Abstract
Here, we leverage a unique collection of 708 prospectively collected autopsied brains to assess the methylation state of the brain's DNA in relation to Alzheimer's disease (AD). We find that the level of methylation at 71 of the 415,848 interrogated CpGs is significantly associated with the burden of AD pathology, including CpGs in the ABCA7 and BIN1 regions, which harbor known AD susceptibility variants. We validate 11 of the differentially methylated regions in an independent set of 117 subjects. Further, we functionally validate these CpG associations and identify the nearby genes whose RNA expression is altered in AD: ANK1, CDH23, DIP2A, RHBDF2, RPL13, RNF34, SERPINF1 and SERPINF2. Our analyses suggest that these DNA methylation changes may have a role in the onset of AD since (1) they are seen in presymptomatic subjects and (2) six of the validated genes connect to a known AD susceptibility gene network.
Introduction
Evidence is emerging that DNA methylation levels at certain CpG dinucleotides can be both highly variable across individuals and stable over time within an individual1,2. This suggests that differences in DNA methylation in certain loci could be correlated with life experiences of a given individual, such as a disease risk factor or a diagnosis3. The most compelling evidence that the epigenome may influence AD comes from the manipulation of histone deacetylases (HDAC) in model systems of AD and in off-label treatment of AD patients with HDAC inhibitors4,5. To date, results of epigenomic studies of AD are sometimes conflicting and have not yet returned robust associations 6-9. Further, there is a gradual increase in methylation at many sites throughout the genome with increasing age that has to be carefully considered when studying AD.8,10-12 In this study, we present the results of a statistically rigorous gene discovery effort to identify regions of the genome that are differentially methylated in relation to the burden of AD neuropathology. We show that 71 discrete CpG dinucleotides in the human genome exhibit altered DNA methylation levels in relation to AD, that these changes are an early feature of AD, that the transcription of genes found in these differentially methylated regions is also independently associated with AD pathology, and that these differentially expressed genes connect to a previously reported genetically defined AD susceptibility network13.
Description of subjects and data
Our data set consists of methylation measures at 415,848 discrete CpG dinucleotides in 708 subjects. These methylation profiles were generated using the Illumina HumanMethylation450 beadset and a sample of dorsolateral prefrontal cortex obtained from each individual. Since we dissected out the gray matter from each sample, we have profiled a piece of tissue composed primarily of different neuronal populations and other parenchymal cells such as glia. These subjects are part of the Religious Order Study (ROS) or the Memory and Aging Project (MAP), two prospective cohort studies of aging that include brain donation at the time of death. Since the subjects are cognitively non-impaired at study entry, we have studied a random selection of the older population. Over the course of the study, some subjects decline cognitively and display a range of amyloid pathology burden at the time of death, with 60.8% of subjects meeting a pathologic diagnosis of AD14 (Supplementary Table 1a). To technically validate the nature of our data, we compared our Illumina-derived data to genome-wide DNA methylation sequence data generated from the same brain DNA samples in four of the subjects (two non-impaired and two AD subjects): in these four subjects, we see a very strong correlation (mean r= 0.97) between the estimated levels of methylation generated by the two technologies, consistent with prior reports15.
Interestingly, when examining the nature of human cortical methylation profiles across our subject population, we note that the mean Pearson correlation of methylation levels for all possible subject pairs is 0.98 (Supplementary Figure 1), suggesting that that majority of CpG sites do not show significant interindividual variation in methylation levels despite the very different life course of each of these older subjects. As expected, we see many more differences in DNA methylation profiles between our cortical samples and lymphoblastic cell lines from HapMap individuals that were profiled for assessments of data quality in our experiment (Supplementary Figure 2).
Discovery study to identify differentially methylated chromosomal regions
Our analytic strategy involves three stages, which are illustrated in Figure 1: Stage 1 is our DNA methylation screen for chromosomal regions in which methylation levels correlate with AD pathology. Details of the analytic model are presented in the Online Methods section. It is followed by Stage 2 in which we replicate the significantly associated CpGs from Stage 1 in an independent set of subjects. In Stage 3, we attempt to functionally validate the role of the differentially methylated regions that are replicated in Stage 2 using mRNA obtained from AD and non-AD subjects. This strategy accomplishes 2 goals: (a) further confirms the role of a given differentially methylated region by showing that a meaningful biological effect (transcriptional change) relates to the disease and (b) helps to narrow down which of the genes near the differentially methylated CpGs are differentially expressed and may therefore be the target gene(s) in a given region.
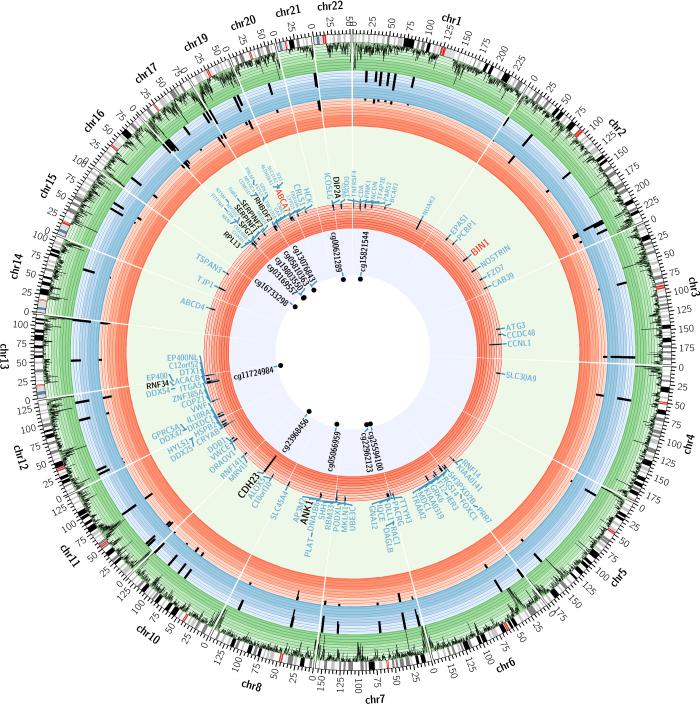
Figure 1. Summary of the genome-wide brain DNA methylation scan for NP burden and its validation using two sets of brain RNA data.
Each sector of this diagram presents summary results of the three different analyses within a chromosome. The perimeter of this circular figure presents the physical position along each chromosome (in Mb). The cytogenetic bands of each chromosome are presented in the first circle, with the centromere highlighted in red. The next circle (green) reports the density of CpG probes successfully sampled by the Illumina beadset that are present in a given genomic segment. The blue circle reports the results of the DNA methylation scan: using a “–log(p-value)” scale, we report the results for each of the 71 associated CpG found in 60 independent differentially methylated regions (DMR) from the analysis relating DNA methylation levels to NP burden. Similarly, the first red circle reports the –log(p-value) for the 71 CpGs in the replication analysis. The large lightly colored circle reports the names of genes found within 50 kb of each associated CpG (light blue letters). The ABCA7 and BIN1 regions, which harbor AD susceptibility alleles, are highlighted in red letters. The subset of the genes with differential mRNA expression in AD in the Mayo clinic dataset are shown in black. The next, red circle reports the results of the association of RNA expression level of these genes to a diagnosis of AD in the Mayo clinic dataset. The central circle reports the set of validated CpGs who also have nearby genes whose expression is altered.
In the primary analysis of our cortical methylation profiles (Stage 1), we identified autosomal CpGs whose level of methylation correlates with the burden of neuritic amyloid plaques (NP), a key quantitative measure of Alzheimer's disease neuropathology. NP burden better captures the state of the brain of a deceased subject since cognitively intact individuals display a range of NP pathology, some of which meet neuropathologic criteria for a diagnosis of AD16,17. Table 1 and Supplementary Table 2 contain the results of the primary genome-wide analysis: 137 CpGs are associated with the burden of NP pathology at a p<1.20×10−7. This threshold of significance accounts for the testing of all 415,848 tested CpGs by imposing a Bonferroni correction on a standard p<0.05. Since the exact number of functionally independent units of methylation in the genome is currently unknown, we have chosen this simple but conservative strategy to account for the testing of multiple hypotheses and correct for the testing of each CpG that was measured. Since the proportion of neurons found in each sample was not related to AD (p=0.08), we did not include this as a term in the primary analysis. However, to focus only on the most conservatively associated CpGs, we performed a secondary analysis that includes the variable that captures the proportion of neurons as well as surrogate variables that capture structure in the methylation data that do not correlate with known confounders and may capture cryptic technical or other artifacts. Of the 137 CpGs discovered in the primary analysis, 71 CpGs remain significant in the more conservative secondary analysis (Table 1, Supplementary Table 2). Some of these 71 CpGs are found in the same chromosomal segment and are highly correlated in their level of methylation. Altogether, the 71 CpGs are found in 60 discrete differentially methylated regions distributed throughout the genome (Figure 1): in 8 of these 60 regions, up to three neighboring CpGs with correlated levels of methylation emerge as significant in our analysis and probably capture the same effect.
