Korthauer U, Kranz DM, Bluestone JA. 2001. Development and applications of surface-linked single chain antibodies against T-cell antigens. J Immunol Methods 248:77–90. doi: 10.1016/S0022-1759(00)00344-6. [DOI] [PubMed] [Google Scholar]
- 45.Malecek K, Zhong S, McGary K, Yu C, Huang K, Johnson LA, Rosenberg SA, Krogsgaard M. 2013. Engineering improved T cell receptors using an alanine-scan guided T cell display selection system. J Immunol Methods 392:1–11. doi: 10.1016/j.jim.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Backhaus K, Heilmann CJ, Sorgo AG, Purschke G, de Koster CG, Klis FM, Heinisch JJ. 2010. A systematic study of the cell wall composition of Kluyveromyces lactis. Yeast 27:647–660. doi: 10.1002/yea.1781. [DOI] [PubMed] [Google Scholar]