Abstract
We developed a high-throughput methodology, termed fluorescent tagging of full-length proteins (FTFLP), to analyze expression patterns and subcellular localization of Arabidopsis gene products in planta. Determination of these parameters is a logical first step in functional characterization of the approximately one-third of all known Arabidopsis genes that encode novel proteins of unknown function. Our FTFLP-based approach offers two significant advantages: first, it produces internally-tagged full-length proteins that are likely to exhibit native intracellular localization, and second, it yields information about the tissue specificity of gene expression by the use of native promoters. To demonstrate how FTFLP may be used for characterization of the Arabidopsis proteome, we tagged a series of known proteins with diverse subcellular targeting patterns as well as several proteins with unknown function and unassigned subcellular localization.
Plants function as a complex network of interacting cell types, tissues, and organs. In turn, each cell represents an equally complex network of morphologically and functionally diverse subcellular structures. A comprehensive, systems-based understanding of plant physiology, morphogenesis, and development is impossible without thorough knowledge of protein expression and localization patterns within these supracellular and subcellular domains. Thus, there is a need for a sensitive high-throughput assay to simultaneously determine native subcellular localization and expression patterns of plant proteins in vivo (Girke et al., 2003).
To visualize localization and expression within individual cells and tissues, proteins are usually labeled with either a reporter or an antigenic tag, or are detected by specific antibodies. Enzyme reporters, such as β-glucuronidase (GUS; Jefferson, 1987; Kertbundit et al., 1998), cannot reflect subcellular protein localization at high resolution or in real time as they are detected indirectly via their chromogenic reaction products (Taylor, 1997). The use of antigenic tags, such as the T7 or HA epitopes, or of specific antibodies, requires labor-intensive and time-consuming techniques like immunocytochemistry and electron microscopy. By contrast, the green fluorescent protein (GFP) overcomes these limitations (Chalfie et al., 1994). Due to their autofluorescence and high quantum yields, GFP (Chalfie et al., 1994) and its spectral variants (Cubitt et al., 1995; Griesbeck et al., 2001) provide sensitive and convenient tools to track biological molecules in real time in yeast (Niedenthal et al., 1996; Huh et al., 2003), animal, and plant systems (reviewed in Hanson and Kohler, 2001).
In most cases, the use of GFP as a protein expression marker has not been fully optimized. For example, GFP is typically fused to the N or C terminus of a target protein (Rolls et al., 1999; Cutler et al., 2000; Huh et al., 2003). This approach may abrogate many targeting signals. Specifically, with N-terminal fusions, endoplasmic reticulum (ER) signal peptides may be masked, or they could become stop transfer sequences, thereby generating localization artifacts. N-terminal fusions will likely also obscure mitochondrial or chloroplast transit peptides. With C-terminal fusions, many proteins, such as CesA1, Pin1, and Sku5, may also mislocalize. The CesA family of proteins encodes the catalytic subunit of cellulose. While C-terminal fusion of GFP to CesA1 failed to localize to the plasma membrane (T. Richmond and C. Somerville, personal communication), an in-frame fusion of three amino acid residues from the N terminus of the closely related CesA7(IRX3) protein rescued the irx3-1 mutation and localized in the same pattern as wild-type protein detected by immunohistochemistry (Gardiner et al., 2003). N- and C- terminal fusions of GFP to the auxin efflux carrier PIN1 both failed to yield functional fusion proteins (K. Palme, personal communication), but an internal fusion is functional and shows the polar localization to the cell cortex (Benková et al., 2003). Sku5 encodes a glycosylphosphotidylinositol-anchored protein (Sedbrook et al., 2002). NeitherC- nor N-terminal fusions to GFP complemented a mutation, sku5-1, that affects root growth pattern (J. Sedbrook, personal communication). An internal fusion, with GFP placed between the predicted signal sequence and the remainder of the polypeptide, rescued the mutant phenotype and localized to the plasma membrane (Sedbrook et al., 2002). N-terminal or C-terminal fusions might also obscure posttranslational modification sites, such as myristylation or farnesylation sites for membrane targeting. In addition, C-terminal fusions could mask stem-loop structures in the 3′ part of the coding sequence and the 3′ untranslated region (UTR) that are essential for the correct localization of some mRNAs (Chartrand et al., 1999). Importantly, the interference with native localization information by N- or C-terminal fusions may not only cause a fusion protein to fail to reach its appropriate location, but can also result in artifactual localization.