Table 1.
CpGs associated with amyloid burden: validated CpGs and CpGs in known AD loci
| CpG | Chr | Position (bp) |
Discovery Study | Replication Study | Genes within 50 kb of associated CpG | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate Np burden Model 1 |
P-value Np burden Model 1 |
Estimate Np burden Model 3 |
P-value Np burden Model 3 |
Estimate AD |
P-value AD |
Estimate Braak score |
P-value Braak score |
||||
| cg11724984 | 12 | 121890864 | 3.02 | 4.76E-09 | 4.48 | 9.75E-11 | 16.7 | 1.27E-07 | 3.40 | 5.44E-06 | RNF34,KDM2B |
| cg23968456 | 10 | 73521631 | 4.97 | 3.97E-10 | 4.79 | 8.97E-09 | 16.8 | 1.45E-05 | 1.62 | 8.27E-06 | CDH23, C10orf105,C10orf54 |
| cg15821544 | 1 | 43473840 | 3.52 | 1.17E-07 | 5.11 | 8.09E-11 | 18.9 | 1.46E-07 | 2.47 | 2.08E-04 | SLC2A1,FLJ32224 |
| cg16733298 | 16 | 19127132 | 2.75 | 5.24E-08 | 3.96 | 1.32E-09 | 12.9 | 9.76E-06 | 3.58 | 2.30E-04 | COQ7,ITPRIPL2 |
| cg22962123 | 7 | 27153605 | 1.7 | 1.12E-07 | 3.08 | 8.79E-13 | 10.8 | 5.56E-08 | 5.13 | 2.79E-04 | HOXA1,HOTAIRM1,HOXA2,AK291164,HOXA3,AK311383,BC035889,HOXA4,LOC100133311,HOXA5,HOXA6,DQ655986,HOXA7,HOXA9,HOXA10 |
| cg13076843 | 17 | 74475294 | 2.35 | 1.68E-09 | 2.29 | 5.99E-08 | 9.32 | 5.81E-07 | 3.02 | 2.99E-04 | UBE2O,AANAT,RHBDF2,AX747521,CYGB,PRCD |
| cg25594100 | 7 | 4786943 | 3.15 | 2.54E-11 | 4.22 | 3.33E-13 | 14.9 | 1.83E-08 | 4.09 | 4.19E-04 | FOXK1,KIAA0415,RADIL |
| cg00621289 | 21 | 47855916 | 3.5 | 6.48E-08 | 5.06 | 2.18E-11 | 17.5 | 3.98E-07 | 2.02 | 4.95E-04 | PCNT,DIP2A |
| cg19803550 | 17 | 1637391 | 4.36 | 1.04E-08 | 4.48 | 1.60E-08 | 19 | 6.81E-07 | 1.51 | 5.61E-04 | PRPF8,TLCD2,MIR22HG,AF070569,MIR22,WDR81,SERPINF2,SERPINF1,SMYD4 |
| cg03169557 | 16 | 89598950 | 4.86 | 3.99E-10 | 4.88 | 3.36E-09 | 18.6 | 1.67E-06 | 1.52 | 6.05E-04 | ANKRD11,SPG7,SNORD68,RPL13,CPNE7 |
| cg05066959 | 8 | 41519308 | 2.69 | 7.13E-14 | 2.78 | 7.56E-13 | 11.2 | 4.88E-10 | 3.45 | 6.48E-04 | AGPAT6,NKX6-3,JA429246,ANK1 |
| cg05810363 | 17 | 74475270 | 2.95 | 3.68E-10 | 2.86 | 8.11E-09 | 11.3 | 3.36E-07 | 2.76 | 7.93E-04 | UBE2O,AANAT,RHBDF2,AX747521,CYGB,PRCD |
| Known AD loci | |||||||||||
| cg22883290 | 2 | 127800646 | 4.41 | 3.73E-08 | 4.44 | 8.97E-08 | 15.9 | 1.83E-05 | 0.96 | 0.0067 | BIN1 |
| cg02308560 | 19 | 1071176 | 2.19 | 3.06E-08 | 3.62 | 2.45E-12 | 13.7 | 1.77E-08 | 3.60 | 0.011 | CNN2,ABCA7,HMHA1,POLR2E,GPX4,SBNO2 |
Table legend: The Np analysis reports the results of a linear regression analysis relating DNA methylation level to the burden of neuritic plaque. “Model 1” refers to the primary analysis, and “Model 3” refers to the secondary that includes a variable for the estimate of neuronal cells and surrogate variables. The “AD” analysis reports the result of a logistic regression relating the level of methylation of a given CpG to a pathologic diagnosis of Ad. The threshold of genome-wide significance is p<1.12×10-7 after accounting for the testing of probes genome-wide in both Model 1 and Model 3. We have listed all genes found within a segment 50 kb upstream and downstream of the associated CpG. In the “Known AD loci”, we have listed those CpGs that are significant in our discovery analysis
Glossary: AD – Alzheimer's disease, Chr – chromosome, NP – neuritic plaque, patho AD – pathologic diagnosis of AD
Individually, any one of the significantly associated CpGs (Table 1, Supplementary Table 2) has a modest effect on the brain's NP burden: on average, each the 71 CpGs explains 5.0% (range 3.7-9.7%) of the variance in NP burden. However, this is greater than the proportion of variance explained by genetic variants associated with AD, with the exception of APOE. For example, in our subjects, the well-validated CR1 susceptibility allele explains just 1% of variance in NP burden18, and all known AD variants and APOE ε4 account for 13.9% of the variance in NP burden. If we consider all 71 CpGs in one comprehensive model, they explain 28.7% of the variance in NP burden, suggesting that methylation levels of certain genomic regions is correlated and that cortical DNA methylation of a large number of discrete regions is strongly correlated with a key measure of AD neuropathology.
Notably, two of the 71 significantly associated CpGs (Supplementary Table 2) are found in loci that harbor known AD susceptibility alleles: cg22883290 in the BIN1 locus (beta=4.44, p=9.00×10−8) and cg02308560 in the ABCA7 locus (beta=3.62, p=2.45×10−12)19-22. cg22883290, is located 5 kb from the 5’ end of the BIN1 gene and 92 kb from the index SNP, rs744373, that best captures the genetic association to AD in this region (Figure 2c)19. The susceptibility variant rs744373 is moderately associated with the level of methylation at cg22883290 (p=0.0003). However, the CpG association with AD pathology is not driven by the variant: adjusting for rs744373 does not meaningfully change the effect size of the CpG association to NP burden (model with rs744373 as a covariate: beta=4.37, p=4.91×10−7). Within our dataset of modest size, rs744373 is not associated with AD susceptibility, and we therefore cannot formally test for mediation of the SNP's association to disease by CpG methylation. In the case of ABCA7, the index SNP (rs3764650) is associated with NP burden23 but has no association (p=0.07) with the level of methylation at cg02308560 which is 25 kb away, so, in both of these regions, SNPs and CpGs appear to have independent effects on AD susceptibility. Overall, risk of AD may therefore be affected by different sources of genomic variation (genetic and epigenetic) that have independent effects on the disease process.
