Abstract
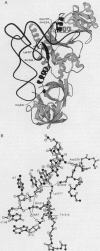
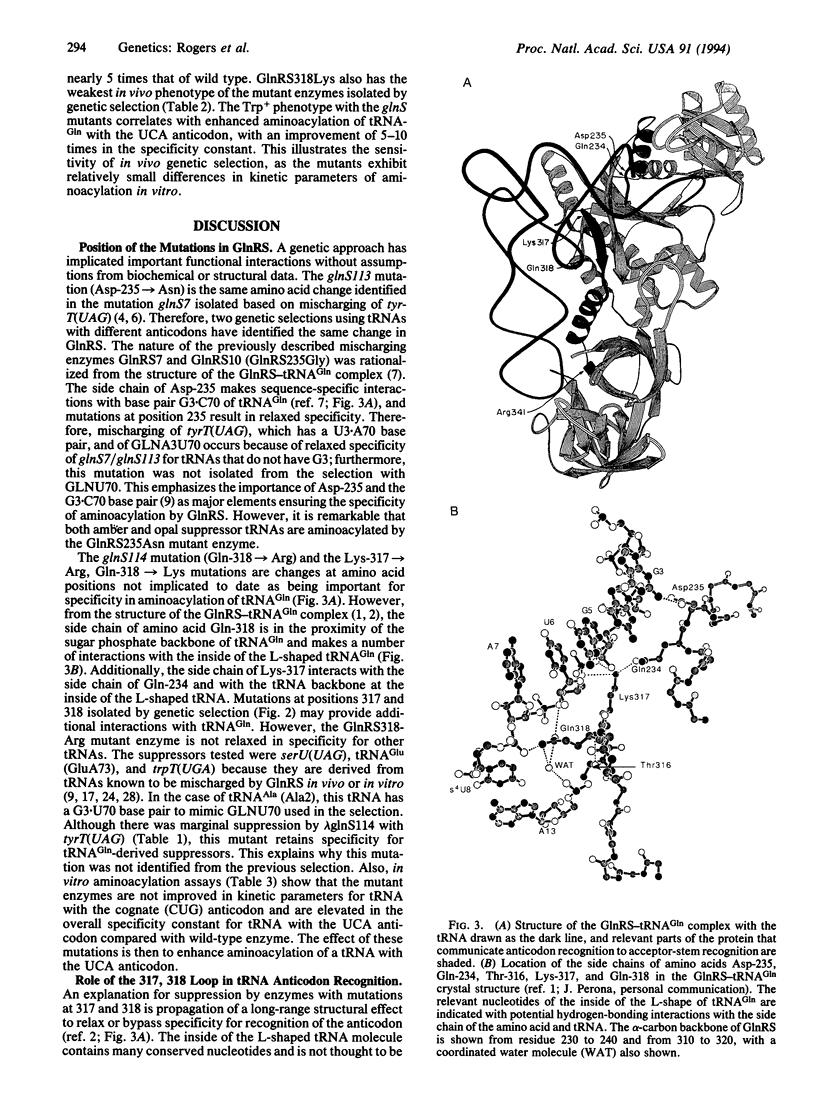
Wild-type Escherichia coli glutaminyl-tRNA synthetase (GlnRS; EC 6.1.1.18) poorly aminoacylates opal suppressors (GLN) derived from tRNA(Gln). Mutations in glnS (the gene encoding GlnRS) that compensate for impaired aminoacylation were isolated by genetic selection. Two glnS mutants were obtained by using opal suppressors differing in the nucleotides composing the base pair at 3.70: glnS113 with an Asp-235-->Asn change selected with GLNA3U70 (GLN carrying G3-->A and C70-->U changes), and glnS114 with a Gln-318-->Arg change selected with GLNU70 (GLN carrying a C70-->U change). The Asp-235-->Asn change was identified previously by genetic selection. Additional mutants were isolated by site-directed mutagenesis followed by genetic selection; the mutant enzymes have single amino acid changes (Lys-317-->Arg and Gln-318-->Lys). A number of mutants with no phenotype also were obtained randomly. In vitro aminoacylation of a tRNA(Gln) transcript by GlnRS enzymes with Lys-317-->Arg, Gln-318-->Lys, or Gln-318-->Arg changes shows that the enzyme's kinetic parameters are not greatly affected by the mutations. However, aminoacylation of a tRNA(Gln) transcript with an opal (UCA) anticodon shows that the specificity constants (kcat/Km) for the mutant enzymes were 5-10 times above that of the wild-type GlnRS. Interactions between Lys-317 and Gln-318 with the inside of the L-shaped tRNA and with the side chain of Gln-234 provide a connection between the acceptor end-binding and anticodon-binding domains of GlnRS. The GlnRS mutants isolated suggest that perturbation of the interactions with the inside of the tRNA L shape results in relaxed anticodon recognition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Brunie S., Schulman L. H. Transition state stabilization by a phylogenetically conserved tyrosine residue in methionyl-tRNA synthetase. J Biol Chem. 1991 Sep 15;266(26):17136–17141. [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971 Jun 14;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Hoben P., Söll D. Glutaminyl-tRNA synthetase of Escherichia coli. Methods Enzymol. 1985;113:55–59. doi: 10.1016/s0076-6879(85)13011-9. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Hoben P., Yamao F., Ozeki H., Söll D. Transfer RNA mischarging mediated by a mutant Escherichia coli glutaminyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5076–5080. doi: 10.1073/pnas.81.16.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi H., Kodaira M., Yamao F., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. II. Duplicate genes for tRNA2Gln. J Mol Biol. 1979 Aug 25;132(4):663–677. doi: 10.1016/0022-2836(79)90381-4. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Kondo K., Yoshimura M., Ozeki H. Mutant of the glutamine transfer RNA gene as UGA suppressor in Escherichia coli. Mol Gen Genet. 1990 Sep;223(3):433–437. doi: 10.1007/BF00264450. [DOI] [PubMed] [Google Scholar]
- Kleina L. G., Masson J. M., Normanly J., Abelson J., Miller J. H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol. 1990 Jun 20;213(4):705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon O., Schulman L. H. Covalent coupling of 4-thiouridine in the initiator methionine tRNA to specific lysine residues in Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1987 Nov 3;26(22):7113–7121. doi: 10.1021/bi00396a037. [DOI] [PubMed] [Google Scholar]