Tagged proteins are often expressed from a strong constitutive rather than from the native promoter, producing the protein in cells or under conditions where it does not normally function. Furthermore, overexpression may disrupt multiprotein complexes or can mask subtle localization patterns when there is an overabundance of the tagged protein, e.g. during plasma membrane targeting.
To circumvent these problems, we designed a high-throughput strategy, termed Fluorescent Tagging of Full-Length Proteins (FTFLP). This procedure tags proteins at a selected internal site and incorporates the native gene regulatory sequences. The tagged proteins are then stably expressed in transgenic Arabidopsis plants to avoid ambiguities in interpretation of data from transient expression. Here, we describe how this experimental approach could be used for characterization of the Arabidopsis proteome.
RESULTS
The FTFLP Technique
Fluorescent tagging of proteins to study their expression and localization patterns should ideally conform to the following three criteria: (1) the fluorescent reporter should be stable over the range of physiological conditions found in different subcellular compartments, such as pH, (2) the insertion of the reporter should minimally disturb the conformation of the target protein and preserve native targeting signals and posttranslational modification sites, and (3) expression of the tagged protein should occur from its native regulatory sequences to faithfully detect its developmental and/or tissue-specific regulation. Our FTFLP approach fulfills these three requirements.
As a fluorescent tag, we used the citrine variant of yellow fluorescent protein (YFP) (Griesbeck et al., 2001), which can be used not only to visualize a single protein but also to study protein-protein interactions in vivo as an energy acceptor in bioluminescence resonance energy transfer (BRET; Xu et al., 1999) and fluorescence resonance energy transfer (FRET) assays (Tsien and Miyawaki, 1998; Pollok and Heim, 1999). Citrine YFP has enhanced photostability, and is much less sensitive to pH and anions, such as chloride, compared to other YFP variants (Griesbeck et al., 2001). The reduced sensitivity to pH allows detection of proteins targeted to the extracellular matrix or to other relatively acidic subcellular compartments, making it more suitable for tagging proteins with a wide range of targeting specificities. Some proteins were also tagged with cyan fluorescent protein (CFP; ECFP, CLONTECH, Palo Alto, CA) for comparison of localization patterns obtained with different tags and for future colocalization and protein interaction studies. Methods and results for CFP were identical to those for YFP.
We flanked the YFP open reading frame with flexible linker peptides (Doyle and Botstein, 1996), minimizing protein folding interference between YFP and the tagged Arabidopsis protein. To avoid placing identical nucleotide sequences on each side of the tag, we used two different linkers: the N terminus of YFP was linked to a Gly-rich peptide, (Gly)5Ala, and the C terminus is linked to an Ala-rich linker peptide, AlaGly(Ala)5GlyAla.
To further avoid effects of the YFP tag on native subcellular localization, the location of the tag relative to the target sequence was determined for each individual protein, based on computer-assisted predictions of protein folding and functional domains. We reasoned that it should be possible to devise a default tag location for most target proteins. The tag should ideally reside within a stretch of hydrophilic residues, outside of any specific protein domain, and near the C terminus. Thus, our default strategy was to insert the YFP tag 30 bp (10 amino acids) upstream of the stop codon. This location minimizes disturbance of the contiguous protein sequence, and protects the activity of membrane anchoring signals typically found within a few C-terminal amino acid residues (Casey, 1995; Zhang and Casey, 1996). Indeed, the addition of a four-amino acid (CaaX) farnesylation motif to the C terminus of GFP is sufficient to direct it to the plasma membrane (D. Jackson, unpublished data) and fusion of a tripeptide peroxisomal targeting signal to the C terminus of a reporter protein also allows specific localization (Lee et al., 1997). In cases where the default tag position overlapped with a predicted protein domain (Mulder et al., 2003), an alternative tag site near the C terminus was selected to meet our criteria of being within a hydrophilic region and not disrupting any predicted domain. Over 93% of the genes we tested had suitable primers within the last 10 amino acid residues from the C terminus.
