Abstract
The variation among sequences and structures in nature is both determined by physical laws and by evolutionary history. However, these two factors are traditionally investigated by disciplines with different emphasis and philosophy—molecular biophysics on one hand and evolutionary population genetics in another. Here, we review recent theoretical and computational approaches that address the critical need to integrate these two disciplines. We first articulate the elements of these integrated approaches. Then, we survey their contribution to our mechanistic understanding of molecular evolution, the polymorphisms in coding region, the distribution of fitness effects (DFE) of mutations, the observed folding stability of proteins in nature, and the distribution of protein folds in genomes.
Introduction
In this review, we highlight the recent results from the theoretical and computational models being developed at the interface of biophysics and evolutionary population genetics. These models integrate the tools from molecular biophysics that have been developed to determine and design properties of proteins, our emerging knowledge of the genotype-phenotype relationship (GPR), and established approaches population genetics. Because these models are built bottom-up, integrating insights from biophysics and cell biology, they provide a robust and mechanistic understanding of the origin of observed genetic and structural variation.
This field is still in its infancy. However, it already offers new insights into the molecular determinants of the rate of protein evolution, the genetic variation in coding regions, the distribution of fitness effects of mutations, and the observed thermodynamic and structural properties of proteins in nature.
Bottom-up and multiscale evolutionary models: the basic elements
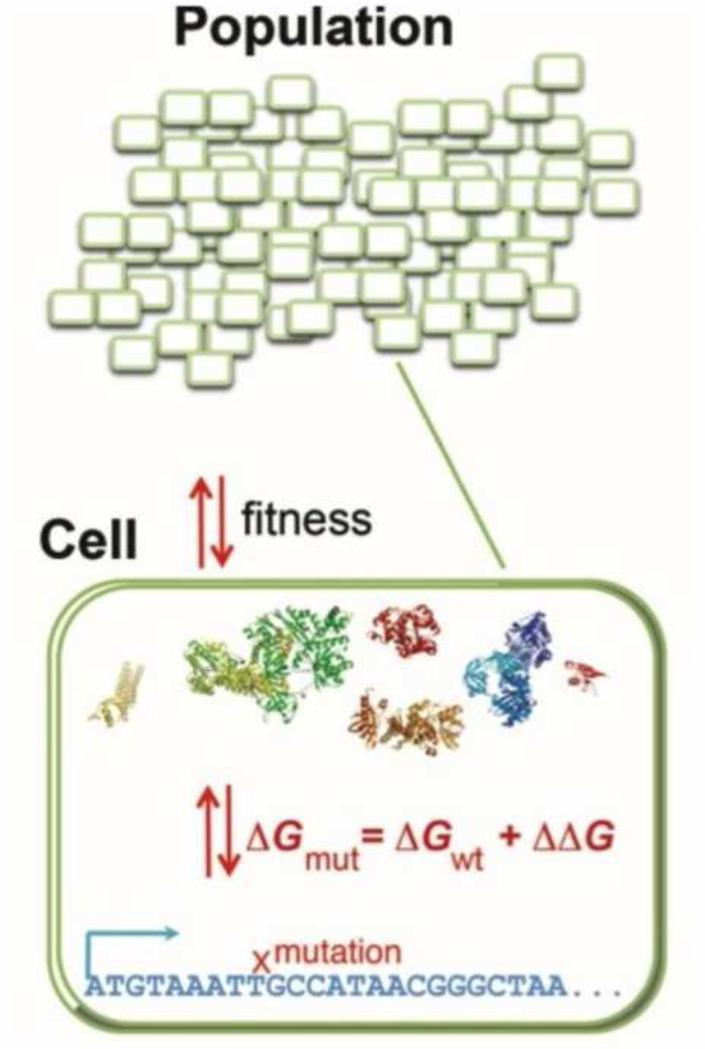
The underlying motivation for these multiscale models is to integrate our accumulated understanding of the mechanism in biological systems and evolutionary population dynamics. There are four elements to these models: (i) the genotype-phenotype relationship (GPR), (ii) the representation of the genomes and the protein products, (iii) the sources of genetic variation either by mutation or recombination, and (iv) the population dynamics and demographic model (Figure 1).
Figure 1. Schema of a bottom-up and multi-scale evolutionary models.