- Masson J. M., Miller J. H. Expression of synthetic suppressor tRNA genes under the control of a synthetic promoter. Gene. 1986;47(2-3):179–183. doi: 10.1016/0378-1119(86)90061-2. [DOI] [PubMed] [Google Scholar]
- Murgola E. J. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A., Risler J. L., Zelwer C., Brunie S. Structural similarities in glutaminyl- and methionyl-tRNA synthetases suggest a common overall orientation of tRNA binding. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2903–2907. doi: 10.1073/pnas.88.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Swanson R. N., Rould M. A., Steitz T. A., Söll D. Structural basis for misaminoacylation by mutant E. coli glutaminyl-tRNA synthetase enzymes. Science. 1989 Dec 1;246(4934):1152–1154. doi: 10.1126/science.2686030. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Swanson R., Steitz T. A., Söll D. Overproduction and purification of Escherichia coli tRNA(2Gln) and its use in crystallization of the glutaminyl-tRNA synthetase-tRNA(Gln) complex. J Mol Biol. 1988 Jul 5;202(1):121–126. doi: 10.1016/0022-2836(88)90524-4. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Chen L., Frankel A. D., Williamson J. R. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Schimmel P. R. Structural organization of complexes of transfer RNAs with aminoacyl transfer RNA synthetases. Nucleic Acids Res. 1977;4(5):1649–1665. doi: 10.1093/nar/4.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Adachi T., Inokuchi H., Söll D. Switching tRNA(Gln) identity from glutamine to tryptophan. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3463–3467. doi: 10.1073/pnas.89.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6627–6631. doi: 10.1073/pnas.85.18.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Inaccuracy and the recognition of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:185–208. doi: 10.1016/s0079-6603(08)60627-3. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman J. M., Rogers K., Rogers M. J., Söll D. Synthetase competition and tRNA context determine the in vivo identify of tRNA discriminator mutants. J Mol Biol. 1992 Dec 20;228(4):1055–1062. doi: 10.1016/0022-2836(92)90314-a. [DOI] [PubMed] [Google Scholar]
- Spanjaard R. A., Chen K., Walker J. R., van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg). Nucleic Acids Res. 1990 Sep 11;18(17):5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Uemura H., Rogers M. J., Swanson R., Watson L., Söll D. Site-directed mutagenesis to fine-tune enzyme specificity. Protein Eng. 1988 Oct;2(4):293–296. doi: 10.1093/protein/2.4.293. [DOI] [PubMed] [Google Scholar]
- Vidal-Cros A., Bedouelle H. Role of residue Glu152 in the discrimination between transfer RNAs by tyrosyl-tRNA synthetase from Bacillus stearothermophilus. J Mol Biol. 1992 Feb 5;223(3):801–810. doi: 10.1016/0022-2836(92)90991-r. [DOI] [PubMed] [Google Scholar]
- Weygand-Durasević I., Schwob E., Söll D. Acceptor end binding domain interactions ensure correct aminoacylation of transfer RNA. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):2010–2014. doi: 10.1073/pnas.90.5.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Inokuchi H., Cheung A., Ozeki H., Söll D. Escherichia coli glutaminyl-tRNA synthetase. I. Isolation and DNA sequence of the glnS gene. J Biol Chem. 1982 Oct 10;257(19):11639–11643. [PubMed] [Google Scholar]
- Yaniv M., Folk W. R., Berg P., Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J Mol Biol. 1974 Jun 25;86(2):245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]