To faithfully reproduce the expression level and pattern of each target gene, our constructs included the 5′ UTR and promoter sequences, the coding region with introns, and the 3′ UTR and downstream sequence. Most intergenic distances in the Arabidopsis genome are around 2 kb, suggesting that the promoter sequences are contained in a relatively small region (The Arabidopsis Genome Initiative, 2000). Thus, we included up to 3 kb upstream of the translation initiation codon and 1 kb downstream of the STOP codon in our constructs.
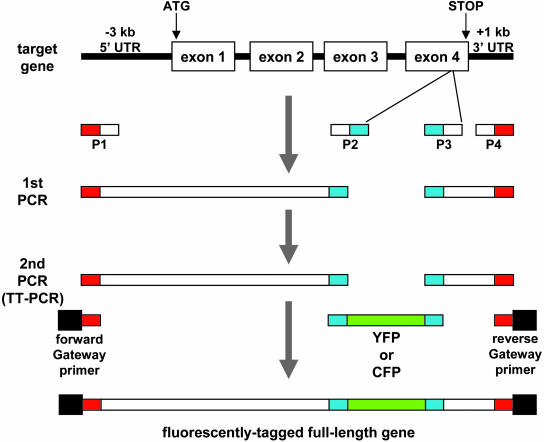
We designed an efficient protocol for generation and in planta expression of tagged genes (Fig. 1). We first amplified the selected gene in two fragments, one from the predicted start of the promoter to the YFP insertion site (i.e. between primers P1 and P2 in Fig. 1), and the second comprising the rest of the coding sequence and 3′sequences (i.e. between primers P3 and P4 in Fig. 1). In addition to gene-specific sequences, primers P1 and P4 were tailed with sequences complementary to the Gateway primers (see below), and P2 and P3 were tailed with sequences complementary to the YFP linkers. We also amplified the YFP coding sequence with flanking linkers (Fig. 1). In a second round of PCR, all three amplified fragments acted as overlapping templates for long flanking homology (LFH) PCR (Wach, 1996). This reaction, which we called triple-template PCR (TT-PCR), used two common primers tailed with the attB1 and attB2 Gateway sequences (Walhout et al., 2000) and complementary at their 3′-end to the P1 and P4 primers (Fig. 1). Thus, TT-PCR introduced the YFP tag into the selected site within the target gene without the need for conventional cloning, and created an internally-tagged full-length gene ready for Gateway recombination cloning.
Figure 1.
FTFLP flowchart. White boxes represent gene-specific sequences, red boxes represent P1 and P2 primer sequences overlapping the forward and reverse Gateway primers, respectively, and blue boxes represent P2 and P3 primer sequences overlapping the fluorescent tag linker sequences. For further details, see text.
For stable expression in transgenic plants, each YFP-tagged gene was cloned and transferred into an Agrobacterium binary vector. For high throughput, we used the Gateway system (Invitrogen, Carlsbad, CA), which is based on bacteriophage λ site-specific recombination (Landy, 1989). The Gateway cloning first produced a donor construct, containing the amplified TT-PCR product in the donor vector, pDONR207 (Invitrogen; Landy, 1989; see also www.invitrogen.com). The tagged gene was next recombined in vitro into the destination vector (Landy, 1989; see also www.invitrogen.com). A binary destination vector was constructed by subcloning the Gateway conversion cassette C.1 (Invitrogen) into the promoter-less T-DNA region of pBIN19, resulting in the pBIN-GW vector. To augment native expression of genes with relatively weak promoters, we also constructed a binary destination vector with tetramerized CaMV 35S enhancers in the T-DNA region of pMN20 (Weigel et al., 2000), resulting in pMN-GW. In our experiments, the efficiency of the first recombination reaction into the donor plasmid was 80% to 90%, whereas recombination into the binary destination vector was 100% efficient (data not shown).