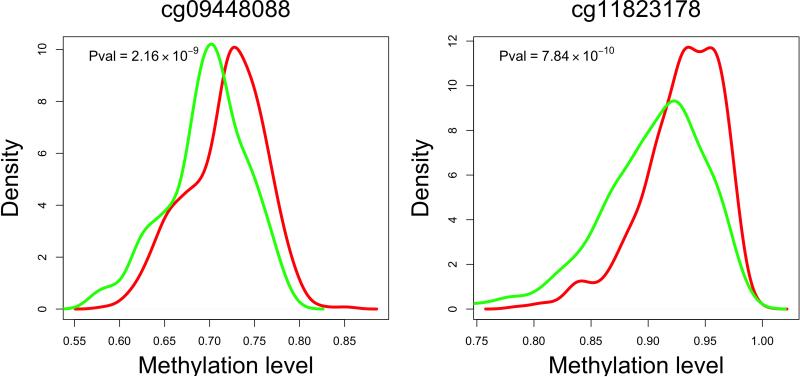
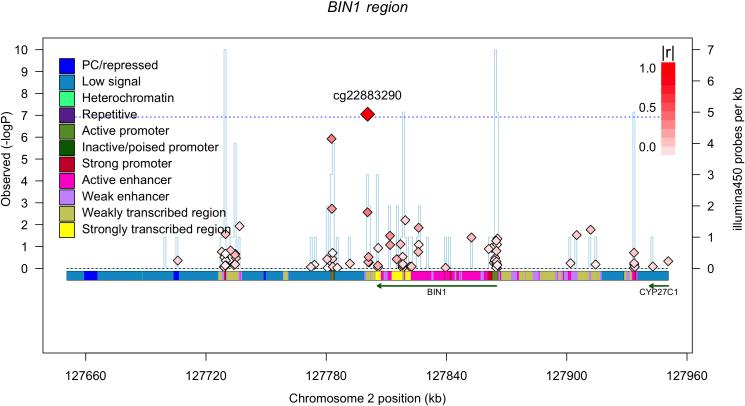
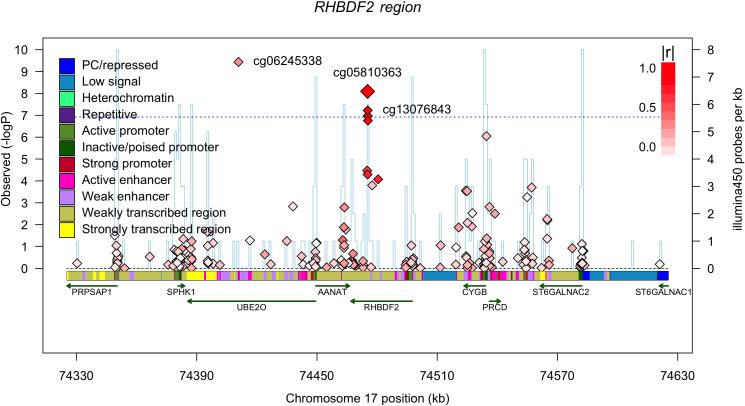
Figure 2. Extent of differences in methylation levels at associated CpGs and regional distribution of associations.
Two of the most AD-associated probes - from the MCF2L (a) and ANK1 (b) differentially methylated regions, Table 1 - are selected to illustrate the increase in methylation levels seen, on average, with a diagnosis of AD in 82% of the CpGs that meet our threshold of significance. Each panel reports data for one CpG, which is listed at the top of the panel. The panel itself is a smoothed histogram presenting the distribution of DNA methylation values at that CpG for subjects classified as having a neuropathologic diagnosis of AD (case, red, n=460) and those subjects that do not meet these diagnostic criteria (control, green, n=263). The scale is truncated at a methylation level of 0.6 since there are no values beyond this point; a methylation value of “1.0” means that all CpG in the sample are methylated. We see that the distribution of AD subjects is statistically significantly different from that of the control subjects. However, the two distributions overlap, and the absolute difference between the two distributions is modest. (c) Regional association plot around cg22883290 in the BIN1 differentially methylated region (DMR) that has previously been associated with AD susceptibility in genome-wide association studies. Each diamond represents on CpG tested in this region; the x-axis reports the physical distance across the region. The y-axis on the left reports the magnitude of the p-value in the analysis relating DNA methylation to NP burden. The horizontal dotted blue line highlights the threshold of significance for this analysis. The vertical blue line reports the density of CpG probes at a given point; values (probes/kb) are reported on the y-axis on the right. The extent to which DNA methylation level at a given CpG correlates with the level of DNA methylation of the best CpG (cg22883290) is reported using the r2 value and visualized using a red,high/white, low scale (see upper right corner of each panel). Finally, above the diagram of the genes found in this DNA segment, we report the chromatin state of the region, as assessed in healthy, unimpaired older individuals with minimal AD-related pathology. The chromatin state is derived in 200 bp bins, and the color key is presented in the upper left corner of each panel. Overall, the BIN1 gene appears to be in an open, transcribed conformation in healthy, older dorsolateral prefrontal cortex, and the associated CpG appears to be located in a region just 3’ to the gene, which is largely in a conformation found on the periphery of actively transcribed regions. (d) Regional association plot around the RHBDF2 DMR, centered on cg13076843, which meets our threshold of significance. Here, we have an associated CpG that is found in close proximity to two genes, and our RNA analyses suggest that it is RHBDF2 that is the target of the DMR since its expression is altered relative to AD (Table 2).
To facilitate the interpretation of our results, we performed a secondary analysis correlating the level of methylation at these 71 CpGs with a post-mortem, neuropathologic diagnosis of AD. 22 of the NP-associated CpGs are also associated with a diagnosis of AD at a genome-wide level of significance (Table 1 and Supplementary Table 2), and all of the CpGs associated with NP burden display at least some evidence of association (p<0.001) with AD. This is not surprising since NP burden is one criterion for a neuropathologic AD diagnosis. We note an interesting polarization in the direction of these associations: 82% of the differentially methylated regions are more methylated in subjects with a diagnosis of AD. As noted above, the increased level of methylation in relation to AD at any one associated probe is modest (Figure 2a and 2b).
Validation of the associated CpG in an independent sample set
To further assess the robustness of our results, we evaluated the 71 significantly associated CpGs in an independent collection of 117 subjects with a different quantitative measure of AD pathology (Braak staging)24. These subjects were profiled in a sample of frontal cortex using the same Illumina Humanmethylation 450 platform; demographic details are presented in Supplementary Table 1b. We imposed a Bonferroni correction in this analysis and find that 12 CpGs are significant in this analysis (Table 1); since two of these CpGs are found in the same differentially methylated region near RHBDF2 (Figure 2D), eleven of the differentially methylated regions from the discovery study are validated (Supplementary Figure 3). Thus, despite the use of a different but related measure of AD pathology and a much smaller sample size, we see robust replication of our discovery screen's results. In addition to the significant CpGs, many other CpGs display suggestive evidence of association: when evaluating the entire set of 71 CpG, we see that the effect size of most of these CpGs is consistent across the two datasets (Supplementary Figure 4), suggesting that most of these CpGs will be validated as larger sample sizes are profiled. Both the BIN1 (p=0.0067) and the ABCA7 (p=0.011) CpGs display suggestive evidence of replication (Table 1).
Cognitively non-impaired subjects display the same alterations in methylation
To begin to explore the question of whether the increased level of DNA methylation in the associated regions is a cause or an effect of the neurodegenerative process of AD, we limited the NP analysis to those subjects who were deemed to be cognitively non-impaired at the time of death (no AD and no mild cognitive impairment). As has been well documented in neuropathological and imaging studies25,26, a large fraction of non-impaired, older individuals demonstrate accumulation of amyloid pathology that is asymptomatic. Within the subset of non-impaired subjects, the p value for the CpG associations is diminished given the reduced sample size (n=237), but the beta values, which capture the magnitude of the association's effect, are not significantly different from the beta values calculated from the entire sample collection (Supplementary Table 3). This suggests that the altered DNA methylation that we have identified in our discovery study is an early feature of AD pathology and occurs in the presymptomatic stage of the disease. These DNA methylation changes are therefore not secondary to the later stages of the dementing process. The question of whether altered DNA methylation contributes to the pathologic process or is an early epiphenomenon of the neurodegenerative process remains open.