A model population consists of N organisms each with explicit genomes that encode proteins. The fitness of an organism is proportional to the folding stability of the proteins in the cytoplasm. The protein products are represented by their 3D structures from the protein databank (PDB)[16*]. When a random mutation occurs in the genome, tools in protein engineering and the 3D structure are used to estimate its effect on folding stability and, consequently, fitness. Alternatively, the proteins can be represented by 3D lattice models that allows for the exact calculation of biophysical properties [65] or for the possibility of a change in fold. The entire population is subject to mutation, drift, and purifying selection.
Traditional models in evolutionary biology assume the distribution of fitness effects (DFE) and then infer the possible dynamics [1,2], or assume the possible dynamics and then infer the DFE [3-5]. Both approaches have potential limitations because demography and the DFE are intrinsically coupled [6]. In contrast, in the bottom-up approach (Figure 1), the DFE is not an assumption but a consequence of the model. The integrated approach also builds on tools in protein folding and engineering, which have matured in the past decade, to estimate the effects of random mutations on proteins. Lastly, the bottom-up approach adds molecular realism to the traditional models in genetics (e.g., site-independence, 2-allele, etc.) by its explicit representation of the genes.
Contribution of biophysics to population and evolutionary genetics: The distribution of fitness effects of mutations in coding regions
In order to function, most proteins (with the obvious exception of intrinsically disordered domains) must maintain their native 3D structure. This requires folded proteins to be sufficiently stable against thermal fluctuations in the cellular environment. Protein folding stability, or the free energy difference between folded and unfolded states is a well-defined measurable sequence-dependent molecular property of proteins [7-9]. Folding stability determines the amount of folded (active) proteins according to the Boltzmann relation in statistical mechanics and it further modulates the protein abundance in cytoplasm by affecting turnover rates [10**]. The GPRs in these models are motivated by the selection for abundance of folded proteins[11,12], toxicity of misfolding proteins [13-15] and metabolic flux [16*]. In all these GPR, folding stability therefore is a key molecular parameter of fitness because it determines the total abundances of unfolded or folded proteins. The main quantity that defines the fate of arising mutations in population genetics is the selection coefficient s:
where bbefore and bafter are the finesses of an organism (often defined in terms of growth or division rates) before and after the mutation respectively [17]. The selection coefficient quantifies the effect of a mutation on the fitness of an organism. In GPR based on protein folding stability, and under the assumption that the protein folding thermodynamics is two-state [7,8,18], the selection coefficient upon a mutation can be approximately expressed as [11,12,15]
| (1) |
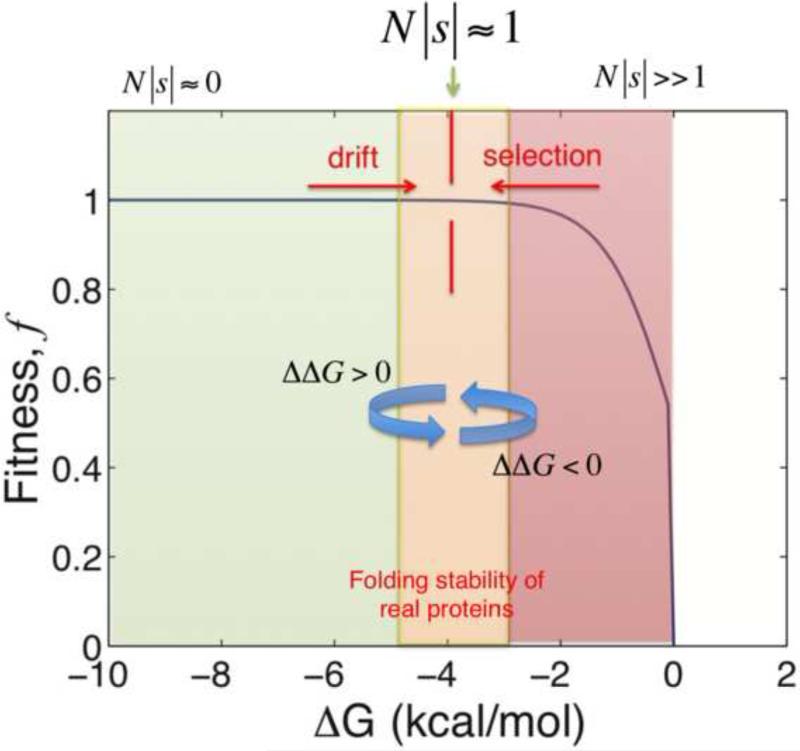
where ΔGwildtype is the folding stability of the protein prior to the mutation, and ΔΔG = ΔGmutant − ΔGwildtype is the change in protein folding stability due to the mutation. The factor β = 1/kBT (where kBT=0.593 kcal/mol at room temperature). This non-linear expression provides a mechanistic interpretation of epistasis in proteins (Fig. 2). The effect of a specific arising random mutation ΔΔG is modulated by the pre-mutation (“background” or wild type) folding stability ΔG.