Following recombination into the donor vector, we completely sequenced all tagged gene clones to estimate the rate of sequence errors introduced by PCR. Each amplified gene carried between zero and three amino acid substitutions, corresponding to average amino acid substitution rate of 0 to 0.3/1 kb of amplified sequence. Additional details and step-by-step experimental protocols for FTFLP can be found on our web site, http://aztec.stanford.edu/gfp/. As proof of concept for the FTFLP approach, we fluorescently tagged a number of Arabidopsis genes whose protein products have diverse, known subcellular locations, such as the peroxisome, vacuole, plasma membrane, ER, Golgi, nucleus, and cytosol. Transgenic plants expressing these genes were produced by Agrobacterium-mediated transformation and examined for expression of the tagged transgenes in various plant organs, such as roots, hypocotyls, and cotyledons. On average, 20 to 40 primary transgenic plants were screened per construct, using a fluorescence dissecting microscope, followed by detailed analysis by confocal laser scanning microscopy. In most cases, 80% to 90% of plants exhibited fluorescent protein expression. In the following sections, we describe selected examples of patterns of subcellular localization and expression of known proteins (reference genes) as well as several proteins with unknown function and unassigned subcellular localization.
Peroxisomal Targeting
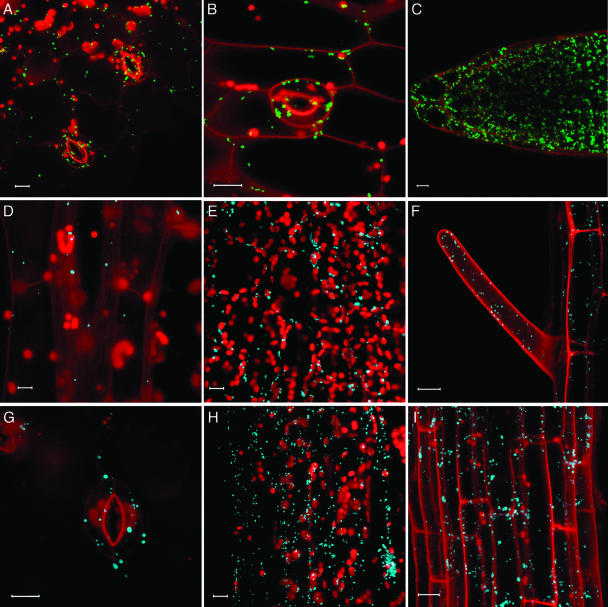
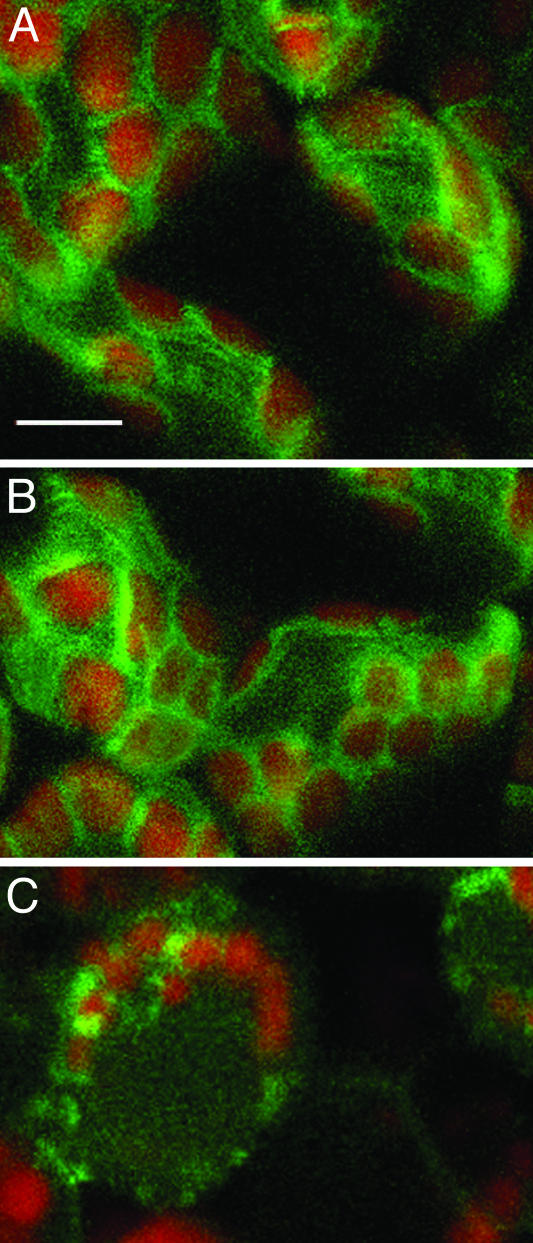
Localization to peroxisomes was exemplified by two Arabidopsis proteins, the multifunctional protein 2 (MFP2) and peroxisomal targeting signal type 1 receptor (Peroxin-5, Pex5; Cutler et al., 2000; Mullen et al., 2001). The corresponding genes were tagged with YFP and expressed from their native promoters with and without the 35S enhancers. Expression was examined by confocal laser scanning microscopy in hypocotyls, leaves, and roots. In all these organs, MFP2 localized to discrete subcellular structures that represent peroxisomes (Fig. 2, A–C). While the 35S enhancers clearly increased the levels of MFP2 expression, they did not alter its subcellular localization (data not shown). Similar expression patterns were obtained with YFP-tagged Pex5 (data not shown).