Distribution of associated CpGs among different chromatin states
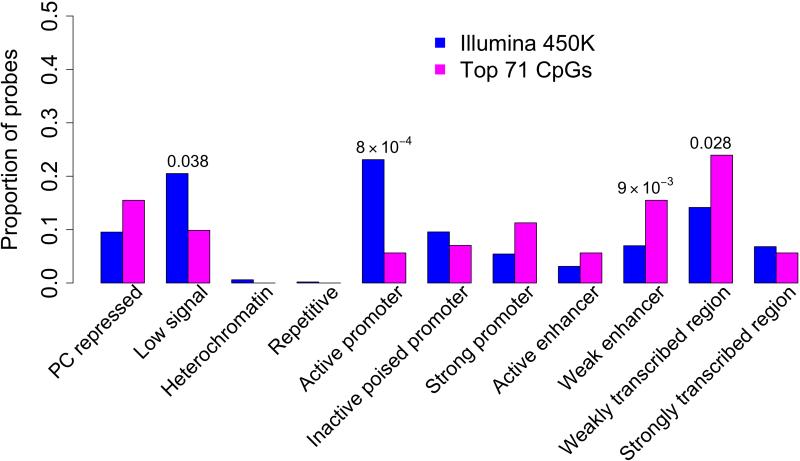
To better understand the functional consequences of the associated CpGs, we interpreted our results in relation to a chromatin map of the dorsolateral prefrontal cortex, generated in collaboration with the Epigenomics Roadmap team (http://www.roadmapepigenomics.org). It is derived from two MAP subjects who were cognitively non-impaired at the time of death and had minimal AD-associated pathology on post-mortem examination. Using histone modification profiles and established methods27, each 200 bp segment of the genome is annotated as being in one of 11 chromatin states (Figure 3a) that capture the transcriptional states and putative regulatory elements found in this tissue. Using this reference map, we see that at least some of the 71 associated CpGs are found in every chromatin state but that there is an enrichment of associated CpGs in regions predicted to be weak enhancers (p=0.0098) or to be in a weakly transcribed chromatin state (p=0.028), (Figure 3b, Supplementary Table 4). Further, we see a strong under-representation in regions displaying a strong promoter profile in the reference chromatin map (p=8×10−4). These data suggest that the chromatin architecture of strong promoters that drive fundamental cellular processes of neurons and glia in the healthy brain may not be strongly altered by AD. Rather, methylation changes appear to primarily affect genomic regions that are weakly transcribed or inactive in the healthy older brain. There are no enrichments noted in different genic features or in different structures defined in relation to CpG islands (Supplementary Figure 5a and 5b).
Figure 3. Distribution of CpGs associated (p<1×10−7) wih NP among 11 chromatin states found in mid-frontal cortex. (A) Chromatin map of the dorsolateral prefrontal cortex.
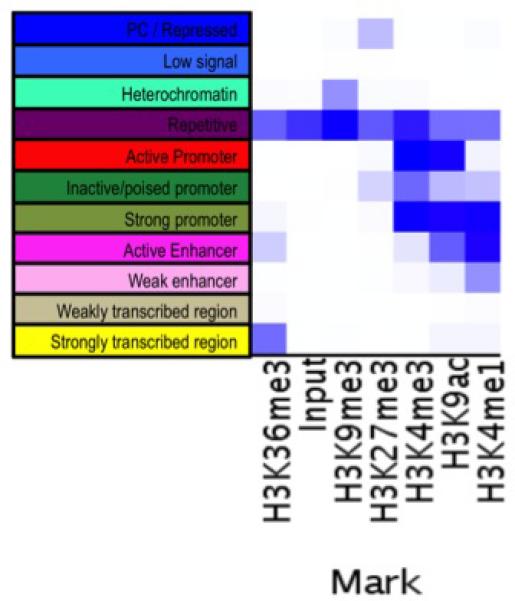
Using data generated by the National Institute of Health's Epigenomic Roadmap effort, we assign each chromosomal segment to one of 11 discrete chromatin states that are listed in a column to the left of the figure. On the X-axis, we list the individual chromatin marks (antigens) targeted in each set of ChiP-Seq data generated from MAP subjects that were cognitively non-impaired at the time of death and have minimal pathology on neuropathological examination. The heatmap (white, low; blue, high) graphically displays the relative abundance of sequences found in a segment of DNA after immunoprecipitation for a particular histone mark. Each chromatin state has a unique complement of histone marks. (B) We use the chromatin map illustrated in panel A to identify the chromatin state within which each of the interrogated CpG dinucleotides are found. The histogram compares the distribution of chromatin states found at those 71 associated CpG dinucleotides whose methylation level is associated with neuritic plaques (Table 1) (pink bars) to the overall distribution of chromatin states found in all 415,848 CpG dinucleotides that were analyzed (blue bars).
Functional validation of the CpG associations
Focusing on the 12 CpGs that have been validated in the replication stage, we evaluated their role in AD by assessing the level of expression of genes found in the vicinity (±50 kb) of these DMRs in an independent set of 202 AD and 197 non-AD individuals assembled by the Mayo Clinic (see Supplementary Table 5 for demographic characteristics) that have RNA data from the temporal cortex. We find that the level of expression of 8 of the 21 selected genes have significant associations (p<0.0023) with AD in these data (Table 2): ANK1, CDH23, DIP2A, RHBDF2, RPL13, RNF34, SERPINF1 and SERPINF2.
Table 2.
Analyses of transcriptional data within loci associated with amyloid burden
| Gene | Mayo Clinic subjects AD diagnosis |
|
|---|---|---|
| Beta | P value | |
| CDH23 | 0.58 | <0.0001 |
| DIP2A | 0.07 | <0.0001 |
| RPL13 | −0.08 | <0.0001 |
| RHBDF2 | 0.22 | 0.0001 |
| SERPINF1 | −0.14 | 0.0004 |
| SERPINF2 | 0.2 | 0.0008 |
| ANK1 | −0.17 | 0.0012 |
| KIAA0145 | 0.05 | 0.0013 |
| SMYD4 | 0.03 | 0.065 |
| PRPF8 | −0.04 | 0.13 |
| SLC2A1 | 0.02 | 0.13 |
| PCNT | 0.03 | 0.15 |
| COQ7 | 0.02 | 0.22 |
| ANKRD11 | −0.02 | 0.24 |
| C10ORF54 | 0.07 | 0.25 |
| FOXK1 | 0.02 | 0.28 |
| UBE2O | −0.02 | 0.28 |
| RNF34 | −0.05 | 0.30 |
| WDR81 | −0.03 | 0.39 |
| SPG7 | 0.04 | 0.54 |
| CYGB | −0.02 | 0.62 |
| AGPAT6 | 0 | 0.9 |
The “Beta” refers to effect size in the linear regression associating gene expression and AD diagnosis, with non-AD being the reference subject group. The significance threshold is p<0.0023 given 22 tests. Results are shown for mean expression level across available probes for each gene. Per probe results are presented in Supplementary Table 6.
Integrating our results with known AD genes
To further evaluate the role of these eight genes in relation to well-validated AD genes, we used the DAPPLE algorithm to evaluate the connectivity of these genes with the network of known AD susceptibility genes. We have previously used this method that requires co-expression of interacting protein pairs and adjusts for gene size, and we reported the existence of an AD susceptibility network derived from protein:protein interaction data13. Here, we use an updated model that includes the latest results from genome-wide association studies and the studies of rare variation. First, we find that the network of susceptibility genes from genome-wide association studies and mendelian AD genes is significant both in terms of direct connectivity (p=0.0072) and indirect connectivity (proportion of susceptibility genes sharing a common interactor, p=0.037)(Supplementary Figure 6). We then repeated the analysis after adding the eight genes found in the validated differentially methylated regions that also display altered RNA expression in AD. As seen in Figure 4, several of the differentially expressed genes found in the differentially methylated regions - ANK1, DIP2A, RHBDF2, RPL13, SERPINF1 and SERPINF2 - connect to the AD susceptibility network derived from genetic studies. The direct (p=0.0072) and indirect (p=0.042) network connectivity remain significant in the iteration of the network analysis that includes the eight genes with altered RNA expression levels.
Figure 4. Genes identified in our DNA methylation screen connect to a network of known AD susceptibility genes.
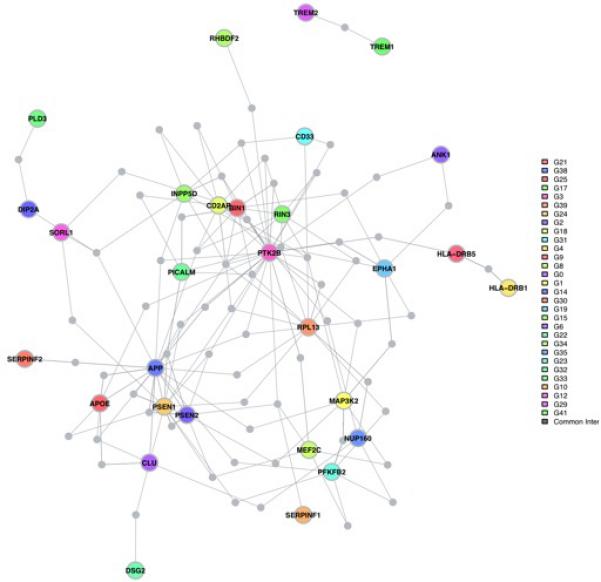
Using protein:protein interaction data, the DAPPLE algorithm evaluated the extent of connectivity among known AD genes (susceptibility and Mendelian genes) and the eight genes found in DMRs that are also differentially expressed relative to AD. The figure displays the results of an analysis allowing for one common interactor protein that is not known to be associated with AD. For example, RHBDF2 is displayed at the top of the figure in green and connects to PTK2B, a protein tyrosine kinase genetically associated with AD susceptibility which has a central role in this network. Interestingly, SERPINF1 and SERPINF2 connect to different elements of the amyloid component of the network (bottom left). Further, DIP2A connects the recently described PLD3 gene that has a rare AD susceptibility allele to SORL1, a gene with a common AD susceptibility allele, that connects to the amyloid precursor protein (APP). These interconnections are consistent with the reported effects of both PLD3 and SORL1 on amyloid biology and implicate DIP2A in the same process. Alternative figures presenting the network with all interacting proteins listed and the result of the network analysis with only the genetically associated loci are found in Supplementary Figure 6. The colored nodes are the proteins encoded by genes implicated in AD (genetic and epigenomic associations); the colors have no meaning. The connecting proteins not known to be associated AD are shown in gray.