Figure 2. Fitness effects of mutations on a protein folding thermodynamic landscape.
The integrated biophysics-population dynamics models typically assume that the fitness of the model organism is proportional to the total number of folded (functional) proteins in the cytoplasm. That is, fitness f ∝1/(1+eβΔG). Under this assumption, equation 2 defines how molecular changes (ΔΔG) map to fitness effect (s) [11,12,15]. In the regime of very stable proteins, the factor eβΔG → 0, thus N|s| ≈ 0 even if ΔΔG values are nonzero. Additionally, because arising mutations are predominantly destabilizing, most mutations that fix in this regime are destabilizing giving rise to a mutational drift of ΔG towards the less stable regime. Conversely, in the regime of unstable proteins, N|s| > > 1 and selection dominates. Hence, in the unstable regime, mutations that fix are predominantly stabilizing. Mutation-selection balance occurs at the folding stability value where N|s| ≈ 1. Altogether, the epistatic interactions mutations on the thermodynamic fitness landscape results in the near neutrality of the fitness effects of fixed substitutions even if their molecular effects (ΔΔG) are non-neutral.
To be more quantitative, we provide an example of the fitness effect values realized from actual simulations[16*]. In the particular simulation, the population size was N=103. A destabilizing mutation of ΔΔG =1 kcal/mol occurring in genes with ΔGpre-mutation = −8 kcal/mol has a fitness effect of 251658240 Ns ≈ −10−4 however, the same mutation occurring in genes with ΔGpre-mutation = −0.5 could be lethal. A stabilizing mutation of ΔΔG = −1 kcal/mol occurring in genes with ΔGpre-mutation = −8 kcal/mol has a fitness effect of 251658240 Ns ≈ +10−4; however, if it occurs in genes with ΔGpre-mutation = −0.5 kcal/mol, the mutation is extremely beneficial 251658240 Ns ≈ +102. Thus, in the regime where proteins are very stable, both destabilizing and stabilizing mutations have Ns < < 1; however, because of the larger supply of destabilizing than stabilizing mutations, most mutations that fix are destabilizing. This imbalance gives rise to a mutational drift of ΔG towards less stable proteins and away from the flatter part of the fitness landscape. In the regime where proteins are less stable, selection for stabilizing and against destabilizing mutations lead to the fixation of a larger fraction of stabilizing mutations. Mutation-selection balance occurs at the folding stability value where stabilizing and destabilizing mutations have equal likelihood of fixation[16*]. This balance indeed occurs in the regime of moderate protein folding stability and gives rise to the observation that proteins are “marginally” stable [19,20]. It is important to note the common misconception that selection for stability must result in very stable proteins and that the observed modest stabilities of proteins (in comparison for example with de novo designed ones [21]) therefore implies a “stability-activity tradeoff’’ [22] or provides the evidence against selection for stability altogether [23*]. As discussed here and in [9,11,19,20,24,25*,26*], selection for folding stability should not lead to most stable proteins. Rather, it is balanced by mutational drift towards destabilization resulting in a mutation selection balance that establishes observable distributions of protein stabilities.
We note that the distribution for the parameters on the right hand side of Eq. 1, ΔGwildtype and ΔΔG, has been well-established experimentally [27]. The distribution of effects of random mutations on folding stability has also been estimated to be universal across several classes of protein folds[28]. Stability-centric models successfully reproduced experimentally observed distributions of protein stabilities [9,24,26*,29] and distribution of fitness effects in viruses [11]. Thus, the distribution of fitness effect s (DFE) of arising random mutations is in principle the convolution of the well-established distributions of ΔGwildtype and ΔΔG.
While the DFE has been measured for viruses [30], its measurement in living organisms is difficult and resolution-limited [31]. Thus, studies on the DFE have largely relied on Bayesian maximum-likelihood approaches to fit population dynamic and demographic models to patterns of polymorphisms and amino acid differences between species [32-35]. A consensus result is that the DFE is characteristically skewed and can be described by a gamma distribution [32,34,35]. To arrive at a more mechanistic understanding of the DFE and polymorphisms, a recent work extended these biophysics-based evolutionary models to the polyclonal regime[16*]. The authors assumed that fitness is proportional to the total metabolic flux of a prototypical metabolic pathway and the total number of misfolded proteins. The PDB structures of proteins representing a prototypical glycolysis pathways were used in model cells in [16*]. They could keep track of all arising mutations, their history, and biophysical properties. More importantly, they could also mimic “population-wide deep sequencing” and compare with real SNPs [16*,36].