Figure 2.
Peroxisomal targeting. A–C, YFP-MFP2 in cotyledon epidermis, hypocotyl epidermis, ansd root tip tissue. D–F, CFP-MFP2 in hypocotyl, leaf mesophyll, and root epidermis. G–I, CFP-Pex5 in cotyledon epidermis, leaf mesophyll, and root axis tissue. YFP signal is indicated in green, CFP signal is indicated in blue, and plastid autofluorescence is in red. To visualize cell outlines, roots were stained with propidium iodide indicated in red. All images are projections of several confocal sections. Bars represent 10 μm.
To demonstrate that the fidelity of subcellular targeting was not limited to protein tagging with YFP, we also tagged MFP2 and Pex5 with CFP. Figure 2 illustrates that the CFP-tagged MFP2 (sections d–F) and Pex5 (sections G–I) exhibited peroxisomal targeting identical to that of YFP-tagged MFP2 and Pex5, in different plant tissues, such as cotyledons, hypocotyls, leaves, and roots. The CFP-tagged genes also showed higher levels of expression but the same subcellular targeting specificities in the presence of the 35S enhancers (data not shown).
Tonoplast Membrane Targeting
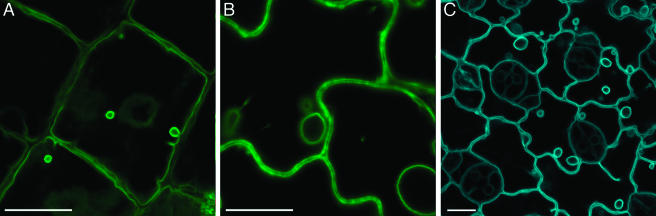
Tonoplast membrane targeting was examined using delta-tonoplast intrinsic protein (TIP), an Arabidopsis tonoplast protein (Cutler et al., 2000) and a homolog of aquaporins found in membranes of storage vacuoles in pea and barley (Jauh et al., 1998; Jauh et al., 1999). Figure 3 shows that YFP-tagged delta-TIP was expressed in hypocotyls (section A), leaves (section B), and roots (not shown), and localized exclusively to the tonoplast membrane. The use of a CFP tag or of the 35S enhancers did not affect the subcellular localization of delta-TIP, but the enhancers slightly elevated its expression level (Fig. 3C). In addition, YFP and CFP signal was observed in bulb-like structures (Fig. 3) previously reported to associate with the tonoplast membrane and to contain another tonoplast protein, gamma-TIP (Saito et al., 2002).