Discussion
Overall, while our study has certain limitations due to the Illumina platform - such as surveying only a fraction of the human genome's CpGs and the array's inability to distinguish two closely related chromatin marks (DNA methylation and DNA hydroxymethylation) we nonetheless report several replicated, functionally validated associations between altered DNA methylation and the presymptomatic accumulation of AD pathology. These changes do not appear to be part of a generalized, genome-wide process: specific differentially methylated regions are targeted in AD and are unlikely to be found in genes that are actively transcribed in the healthy older brain. Instead, the associated regions are more likely to be in a poorly transcribed chromatin conformation in the older healthy brain (Figure 3b). Altered DNA methylation (Table 1) and enhanced mRNA expression (p=1.09×10−4) of the BIN1 gene in AD in the Mayo clinic dataset links our study to a well-validated AD susceptibility locus and a recent report of enhanced BIN1 expression in AD.28 The BIN1 cg22883290 association is significant in our discovery study, and, while the results are suggestive (p=0.0067) in the small replication analysis, the direction of the effect is consistent (Table 1), suggesting that the association is likely to validate with additional subjects. Our results therefore refine our understanding of the BIN1 locus and suggest that different types of genomic variation (SNP and CpG) can have independent effects that integrate on the expression of BIN1 and influence AD susceptibility. Similarly, cg02308560 in the ABCA7 locus is associated to AD pathology, and this association is independent of the susceptibility allele found in its vicinity. These two loci illustrate the point that, while genetic variation can drive differences in DNA methylation for certain CpGs3, the associations that we report are not driven by genetic associations with AD pathology: we have also recently completed a genome-wide SNP association study with the same trait in the same subjects and found no significant genetic associations23. Thus, our CpG associations are not driven by SNP associations.
The colocation of genetic susceptibility with CpG associations to AD pathology, along with the presence of the CpG associations in cognitively non-impaired subjects (Supplementary Table 3) and the connection of six of the differentially methylated genes (Figure 4) to an existing AD susceptibility network (Figure 4), suggest that DNA methylation changes play a role in the onset of AD. However, as with genetic studies, our epigenome-wide scan only reports associations with a trait. Thus, we cannot state that the observed changes in methylation are causal: given the plasticity of the epigenome, it is possible that these changes are an early consequence of AD pathology. Further experimental work needs to be conducted to resolve this important question.
Looking at the AD network map, DIP2A connects directly to the known SORL1 susceptibility gene29 and indirectly to PLD3, a recently reported AD susceptibility gene that is otherwise not connected to the AD susceptibility network. DIP2A may function as a cell surface receptor protein30 and, given the putative role of PLD331 and SORL1 in amyloid processing, its relation to the burden of NP pathology may well be related to a direct effect on amyloid processing. SERPINF1 and SERPINF2 also connect to elements of the amyloid machinery. Interestingly, SERPINF1 mRNA expression is reduced in AD (Table 2), and its knockdown in an in vitro system leads to reduced neurite outgrowth32, suggesting one potential effect of this gene.
On the other hand, ANK1 and RHBDF2 connect to PTK2B, an AD gene that is a key element of the signaling cascade involved in modulating the activation of microglia and infiltrating macrophages. Several other AD genes, such as CD33 and EPHA1, connect to this molecule as well. While little is known on the potential role of ANK1, the connection of RHBDF2 with PTK2B is consistent with the known role of this molecule in myeloid cells: it is necessary for the transport of TNFα converting enzyme (TACE, also called ADAM17), which releases TNFα from the cell surface33. Absence of RHBDF2 in mice impacts the normal release of TNFα from the cell surface34 and impairs systemic immune responses to pathogens35. In vitro work also suggests that RHBDF2 may function in regulating the substrate specificity of the TACE/ADAM17 protease which functions in the release of TNFα but also of other proteins such as epidermal growth factor (EGF)36 Its exact role in AD is not clear at this point, but our data suggests that RHBDF2 expression is increased in the context of AD (Table 2). Its connection to PTK2B further suggests that it may be involved in the role of microglia and infiltrating macrophages in the AD pathophysiological process. Consistent with this, adjusting for an estimate of the number of microglial cells using an RNA-based model seems to account for the AD-associated differences in RHBDF2 mRNA expression (Supplementary Table 7a and 7b).
Such cell type adjustment using surrogate markers for different cell types37-39 are crude analyses but are helpful to begin to assign some of the transcriptional alterations to certain cell types. For example, using GFAP expression as surrogate marker for astrocytes, we see that adding a term for GFAP expression in our assessment of CDH23 RNA expression largely abrogates the association of CDH23 RNA expression with AD (Supplementary Tables 7b). Since GFAP expression is enhanced with astrocyte activation40,41, we cannot distinguish whether the alteration of CDH23 RNA expression (and presumably its altered DNA methylation) is caused by an increased number of astrocytes near neuritic plaques, the activation of these astrocytes, or a combination of both effects. Regardless of the exact mechanism, in the case of CDH23, our DNA methylation screen has uncovered a robust alteration in methylation that can now be dissected mechanistically.
The human cortex has a complex architecture that includes many different types of neurons, glia, and other cells such as microglia, peripheral immune cells, and endothelial cells from cortical capillaries. The changes in DNA methylation that we report most likely represent the altered methylation state of a subset of cells within our cortical sample since AD pathology accumulates over several decades and only a small number of cells will be affected at a given time. It is too early to confidently differentiate between three possibilities that could explain these modest but robust changes in methylation that occur in relation to AD pathology: (1) a fraction of the constituent cortical cells are changing, such as activated astrocytes in the vicinity of neuritic plaques that overexpress CDH23, (2) the relative proportion of the constituent cell populations of the cortex is changing as some populations such as neurons are lost, or (3) there is a modest influx of immune cells from the systemic circulation that alters the relative abundance of the different cortical cell populations. It is also likely that more than one of these or other, unsuspected processes may be at work.
Overall, the replication of our study's results by an independent study24 and its functional validation with RNA data makes our strategy for investigating the brain's epigenome more broadly relevant to other epigenomic epidemiology studies. With clear effect sizes in hand, our results can be used to calibrate the design of future human studies in the brain or other organs. We have also made the important observation that these epigenomic changes are happening early in the pathologic process: while subjects display no cognitive impairment but have accumulated amyloid pathology. Going forward in the aging brain, we clearly need to more precisely map the alterations of chromatin structure that contribute to AD pathophysiology and to assess, using model systems, whether remodeling the epigenome is a fruitful goal for the development of AD therapies.
Online Methods
Subjects and genotypes
The analyses in this manuscript included deceased subjects from two large, prospectively followed cohorts maintained by investigators at Rush University medical Center in Chicago, IL: the Religious Orders Study (ROS) and the Memory and Aging Project (MAP). The ROS cohort, established in 1994, consists of more than 1,100 Catholic priests, nuns, and brothers from 40 groups in 12 states who were at least 55 years of age and free of known dementia at the time of enrollment. The MAP cohort, established in 1997, consists of more than 1600 men and women primarily from retirement facilities in the Chicago area who were at least 53 years of age and free of known dementia at the time of enrollment. All participants in ROS and MAP sign an informed consent agreeing to annual detailed clinical evaluations and cognitive tests, and the rate of follow-up exceeds 90%. Similarly, participants in both cohorts signed an Anatomical Gift Act donating their brains at the time of death. The overall autopsy rate exceeds 85%. As in previous manuscripts17, we analyzed the ROS and MAP cohorts jointly since they were designed to be combined, are maintained by a single investigative team, and a large set of phenotypes collected are identical in both studies. All aspects of these studies were approved by the Institutional Review Boards of Rush University Medical Center and Partners Healthcare. More detailed information regarding the two cohorts can be found in previously published literature42,43. Genotypes were available from prior studies and were derived from Affymetrix GeneChip 6.0 or Illumina Omni1-Quad genotypes and imputation using the HapMap reference, as previously described.23
The replication DNA methylation analysis uses samples of prefrontal cortex (PFC) obtained from 117 individuals archived in the MRC London Neurodegenerative Disease Brain Bank (http://www.kcl.ac.uk/iop/depts/cn/research/MRC-London-Neurodegenerative-Diseases-Brain-Bank/MRC-London-Neurodegenerative-Diseases-Brain-Bank.aspx). All samples were dissected by trained specialists, snap-frozen and stored at -80°C. Genomic DNA was isolated from ~100mg of each dissected brain region using a standard phenol-chloroform extraction method and tested for degradation and purity prior to analysis.