A major contribution from this work is its recapitulation “from first principles” of the DFE derived using the maximum Bayesian approaches. The DFE observed in simulations is skewed and can be well fitted to a gamma distribution, in agreement with empirical studies that estimated the DFE using maximum likelihood methods in human [32,34,35] and in flies[37] .
The near-neutrality of the resulting DFE from this mechanistic approach also shows that the near neutral theory of Ohta [38] should not be taken simply as a postulate, but rather as a robust consequence of the interplay between biophysics and evolutionary dynamics. Additionally, this mechanistic models shows that the patterns of polymorphisms, when framed in very direct observables such as changes in folding stability, supports the argument for a predominantly non-adaptive tempo of evolution at least for the coding region of the human genome (see Fig. 6 in ref. [16*]).
Contribution of population genetics to molecular biophysics: Environmental determinants of the evolution of protein folding stability
Under the assumption of mutation-selection balance, the selection coefficient of fixed mutations would be N|s|~1 (Fig. 2) [39]. Specifically, under the assumption that proteins are under selection to avoid the cytotoxic effects of misfolding, the selection coefficient is s ≈ AeβΔGpre-mutation (Eq. 2, ref. [40]) where A stands for the protein copy number in cytoplasm. This expression for s translates to (ref. [40])
| (Eq. 2) |
Equation 2 is significant because it quantifies the direct effect of Darwinian selection on folding stability through its dependence on the effective population size Ne [11,40]. In particular, weak selection in low population sizes is predicted to lead to the evolution of less stable proteins; conversely, stronger selection in large population sizes will lead to the evolution of more stable proteins. Equation 2 also quantitatively defines the contribution of protein cellular abundance (A) on folding stability. Highly abundant proteins are predicted to be more stable than proteins with low copy number in the cell. The third term in Equation 2 gives the distribution of changes on protein folding stability (ΔΔG) due to random mutations. This distribution is approximately a Gaussian with mean ΔΔGmean=1 kcal/mol and standard deviation ΔΔGsd = 1.7 kcal/mol [9]. Both parameters are estimated from empirical measurements of folding stability changes due to single point mutations (ProTherm database[27]). Altogether, Equation 2 quantifies the direct and nonnegligible contribution of non-biophysical parameters (population size and abundance) on the evolution of protein folding stability.
The magnitude of the contribution of different selection and cellular factors can be quantified in terms of resulting statistical variation of protein stabilities suggested by Eq.2. Protein cellular abundances span ~10 to ~106 copies per cell (as shown in yeast [41] and E. coli [42]), which is equivalent to variation of ~7 kcal/mol in protein stability (Eq[2]). Effective population sizes in nature range from 104 (vertebrates) to 109 (bacteria)[43], which could impose a ~6 kcal/mol spread in folding stability. Thus, the variation of protein folding stability in nature could be largely due to protein abundance and population size, however, this requires more proteomic measurements to prove.
Meanwhile, the range in abundance should systematically manifest itself in the structural properties of proteins across a genome. The observation that highly abundant, slow evolving proteins and proteins from thermophilic bacteria share similar amino acid composition [44] lends support to the dependence of stability on abundance. To demonstrate this prediction more unambiguously, it was shown that protein domains in yeast that are highly abundant in the cell show more favorable van der Waals interaction energy and more extensive hydrogen bond network [40].
As noted in [11,25*,45] population size may affect the cellular distribution of important signatures of folding stabilities such that organisms with small effective population sizes (e.g., endosymbiotic parasites that undergo episodic bottle-necking) will evolve less thermodynamically stable proteins, simply because deleterious mutations will fix at a higher probability in smaller population sizes. On the contrary, organisms with higher population sizes, which experience stronger purifying selection, are predicted to evolve more stable proteins. Additionally, assuming that all other things are equal, vertebrates (with effective population sizes of 104–105 [46] are predicted by Eq. 2 to evolve proteins that are on average 6 kcal/mol less stable than proteins in prokaryotes (whose population sizes are ~108 [46]). Interestingly, protein structures of viruses, which undergo episodic bottlenecking (and hence have a low effective population size), show weak van der Waals interaction and low hydrogen-bond contact densities [47]. Large variations in effective population sizes also occur even among closely related species. In bacteria, species that are endosymbiotic have lower effective population sizes compared to the free-living counterparts. Mendez and co-workers argued that the bias towards higher AT (adenine and thymine) content among obligate endosymbiotic bacteria could be the response against less effective purifying selection against protein misfolding [48]. These bioinformatic studies strongly support the coupling between biophysical properties and evolutionary population variables, but a systematic survey of biophysical properties of proteins (such as folding stability) in genomes should be an exciting subject of future experimental work.