Figure 3.
Tonoplast targeting. A and B, YFP-delta-TIP in hypocotyl and leaf epidermal cells. C, CFP-delta-TIP in leaf epidermal cells. YFP signal is indicated in green, and CFP signal is indicated in blue; plastid autofluorescence was filtered out. All images are projections of several confocal sections. Bars represent 10 μm.
Plasma Membrane Targeting
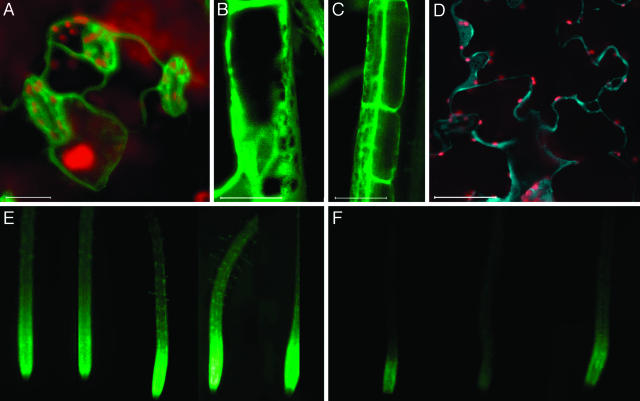
Plasma membrane intrinsic protein 2A (PIP2A) of Arabidopsis, a channel protein (Cutler et al., 2000), was used to visualize protein targeting to the plasma membrane. YFP-tagged PIP2 expressed from its native promoter in the absence (not shown) or in the presence of 35S enhancers localized exclusively at the extreme cell periphery in all plant tissues, including hypocotyls (Fig. 4A), cotyledons (data not shown), and roots (Fig. 4B). This localization pattern could be interpreted as either plasma membrane or cytosolic because the cytoplasm is typically displaced to the cell periphery by the large central vacuole. However, the cytosolic compartment is characterized by transvacuolar strands, variations in cytosol thickness at the cell cortex, and rapid remodeling over time due to cytosolic streaming (Cutler et al., 2000). The absence of these patterns, together with the uniform and sharp pattern of peripheral labeling by YFP-PIP2A, indicates that the localization was in fact at the plasma membrane (also compare with cytosolic patterns in Fig. 7). We also distinguished plasma membrane targeting from cell wall localization by plasmolysis experiments, where the YFP-PIP2A fluorescent signal associated with the displaced membrane in plasmolysed cells (Fig. 4C).
Figure 4.
Targeting to plasma membrane, cell wall, and plasmodesmata. A and B, YFP-PIP2 in hypocotyl and root epidermal cells. C, YFP-PIP2 in plasmolysed hypocotyl cells. D, CFP-PRP2 in hypocotyl epidermal cells. E, YFP-TVCV MP in root epidermal cells. Arrows indicate sites of punctate fluorescence. YFP signal is indicated in green, and CFP signal is indicated in blue. All images are single confocal sections. Bars represent 20 μm.
Figure 7.
Cytosolic localization and light-regulated expression. A, YFP-GSR1 in leaf cells. B and C, YFP-GSR1 in root epidermal cells. D, CFP-GSR1 in leaf cells. YFP signal is indicated in green, CFP signal is indicated in blue, and plastid autofluorescence is in red. All images are projections of several confocal sections. Bars represent 10 μm. E and F, Epifluorescence images of YFP-GSR1-expressing roots grown in the light and in the dark, respectively.