Temporal cortex expression levels for the autopsied Mayo Clinic subjects were obtained as part of a recently published brain expression GWAS (eGWAS), where the methodology is described in detail (eGWAS)44,45. Briefly, expression levels of 24,526 transcripts were measured from the temporal cortex of autopsied brains from subjects with pathologic AD (n=202) and those with other brain pathologies (non-AD, n=197). Total RNA extraction and QC were done using the Ambion RNAqueous kit and Agilent 2100 Bioanalyzer, respectively, according to published methods. Whole genome DASL expression microarrays (Illumina, San Diego, CA) were used for the transcriptome measurements of RNA samples that were randomized across chips and plates using a stratified approach to ensure balance with respect to diagnosis, age, sex, and RNA Integrity Numbers (RIN). Raw probe-level expression data exported from Genome Studio software (Illumina Inc.) were preprocessed with background correction, variance stabilizing transformation, quantile normalization and probe filtering using the lumi package of BioConductor46,47. Preprocessed probe transcript levels were used in the downstream analysis.
Phenotypes
Our primary phenotype of interest in this manuscript was the burden of neuritic plaques, a quantitative measure of the amount of AD neuropathology in the brain at the time of death. Brain autopsies in ROS and MAP were performed across the US, as described in detail elsewhere42,43 Bielschowsky silver stain was used to visualize neuritic plaques within tissue sections from five brain regions: the midfrontal, middle temporal, inferior parietal, and entorhinal cortices, and the hippocampal CA1 sector. As in prior publications17 a quantitative composite score of neuritic plaque burden was then computed for each individual by dividing the subject's raw count in each of the 5 regions by the population standard deviation in that same region, and then taking the average of the standardized counts across the 5 regions. Because the distribution of these average standardized counts is skewed, we used the square-root transformed values in our statistical analyses.
To put the associations with neuritic plaque burden in the context of an AD diagnosis, we also assessed for associations with a neuropathologic diagnosis of AD, which is determined on post-mortem examination. Specifically, subjects were classified as having a pathologic diagnosis AD if they had intermediate or high likelihood of AD based on the National Institute on Aging (NIA)-Reagan criteria. The NIA-Reagan criteria, which integrates both the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) estimates of neuritic plaque density and Braak staging of neurofibrillary tangle pathology, was implemented as reported48. These diagnoses are made by board-certified neuropathologists without access to the clinical data collected during the study. All neuropathologic data is collected in a blinded fashion (relative to clinical diagnosis) by the neuropathology staff.
Experimental Protocol for DNA Extraction from Post-mortem Brain
100 mg sections of frozen dorsolateral prefrontal cortex were obtained from each of 761 deceased subjects from the ROS and MAP studies based at the Rush Alzheimer's Disease Center. These sections were thawed on ice, and the gray matter was carefully dissected from the white matter. DNA extraction was performed using the Qiagen (cat: 51306) QIAamp DNA mini protocol. The Qubit 2.0 Fluorometer was used to quantitate the DNA. 16uL of DNA at a concentration of 50ng/uL as measured by PicoGreen, was used by the Broad Institute's Genomics Platform for data generation by the Illumina InfiniumHumanMethylation450 bead chip assay. The platform produces a data file by implementing the recommended procedures of the proprietary Illumina GenomeStudio software, which includes color channel normalization and background removal. All data generation was conducted by laboratory personnel who were blinded as to the clinical and neuropathological phenotypes of each subject
Subject and Probe Quality Control
For the initial quality check of the data, we used the detection p-value criteria recommended by Illumina. These p-values represent the quality of the probes compared to background noise. We selected probes, which have detection p-value < 0.01 for all samples to ensure the use of good quality probes. Using these criteria, we selected 470,913 out of a total of 485,577 tested probes for further analysis. Of these 470,913 probes, 460,045 are found in one of the autosomes.
However, not all probes are unique: some probes are predicted to cross-hybridize with the sex chromosomes49 based on sequence alignment. Many of these probes showed strong association to gender in a recent report that recommends discarding probes in which 47/50 nucleotides match the sex chromosome sequence during sequence alignment using BLAT49. Specifically, the authors recommend discarding 29,233 probes from the Illumina InfiniumHumanMethylation450 bead chip assay that meet this sequence match criterion. We implemented this recommendation and removed the 29,233 probes from our probe list for downstream analysis. Further, as noted below, we adjusted for gender in our analytic model given the well-described influence of sex on methylation levels.
In addition to cross-reactive probes, a substantial fraction of CpG probes also overlap with known Single Nucleotide Polymorphic sites (SNPs) based on the 1000 genome database49. Methylation level of these CpGs could be affected by a subject's genotype. However, SNPs with very low minor allele frequencies should not have major effect on the methylation data given our sample size. Therefore, we removed CpGs in which a SNP with a minor allele frequency (MAF) ≥0.01 exists within 10 base pairs upstream or downstream of the CpG site, where single base extension occurs. Based on the analysis reported by Chen and colleagues49, we found a total of 14,964 autosomal, polymorphic CpGs with 0.01 ≤ MAF (EU) ≤ 0.99 in subjects of European ancestry, such as our subjects. These CpGs were removed from consideration, leaving 415,848 autosomal CpGs for downstream analysis.
We also evaluated the “per subject” quality of the data after the initial CpG data cleaning. The first step in this component of our quality control pipeline was to remove subjects with poor quality data from further consideration. Specifically, we used Principal Component Analysis (PCA) to select subjects that are within ± 3 SD from mean of a PC for PC1, PC2 and PC3. PCA was performed using a random selection of 50,000 autosome probes for all of the samples. Using these criteria, we removed 13 subjects from a total of 761 subjects for which the Illumina assay was attempted. We excluded an additional 8 samples having for poor bisulfite conversion (BC) efficiency, which is defined as having at least 2 of the 10 BC control probes that fail to reach a value of 0.8. Hence, after our “per subject” quality check, a total 708 samples were selected for downstream analyses.
Normalization of the data
We observed a strong batch effect in our data due to the use of two different thermocyclers during data generation. This batch effect was also confirmed by our PCA analysis. We evaluated different approaches to normalize our data, including COMBAT50 and independent component analysis (ICA). However, when applying these approaches to our data and comparing them with an approach in which we adjust for given confounding variables (such as batch number and age) that we find to be associated with principal components within our data, these approaches (COMBAT and ICA) are overly conservative in normalizing our data. In particular, ICA includes adjustments for many unknown variables that capture structure within a set of data and are not associated with known confounders. Thus, such ICA-derived variables could be related to elements of disease pathophysiology that is not yet understood.
ICA is a matrix factorization technique that separates a matrix into statistically independent, non-Gaussian factors using the R-package “fastICA” developed by Marchini and colleagues51. We applied ICA to the matrix of methylation beta values to infer number of statistically independent components k iteratively from 1 to 40. In each instance, all samples are assigned k surrogate variables representing k entries of the mixing matrix. We then performed a refinement step similar to that used in “iSVA”52 where for each independent component i (1 <= i <=k) we created a sub-matrix of the beta values populated only by methylation probes that were significantly associated with component i. We then performed ICA on this sub-matrix and selected the independent component most correlated with component i, and used those refined components as the surrogate variables. To set an optimal k we explored the average variance explained in the methylation matrix by each value of k, and determined that 7 was a conservative number of components that explained an appreciable percentage of the variance. These 7 surrogate variables were then used in our evaluation of normalization methods.