A recent bioinformatics analysis highlighted the important role of selection for protein stability [49*]. In this work the adaptation in catecaens to changing environment was linked to molecular events in evolution of their Myoglobins (Mb) through ancestor sequence reconstruction on the branches of the Mb phylogenetic tree. Seven positively selected sites were identified which contribute to protein stability according to experimental measurements and computational predictions. Furthermore, the authors noted correlated evolution of stability and abundance of Mb lending empirical support to GPR assumptions of integrated evolutionary models. A recent study [50*] provided a similar phylogenetic analysis of evolution of stability and activity for another important protein RUBISCO - a classic model to study chaperonin-dependent folding [51].
Application of the integrated approaches to the evolution of protein folds
The explosion of genomics data also led to notable observations of the distribution of protein folds in nature. First, it is finite and small, numbering only less than 10,000 [52]. Second, some folds are highly represented while others are rare, giving rise to a distribution of usage of protein folds in a genome that is skewed and uneven [53]. This uneven distribution resembles a power law [54,55], an observation that is robust to the details associated with defining protein folds. The universality of the power law distribution suggests fundamental features of protein and genome evolution.
The models that explain the power law distribution can be broadly classified into two classes. First class of models posits that observed distributions of protein folds reflects certain biophysical properties of proteins such as, e.g. their designability [56-58], propensity to participate in protein-protein interactions [59*,60] or folding rates [61]. Another class of models posits that the observed distribution is simply a consequence of the duplication-divergence dynamics of emergence of new folds without biases due to individual properties of proteins [55,62]. Nevertheless, certain biases are introduced in phenomenological duplication and divergence models just to fit the empirical observations. Additionally, these models should be cognizant of the fact that selection acts at the level of populations of organisms and not individual genes or proteins. This detail is crucial because numerous observations in the genome architecture (beyond the distribution of protein folds), could also be explained by a largely nonadaptive mode of evolution [63,64]; in this view, population size is a crucial parameter.
Many of these works have been recently reviewed [65-67]. Thus, here we instead focus on the studies to reconcile these two classes of models. These models also need to be consistent with evolutionary dynamics arising from neutral drift and Darwinian selection. Indeed, a more robust understanding of fold evolution can only arise by providing molecular and mechanistic details to the phenomenological duplication and divergence schemes.
Zeldovich et al. explicitly modeled the emergence of new folds in a multiscale model with explicit representation of proteins and selection acting at the organism level[68]. Their model cells contained variable number of genes that encoded model lattice proteins. They assumed that the fitness of the organism is a function of the folding stability of the encoded proteins; in particular, the death rate of the organism is a function of the least stable protein in the cell. From simulations that started with random sequences, they observed that once favorable sequence–structure combinations are discovered, the population grows exponentially, and the initially diverse structural repertoire collapses into limited number of selected fold architectures. This repertoire remained stable and abundant at timescales greater than organismal lifetime. The emergence of protein families and superfamilies and ensuing power law distributions that match distributions for real proteins arise as a consequence of properties of the physical model, which suggests new folds from dominant folds by satisfying energetically favored native conformations. Cuypers et al. [69] also modeled genome evolution using a population of virtual cells evolving to maintain homeostasis. Although the work was not aimed at explaining fold distributions (no details on the protein folds is included), they nonetheless observed that an initial rapid expansion of the genome was followed by a prolonged phase of mutational load reduction. This load reduction was achieved by the deletion of redundant genes, generating a streamlining pattern. This integrated biophysics-population dynamics model of fold evolution could potentially explain the dependence of the power law exponent on genome size, [70,71]. We note that there is a well-known correlation between genome size and population size [43], and that the decreasing power-law exponent could be partly due to the population size variation. This conjecture remains to be proven explicitly.