Cell Wall Targeting
Proline-rich protein 2A (PRP2) resides in the cell wall (Fowler et al., 1999). This specific subcellular localization was faithfully reproduced using the PRP2 gene tagged with CFP and expressed with 35S enhancers in hypocotyls (Fig. 4D), cotyledons, and roots (data not shown). The cell wall localization of the fluorescent signal remained after plasmolysis (data not shown), and was also observed using the YFP-tagged PRP2 expressed with and without 35S enhancers (data not shown).
Targeting to Plasmodesmata
Plasmodesmata play essential roles in cell-to-cell transport and communication (Citovsky and Zambryski, 2000; Jackson, 2000; Tzfira et al., 2000; Barton, 2001). However, little is known about the proteins comprising these channels, which connect cells and regulate traffic of molecules as large as viral genomes. Because native plasmodesmal proteins have not been definitively identified, we used a plant viral protein, the cell-to-cell movement protein of turnip vein clearing virus (TVCV MP), as a plasmodesmal marker. Viral cell-to-cell movement proteins (MPs) target to plasmodesmata, and represent a classical tool in studies of plasmodesmal transport (reviewed in Citovsky, 1993; Lazarowitz and Beachy, 1999; Tzfira et al., 2000). Thus, we tagged TVCV MP with YFP and expressed it from the full 35S promoter. Figure 4E shows the characteristic punctate pattern of the tagged protein expressed in root tissues and accumulated at the cell periphery, presumably in the cell wall. Previously, such puncta have been shown to represent plasmodesmal accumulation of MP (Heinlein et al., 1995; Oparka et al., 1997; Boyko et al., 2000; Crawford and Zambryski, 2001; Kotlizky et al., 2001; Roberts et al., 2001).
Targeting to Cytoskeletal Elements
Arabidopsis fimbrin 1 (Fim1; Kovar et al., 2000) cross-links filamentous actin, producing a fine reticulate network of filaments (Kovar et al., 2001).We observed evidence of fine filaments of YFP-tagged Fim1 mainly around chloroplasts in leaf mesophyll cells (Fig. 5, A and B). The fluorescence signal was stronger in plants with the 35S enhancers constructs, but the localization pattern was unchanged (data not shown). Treatment with Latrunculin A, a drug that disrupts actin filaments (Spector et al., 1983), resulted in disruption of the filaments (Fig. 5C), supporting the conclusion that the YFP-Fim1 was associated with actin. The pattern of Fim1 localization in green tissues was remarkably similar to that observed with another filamentous actin binding protein, CHUP1, which is thought to participate in chloroplast positioning (Oikawa et al., 2003).
Figure 5.
Targeting to cytoskeletal elements. A and B, YFP-Fim1 in leaf mesophyll. C, YFP-Fim1 in leaf mesophyll treated with 0.2 μm Latrunculin A. Sections A and B are projections of several confocal sections; section C is a single confocal section. Bar represents 5 μm.
Targeting to the Nuclear Membrane and Proplastids
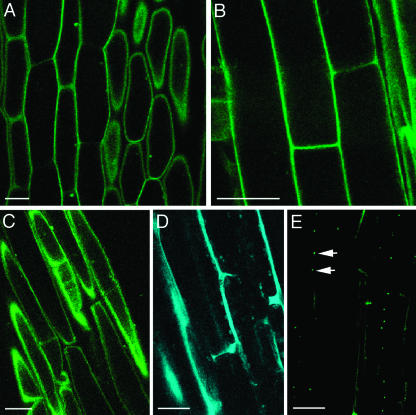
Arabidopsis MFP1 associated factor (MAF1) preferentially localizes to the nuclear envelope and to proplastids (Gindullis et al., 1999; Jeong et al., 2003). YFP-tagged MAF1 exactly reproduced this subcellular localization pattern in roots (Fig. 6A) and leaves (data not shown), accumulating in a distinct perinuclear ring as well as associating with proplastid-like structures. Again, expression from the construct with 35S enhancers did not alter the subcellular targeting of the protein (data not shown).