In our comparison of the results obtained using the two different approaches (including known technical confounders such as batch vs. COMBAT and ICA normalization), we find that the top 70 CpG in the primary analysis remain significant regardless of the approach used and that, while the p-value of the other 67 CpGs that are significant in our primary analysis fluctuates below our threshold of significance, they retain strongly suggestive evidence of association to the neuritic amyloid trait (95% remain at a p<5×10−5). As a result, and given our strategy of validating the CpG analysis with two rounds of RNA analyses to minimize false positives (Figure 1), we opted to use the batch variable approach to directly address the source of technical variation and avoid over-correcting our data for variables that are not demonstrated to be associated with technical or demographic confounders. Thus, we prefer to minimize the risk of false negative results and minimize false positives with two rounds of validation.
Accounting for differences in the proportion of cell types in our tissue
Since the proportion of neurons and other cell types found in the human cerebral cortex can change with AD, we evaluated a technique to account for the possible difference in the proportion of neuronal cells in our brain samples. As discussed in the main text, we used an R package53 to quantify the proportion of NeuN+ cells (primarily neurons) in each brain sample using DNA methylation data. We used the data from the NeuN+ nuclei found in this package to create convex combinations of purified profiles using non-linear least squares. This yielded an estimate of the proportion of NeuN+ nuclei in each of our samples. However, the resulting measure is not significantly associated with a pathologic diagnosis of AD (p=0.08), and we therefore have not included it as a covariate in our primary analysis. However, we add it as a covariate in a secondary analysis that leads us to identify the most conservatively associated subset of 71 CpG.
Data analysis and statistical modeling
For our primary and secondary analyses, we used the β-values reported by the Illumina platform for each probe as the methylation level measurement for the targeted CG site in a given sample instead of M-values (logistically transformedβ-values)52. While M-values have certain favorable statistical properties relative to β-values, they are less biologically interpretable than β-values54. We therefore have opted to use β-values which range from 0 (no methylation) to 1 (100% methylation), and show good correlation to estimates of DNA methylation derived from whole genome bisulfite sequencing approaches (see below for details). Any missing β-value was imputed using a K-nearest neighbor algorithm for k=100.
In order to perform our methylome-wide association study (MWAS) and discover differentially methylated regions associated with neuritic plaque pathology, we used a linear model, adjusting for age at death, sex, study (ROS or MAP), experimental batch and bisulfite conversion efficiency. A logistic regression was used for the AD analysis, using the same covariates. As noted in the section above, we considered but elected not to include terms for (1) the proportion of neurons in each sample or (2) surrogate variables that displayed no correlation with available technical or demographic variables. To account for the testing of multiple hypotheses, we used a Bonferroni correction which yields a p<1.20 ×10−7 as the threshold for genome-wide significance given the 415,848 autosomal CpG probes tested in our analysis. For annotation of the CpG probes, we used the hg19 human reference genome.
The bisulfite conversion efficiency term is calculated using the bisulfite conversion control probes, based on Illumina guidelines. Ten CpG sites designated by Illumina as control sites (6 CpGs targeted by type I probes and 4 CpGs targeted by type-II probes), where we expect each CpG to be 100% methylated, are used to control for non-complete bisulfite conversion. The bisulfite conversion efficiency term used in the primary analysis is the median methylation estimate from the 10 control sites. The bisulfite conversion term is calculated by taking the median value of the probes that Illumina provides to estimate bisulfite conversion efficiency.
To assess whether changes in DNA methylation are an early feature of AD, we compared the results of our association analysis obtained from the full set of subjects (n=708) to those obtained from the subset of subjects who were cognitively non-impaired at the time of death (n=217) by assessing, for a given CpG, whether the beta estimate from the non-impaired subset was different from the beta estimate derived from the entire cohort. We used the fact that the non-impaired subjects are a subset of the entire cohort and tested, using a 1 sample test and the t distribution, whether the results from this subset, specifically the beta estimate taking into account the standard error, were different from those measures obtained from the entire population (i.e. the total cohort of 708 subjects) from which the subset was drawn.
To assess mediation by a third factor (as in the analyses presented in Figure 3), we compared models with and without that third factor. For example, to test if a mRNA expression mediates the association between a CpG and NP burden, we compared the beta estimate of the CpG from the model with only the CpG and our standard confounders to the beta estimate of the CpG from the model with CpG, our standard confounders and a term for the mRNA. If the beta estimate from the model with the mRNA term is >10% less that the model without the mRNA term we say that the mRNA mediates the effect between CpG and NP.
Bisulfite-sequencing data generation and analysis
For four individuals (two with AD and two non-impaired, each pair consists of a man and a woman), we used the same DNA sample profiled using the Illumina HumanMethylation Array to perform whole genome bisulfite sequencing. Genomic DNA was fragmented to 100-500 bp using a Covaris S2 sonicator. DNA fragments were end-repaired, A-tailed, and ligated with methylated paired-end adapters (purchased from ATDBio). Whole genome bisulfite sequencing (WGBS) was performed as previously described55. In short, Illumina genomic DNA adapters were added to the fragments, and the adapter-ligated fragments were size-selected prior to two rounds of 5-h bisulfite treatments using the Epitect Bisulfite kit (Quiagen). Libraries were then purified and run on the Illumina HiSeq2000 using a standard 36 base protocol55.
The WGBS libraries were aligned using BSMap 2.756 to the hg19/GRCh37 reference assembly. Subsequently, CpG methylation calls were made using custom software57, excluding duplicate, low quality reads as well as reads with more than 10% mismatches. Only CpGs covered by ≥5x reads were considered for further analysis.
Methylation profile from 4 samples (2 with AD; 2 non-AD) generated from Illumina450k as well as Bisulfite-sequencing, were compared by selecting a random 50,000 autosomal CpG sites that are present in both datasets for each individual. In each subject, the estimated level of methylation at each CpG measured by the two technologies was compared using a student t-test. The results of the comparison of the technical replicates (Illumina vs. WGBS) are as follows: subject # ROS20963578, r=0.975; subject # MAP5797875, r=0.969; subject # MAP50403446, r=0.971; and subject # ROS20214850, r=0.972. Across the 4 subjects, the mean r=0.972.
Chromatin State Map
In collaboration with the Broad Institute's Roadmap Epigenomic Mapping Center, chromatin immunoprecipitation using antibodies targeting six different chromatin marks (H3K36me3, H3K27me3, H3K4me1, H3K4me3, H3K9ac, and H3K9me3) was performed independently for two MAP subjects who were cognitively non-impaired at the time of death and had minimal evidence of AD, vascular and lewy body pathology on neuropathological examination. In both cases, samples were obtained from the dorsolateral prefrontal cortex that is also sampled in our DNA methylation scan. Chromatin was extracted from each of the cortical samples. Library construction and sequencing were performed as previously reported58. Chromatin state maps were then learned from the sequence data (available at http://www.roadmapepigenomics.org). The H3K36me3 and H3K27me3 data were from donor id 149 while the other data sets were from donor id 112. The data were dichotomized using the default settings of the “BinarizeBed” command of ChromHMM59 except and applying the ‘-center’ option to the already signal extended bed files. The models were trained by applying the “LearnModel” command with default settings from ChromHMM. We selected a model with 11 distinct chromatin states as the optimal model for our tissue sample.
To assess whether certain chromatin states are enriched for associations in our analysis, we compared the distribution of the 137 associated CpGs across the 11 chromatin states to that of all 415,848 probes tested using a chi-square test.
RNA data & analysis
A detailed description of the RNA data from the Mayo Clinic samples has been reported elsewhere44,45 Since the analysis here explored the relation of mRNA levels to an AD diagnosis, a multivariable linear regression model was implemented, adjusting. covariates to correct for technical or biological variables, including age at death, sex, PCR plate, RIN and (RIN-RINmean)2. For those analyses which corrected for the expression levels of genes that are specific for the main five cell types present in the central nervous system (CNS), the following probes were used as covariates: ENO2 for neurons (ILMN_1765796), GFAP for astrocytes (ILMN_1697176), CD68 for microglia (ILMN_2267914), OLIG2 for oligodendrocytes (ILMN_1727567) and CD34 for endothelial cells (ILMN_1732799). Some or all of these 5 expression levels were included to account for neuronal loss, gliosis and/or vascular tissue in the assessed brain regions, where indicated.