Conclusion and Outlook
We have shown in this review that an approach that integrates molecular biophysics and evolutionary population genetics provides more mechanistic insights into the origin of protein fold and sequence variation in nature. These works have largely focused on protein folding stability for reasons that are both scientific (folding stability is the most universal of protein biophysical properties) and pragmatic (there are available biophysical tools to estimate the effects of random mutations on protein folding stability). In the near future, together with developments in protein folding and engineering and drug discovery, we will be able to include in the evolutionary models the effects of random mutations on enzymatic activity or protein-protein interactions using realistic protein structures. Additionally, because the approach is bottom-up, it can be coupled to the current efforts that build comprehensive cellular model of the genotype-phenotype relationship [72]. Lastly, in very well defined biological systems such as viral evolution and development of antibiotic resistance, this integrated molecular biophysics and population dynamics approach offers the possibility of predicting near term evolutionary trajectories.
An important direction of current and future research is to establish more realistic and robust GPR. The progress towards this goal requires synergistic experimental and theoretical efforts. “Top down’’ directed evolution approaches aim to evolve a particular phenotype first and subsequently determine a genomic variation that caused phenotypic changes. However the major challenge here is to establish a causal link between the evolved phenotype and genomic changes given ensuing massive genomic variation (the “passenger-driver problem’’ [73]). An alternative approach is “bottom up’’ where genetic variation is introduced either rationally by genomic editing [10**][74] or via targeted random or saturating mutagenesis [75**,76] with subsequent analysis of fitness changes. The latter can be evaluated either from competition assays or direct measurements of growth rates [10**,74] or through deep sequencing approaches [76-78]. The “bottom up’’ approach provides, in principle, a direct link between mutational changes in molecular properties of proteins and phenotypic change. In practice the relation can be quite complex due to many intervening factors such as protein homeostatsis in cellular milieu mitigating the molecular effects of mutations [10**,79-81]. With the advent of new CRISPR-based [82] and other new tools of genomic editing we will witness major progress in our understanding of GPR in many organisms which in turn will lead to the development of new generation of more accurate and predictive microscopic multiscale evolutionary models.
Highlights.
The natural variation of proteins is a consequence of both biophysics and population dynamics.
Recent theoretical and computational efforts integrate biophysics and evolutionary population biology.
These integrated approaches provide more mechanistic insights into fundamental questions in biophysics and evolution.
Acknowledgments
We acknowledge stimulating discussion with previous and current colleagues and collaborators from our lab Konstantin Zeldovich, Muyuong Heo, Pequi Chen, Boris Shakhnovich, Pouria Dasmeh, Scott Wylie, Amy Gilson and Shimon Bershtein. This work is supported by the NIH grant GM068670 and DARPA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-3:127–144. [PubMed] [Google Scholar]
- 2.Desai MM, Fisher DS. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd MJ, Walke S, Lorimer G, Truscott K, Scopes RK. The single-ring Thermoanaerobacter brockii chaperonin 60 (Tbr-EL7) dimerizes to Tbr-EL14.Tbr-ES7 under protein folding conditions. Biochemistry. 1995;34:14932–14941. doi: 10.1021/bi00045a038. [DOI] [PubMed] [Google Scholar]
- 4.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 5.Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 6.Silander OK, Tenaillon O, Chao L. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 2007;5:e94. doi: 10.1371/journal.pbio.0050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Privalov PL. Stability of proteins: small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 8.Shakhnovich EI, Finkelstein AV. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989;28:1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- 9.Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci U S A. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 2013;49:133–144. doi: 10.1016/j.molcel.2012.11.004. [In this work the authors use genome editing approach to introduce rational variation into E. coli gene encoding DHFR proteins and dissect relative contributions of molecular effect of mutaions and homeostatic response to fitness effect of mutations. The authors provide a model that accounts for observed effects.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein RA. The evolution and evolutionary consequences of marginal thermostability in proteins. Proteins. 2011;79:1396–1407. doi: 10.1002/prot.22964. [DOI] [PubMed] [Google Scholar]
- 13.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobkovsky AE, Wolf YI, Koonin EV. Universal distribution of protein evolution rates as a consequence of protein folding physics. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2983–2988. doi: 10.1073/pnas.0910445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serohijos AW, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cell Reports. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Serohijos AW, Shakhnovich EI. Contribution of selection for protein folding stability in shaping the patterns of polymorphisms in coding regions. Mol Biol Evol. 2014;31:165–176. doi: 10.1093/molbev/mst189. [This paper presents a detailed integrated biophysics-based model of evolution and adaptation of populations in polyclonal regime that takes into account realistic protein structures and biophysically realsitic estimate of mutational effects on protein stability.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartl DL, Clark AG. Principles of population genetics. 4th ed. Sinauer; Sunderland, MA: 2007. [Google Scholar]
- 18.Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- 19.Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 20.Bloom JD, Raval A, Wilke CO. Thermodynamics of neutral protein evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 22.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 23*.Hingorani K, Gierasch LM. Comparing protein folding in vitro and in vivo: foldability meets the fitness challenge. Current Opinion in Structural Biology. 2014;24:81–-90. doi: 10.1016/j.sbi.2013.11.007. [An insightful review of recent studies of the link between protein folding in cells and fitness.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P, Shakhnovich EI. Lethal mutagenesis in viruses and bacteria. Genetics. 2009;183:639–650. doi: 10.1534/genetics.109.106492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Goldstein RA. Population size dependence of fitness effect distribution and substitution rate probed by biophysical model of protein thermostability. Genome Biol Evol. 2013;5:1584–1593. doi: 10.1093/gbe/evt110. [A lucid analysis of population size effects in stability-centered evolutionary models.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Serohijos AW, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cell Rep. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [Analytical theory and simulation analysis show how biophysical factor of sequence depletion at high stabilities explains the observed correlation between protein abundance and evolutionary rates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar MD, Bava KA, Gromiha MM, Prabakaran P, Kitajima K, Uedaira H, Sarai A. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34:D204–206. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 29.Renzette N, Caffrey DR, Zeldovich KB, Liu P, Gallagher GR, Aiello D, Porter AJ, Kurt-Jones EA, Bolon DN, Poh YP, et al. Evolution of the Influenza A Virus Genome during Development of Oseltamivir Resistance In Vitro. J Virol. 2014;88:272–281. doi: 10.1128/JVI.01067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjuan R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 32.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawyer SA, Parsch J, Zhang Z, Hartl DL. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 35.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rospert S, Glick BS, Jeno P, Schatz G, Todd MJ, Lorimer GH, Viitanen PV. Identification and functional analysis of chaperonin 10, the groES homolog from yeast mitochondria. Proc Natl Acad Sci U S A. 1993;90:10967–10971. doi: 10.1073/pnas.90.23.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loewe L, Charlesworth B, Bartolome C, Noel V. Estimating selection on nonsynonymous mutations. Genetics. 2006;172:1079–1092. doi: 10.1534/genetics.105.047217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 39.Crow JF, Kimura M. An Introduction to Population Genetics Theory. Harper & Row; New York: 1970. [Google Scholar]
- 40.Serohijos AW, Lee SY, Shakhnovich EI. Highly abundant proteins favor more stable 3D structures in yeast. Biophys J. 2013;104:L1–3. doi: 10.1016/j.bpj.2012.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 42.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch M. The origins of genome architecture. Sinauer Associates; Sunderland, Mass.: 2007. [Google Scholar]
- 44.Cherry JL. Highly expressed and slowly evolving proteins share compositional properties with thermophilic proteins. Mol Biol Evol. 2010;27:735–741. doi: 10.1093/molbev/msp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wylie CS, Shakhnovich EI. Mutation induced extinction in finite populations: lethal mutagenesis and lethal isolation. PLoS Comput Biol. 2012;8:e1002609. doi: 10.1371/journal.pcbi.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 47.Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS. Do viral proteins possess unique biophysical features? Trends Biochem Sci. 2009;34:53–59. doi: 10.1016/j.tibs.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Mendez R, Fritsche M, Porto M, Bastolla U. Mutation bias favors protein folding stability in the evolution of small populations. PLoS Comput Biol. 2010;6:e1000767. doi: 10.1371/journal.pcbi.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Dasmeh P, Serohijos AW, Kepp KP, Shakhnovich EI. Positively selected sites in cetacean myoglobins contribute to protein stability. PLoS Comput Biol. 2013;9:e1002929. doi: 10.1371/journal.pcbi.1002929. [A phylogeny-based ancestral reconstruction analysis that relates evolution of protein stabilities and abundances to evolution of animals lifestyle.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Studer RA, Christin PA, Williams MA, Orengo CA. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1310811111. 101073/pnas.1310811111. [A phylogenetic-based anlysis of evolution of biophysical properties of an important protein that highlights the complementarity of selection for stability and function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 52.Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 53.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 54.Huynen MA, van Nimwegen E. The frequency distribution of gene family sizes in complete genomes. Mol Biol Evol. 1998;15:583–589. doi: 10.1093/oxfordjournals.molbev.a025959. [DOI] [PubMed] [Google Scholar]