Pathway analysis
We constructed a protein-protein interaction (PPI) network using the online tool DAPPLE 60 in order to determine whether the genes identified in our DNA methylation study significantly interact with known AD associated proteins discovered in genetic studies. We compiled a list of genes associated with AD, including 25 late onset AD GWAS genes, 3 early onset AD associated genes, and TREM1 (Replogle et al., submitted). We produced a PPI network with a cutoff of 2 interacting binding degrees. DAPPLE creates direct and indirect networks of connected proteins using evidence of physical interaction from the InWeb database, which contains 169,810 high-confidence pair-wise interactions involving 12,793 proteins 61. To assess the statistical significance of PPI networks, DAPPLE applies a within-degree node-label permutation strategy to build random networks that mimic the structure of the original network and evaluates four network connectivity parameters on these random networks to generate empirical distributions for comparison to the original network. Our genetically defined AD network was significantly connected based on its Direct Edge Count (p=0.0072) and Seed Common Interactors Degrees Mean (p=0.037). Once the nine genes that emerge from the RNA functional validation analysis are added to the list of genes to be considered in the analysis, the PPI network is expanded and includes six of these genes. In this iteration of the analysis, both the Direct Edeg Count (=0.0072) and Common Interactors Degrees Mean (p=0.042) measures remain significant.
Supplementary Material
Acknowledgements
Support for this research was provided by the National Institutes of Health grants: R01 AG036042, R01AG036836, R01 AG17917,R01AG15819, R01 AG032990, R01 AG18023, RC2 AG036547, P30 AG10161, P50 AG016574, U01 ES017155, KL2 RR024151, K25 AG041906-01. We also thank the Siragusa Foundation for their support of NET, and the Robert and Clarice Smith and Abigail Van Buren Alzheimer's Disease Research Program for their support of NET, SGY, and FZ. This work was funded by NIH grant AG036039 to JM and an Equipment Grant from Alzheimer's Research UK. We thank the National Institute for Health (NIHR) Biomedical Research Unit in Dementia in the South London and Maudsley NHS Foundation Trust (SLaM), Brains for Dementia Research (Alzheimer Brain Bank UK) and the donors and families who made this research possible. We also would like to thank the participants of the ROS and MAP studies for their participation in these studies.
Footnotes
Competing financial interests
None of the authors have a financial interest that relates to the work described in this manuscript.
Online Method
- 1.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. doi:10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Science translational medicine 2. 2010:49ra67. doi: 10.1126/scitranslmed.3001262. doi:10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nature biotechnology. 2013;31:142–147. doi: 10.1038/nbt.2487. doi:10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends in neurosciences. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. doi:10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. doi:10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chouliaras L, et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol Aging. 2013 doi: 10.1016/j.neurobiolaging.2013.02.021. doi:10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakulski KM, et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. Journal of Alzheimer's disease : JAD. 2012;29:571–588. doi: 10.3233/JAD-2012-111223. doi:10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. doi:10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbarian S, Beeri MS, Haroutunian V. Epigenetic Determinants of Healthy and Diseased Brain Aging and Cognition. JAMA neurology. 2013:1–8. doi: 10.1001/jamaneurol.2013.1459. doi:10.1001/jamaneurol.2013.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez DG, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. doi:10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell JT, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. doi:10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome biology. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. doi:10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raj T, et al. Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am J Hum Genet. 2012;90:720–726. doi: 10.1016/j.ajhg.2012.02.022. doi:10.1016/j.ajhg.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett DA, et al. Selected findings from the Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer's disease : JAD. 2013;33(Suppl 1):S397–403. doi: 10.3233/JAD-2012-129007. doi:10.3233/JAD-2012-129007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature biotechnology. 2010;28:1106–1114. doi: 10.1038/nbt.1681. doi:10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negash S, Bennett DA, Wilson RS, Schneider JA, Arnold SE. Cognition and neuropathology in aging: multidimensional perspectives from the Rush Religious Orders Study and Rush Memory And Aging Project. Current Alzheimer research. 2011;8:336–340. doi: 10.2174/156720511795745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett DA, Wilson RS, Boyle PA, Buchman AS, Schneider JA. Relation of neuropathology to cognition in persons without cognitive impairment. Ann Neurol. 2012;72:599–609. doi: 10.1002/ana.23654. doi:10.1002/ana.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chibnik LB, et al. CR1 is associated with amyloid plaque burden and age2 related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. doi:10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 43:436–441. doi: 10.1038/ng.801. doi:ng.801 [pii] 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seshadri S, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. doi:10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. doi:10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. doi:10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 23.Shulman JM, et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA neurology. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. doi:10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunnon K, Smith R, hannon E, De Jager P, Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, MacDonald R, Condliffe D, Harries LW, Katsel P, Haroutunian V, Kaminsky Z, Joachim C, Powell J, Lovestone S, Bennett DA, Schalkwyk, Mill J. Cross-tissue methylomic profiling strongly implicates a role for cortex-specific deregulation of ANK1 in Alzheimer's disease neuropathology. Nature neuroscience. 2014 doi: 10.1038/nn.3782. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann Neurol. 2009;66:200–208. doi: 10.1002/ana.21706. doi:10.1002/ana.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperling RA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. doi:10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. doi:10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapuis J, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.1. doi:10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Alzheimer's Disease I, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. doi:10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, et al. DIP2 disco-interacting protein 2 homolog A (Drosophila) is a candidate receptor for follistatin-related protein/follistatin-like 1--analysis of their binding with TGF-beta superfamily proteins. The FEBS journal. 2010;277:4278–4289. doi: 10.1111/j.1742-4658.2010.07816.x. doi:10.1111/j.1742-4658.2010.07816.x. [DOI] [PubMed] [Google Scholar]
- 31.Cruchaga C, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. doi:10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roet KC, et al. A multilevel screening strategy defines a molecular fingerprint of proregenerative olfactory ensheathing cells and identifies SCARB2, a protein that improves regenerative sprouting of injured sensory spinal axons. J Neurosci. 2013;33:11116–11135. doi: 10.1523/JNEUROSCI.1002-13.2013. doi:10.1523/JNEUROSCI.1002-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. doi:10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siggs OM, et al. iRhom2 is required for the secretion of mouse TNFalpha. Blood. 2012;119:5769–5771. doi: 10.1182/blood-2012-03-417949. doi:10.1182/blood-2012-03-417949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIlwain DR, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. doi:10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maretzky T, et al. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc Natl Acad Sci U S A. 2013;110:11433–11438. doi: 10.1073/pnas.1302553110. doi:10.1073/pnas.1302553110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper-Knock J, et al. Gene expression profiling in human neurodegenerative disease. Nature reviews. Neurology. 2012;8:518–530. doi: 10.1038/nrneurol.2012.156. doi:10.1038/nrneurol.2012.156. [DOI] [PubMed] [Google Scholar]
- 38.Simpson JE, et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer's pathology and APOE genotype. Neurobiol Aging. 2011;32:1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013. doi:10.1016/j.neurobiolaging.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Linnertz C, et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One. 2009;4:e7480. doi: 10.1371/journal.pone.0007480. doi:10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renner NA, et al. Transient acidification and subsequent proinflammatory cytokine stimulation of astrocytes induce distinct activation phenotypes. Journal of cellular physiology. 2013;228:1284–1294. doi: 10.1002/jcp.24283. doi:10.1002/jcp.24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraft AW, et al. Attenuating astrocyte activation accelerates plaque pathogenesis in APP/PS1 mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:187–198. doi: 10.1096/fj.12-208660. doi:10.1096/fj.12-208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett DA, et al. Overview and findings from the rush Memory and Aging Project. Current Alzheimer research. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Current Alzheimer research. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen M, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–228. doi: 10.1212/WNL.0b013e3182605801. doi:10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou F, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. doi:10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. doi:10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 47.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic acids research. 2008;36:e11. doi: 10.1093/nar/gkm1075. doi:10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett DA, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6. doi:10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- 49.Chen YA, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics : official journal of the DNA Methylation Society. 2013;8:203–209. doi: 10.4161/epi.23470. doi:10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. doi:10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 51.Marchini J, Heaton C, Ripley BD. fastICA: FastICA algorithms perform ICA and projection pursuit. R package. 2007 [Google Scholar]
- 52.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. doi:10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics : official journal of the DNA Methylation Society. 2013;8:290–302. doi: 10.4161/epi.23924. doi:10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bock C. Analysing and interpreting DNA methylation data. Nature reviews. Genetics. 2012;13:705–719. doi: 10.1038/nrg3273. doi:10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 55.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. doi:10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. doi:10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. doi:10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. doi:10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. doi:10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. doi:10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature biotechnology. 2007;25:309–316. doi: 10.1038/nbt1295. doi:10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.