- 55.Qian J, Luscombe NM, Gerstein M. Protein family and fold occurrence in genomes: power-law behaviour and evolutionary model. J Mol Biol. 2001;313:673–681. doi: 10.1006/jmbi.2001.5079. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Helling R, Tang C, Wingreen N. Emergence of preferred structures in a simple model of protein folding. Science. 1996;273:666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- 57.England JL, Shakhnovich EI. Structural determinant of protein designability. Phys Rev Lett. 2003;90:218101. doi: 10.1103/PhysRevLett.90.218101. [DOI] [PubMed] [Google Scholar]
- 58.Shakhnovich BE, Deeds E, Delisi C, Shakhnovich E. Protein structure and evolutionary history determine sequence space topology. Genome Res. 2005;15:385–392. doi: 10.1101/gr.3133605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Dixit PD, Maslov S. Evolutionary capacitance and control of protein stability in protein-protein interaction networks. PLoS Comput Biol. 2013;9:e1003023. doi: 10.1371/journal.pcbi.1003023. [This work provides a mechanism on how binding partners in the PPI network allows a protein to explore regions of the sequence space that correspond to less stable proteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heo M, Maslov S, Shakhnovich E. Topology of protein interaction network shapes protein abundances and strengths of their functional and nonspecific interactions. Proc Natl Acad Sci U S A. 2011;108:4258–4263. doi: 10.1073/pnas.1009392108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debes C, Wang M, Caetano-Anolles G, Grater F. Evolutionary optimization of protein folding. PLoS Comput Biol. 2013;9:e1002861. doi: 10.1371/journal.pcbi.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dokholyan NV, Shakhnovich B, Shakhnovich EI. Expanding protein universe and its origin from the biological Big Bang. Proc Natl Acad Sci U S A. 2002;99:14132–14136. doi: 10.1073/pnas.202497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peterson GJ, Presse S, Peterson KS, Dill KA. Simulated evolution of protein-protein interaction networks with realistic topology. PLoS One. 2012;7:e39052. doi: 10.1371/journal.pone.0039052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez A, Lynch M. Non-adaptive origins of interactome complexity. Nature. 2011;474:502–505. doi: 10.1038/nature09992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeldovich KB, Shakhnovich EI. Understanding protein evolution: from protein physics to Darwinian selection. Annu Rev Phys Chem. 2008;59:105–127. doi: 10.1146/annurev.physchem.58.032806.104449. [DOI] [PubMed] [Google Scholar]
- 66.Deeds EJ, Shakhnovich EI. A structure-centric view of protein evolution, design, and adaptation. In: Toone EJ, editor. Advances in Enzymology and Related Areas of Molecular Biology, Vol. 75: Protein Evolution. Wiley; 2007. pp. 133–191. [DOI] [PubMed] [Google Scholar]
- 67.Wilke CO. Bringing molecules back into molecular evolution. PLoS Comput Biol. 2012;8:e1002572. doi: 10.1371/journal.pcbi.1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeldovich KB, Chen P, Shakhnovich BE, Shakhnovich EI. A first-principles model of early evolution: emergence of gene families, species, and preferred protein folds. PLoS Comput Biol. 2007;3:e139. doi: 10.1371/journal.pcbi.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cuypers TD, Hogeweg P. Virtual genomes in flux: an interplay of neutrality and adaptability explains genome expansion and streamlining. Genome Biol Evol. 2012;4:212–229. doi: 10.1093/gbe/evr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 71.Maslov S, Krishna S, Pang TY, Sneppen K. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc Natl Acad Sci U S A. 2009;106:9743–9748. doi: 10.1073/pnas.0903206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Jr., Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bershtein S, Mu W, Shakhnovich EI. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc Natl Acad Sci U S A. 2012;109:4857–4862. doi: 10.1073/pnas.1118157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75**.Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DN. Latent effects of Hsp90 mutants revealed at reduced expression levels. PLoS Genet. 2013;9:e1003600. doi: 10.1371/journal.pgen.1003600. [A joint experimental and theoretical study that explores the dependence of fitness effect of mutations on proteins expression level supporting the generality of “elasticity curve”.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hietpas RT, Bank C, Jensen JD, Bolon DN. Shifting fitness landscapes in response to altered environments. Evolution. 2013;67:3512–3522. doi: 10.1111/evo.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adkar BV, Tripathi A, Sahoo A, Bajaj K, Goswami D, Chakrabarti P, Swarnkar MK, Gokhale RS, Varadarajan R. Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure. 2012;20:371–381. doi: 10.1016/j.str.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 79.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 80.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 81.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 82.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]