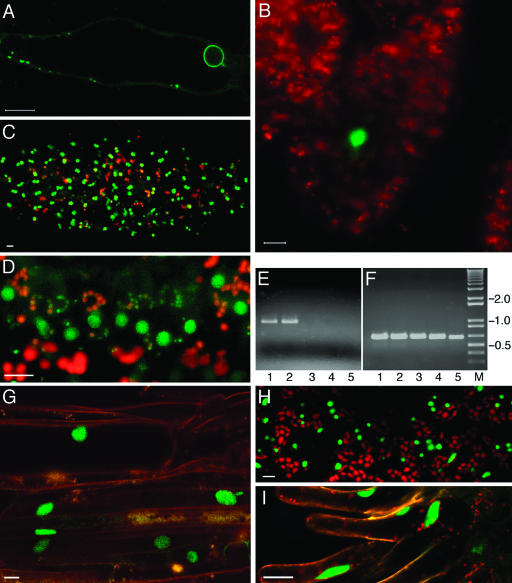
Figure 6.
Proplastid, nuclear membrane, and intranuclear targeting. A, YFP-MAF1 in a root hair cell. B, YFP-VIP2 in megasporocytes. C and D, YFP-VIP2 in tapetal cells. E, Quantitative RT-PCR analysis of VIP2 expression in different plant organs. F, Detection of actin RNA-specific RT-PCR product in the same samples shown in section E. Lane 1, mature flowers; lane 2, flower buds; lane 3, leaves; lane 4, stem; lane 5, roots; lane M, molecular size markers. Numbers on right indicate DNA sizes in kilodaltons. G–I, YFP-VIP1 in root epidermis, leaf epidermis, and stigmatic papillae, respectively. Propidium iodide counter-stain of root cells is in red, YFP signal is indicated in green, and plastid autofluorescence is in red. All images are single confocal sections. Bars represent 10 μm.
Nuclear Targeting and Cell-Specific Expression
VirE2-interacting protein 2 (VIP2, GenBank accession no. AF295433) of Arabidopsis is homologous to yeast Not2p and Drosophila Rga proteins, which are thought to mediate intranuclear interactions between chromatin proteins and the transcriptional complex (Frolov et al., 1998). Indeed, GUS fusions to VIP2 accumulate in the nucleus of tobacco and Arabidopsis cells (V. Citovsky, unpublished data). Here, our analysis of transgenic Arabidopsis expressing YFP-tagged VIP2 confirmed the nuclear targeting of this protein (Fig. 6, B and C). Virtually no signal was detected in the cytoplasm, indicating efficient nuclear import. Indeed, because the predicted size of the citrine-VIP2 fusion protein (approximately 85 kD) is substantially larger than the size exclusion limit of the nuclear pore (approximately 60 kD; reviewed in Dingwall and Laskey, 1991; Garcia-Bustos et al., 1991), its accumulation within the nucleus must result from the active process of nuclear import.
The YFP-tagged VIP2 gene had an intriguing cell-specific pattern of expression. Figure 6 shows that the tagged protein was detected only in megasporocytes (section B) and tapetum cells (sections C and D), but it was not detected in seedling roots, leaves, or hypocotyls (data not shown). This specificity of VIP2 expression was confirmed by reverse transcription (RT)-PCR, which detected VIP2 transcripts in the flower buds (lane 1) and mature flowers (lane 2), but not in roots, leaves, or stems (lanes 3–5, respectively) of wild-type Arabidopsis (Fig. 6E). In control experiments, analysis of actin-specific transcripts generated similar amounts of PCR products in all samples, indicating equal efficiencies of the RT-PCR reactions (Fig. 6F). Thus, transgenic expression of the YFP-tagged VIP2 revealed novel information about the native cell-specific expression pattern of this gene as well as subcellular localization of its protein product.
Another example of an Arabidopsis nuclear protein is the VirE2-interacting protein 1 (VIP1), a basic Leu zipper protein thought to function in transcriptional complexes and, during Agrobacterium infection, facilitate nuclear import of VirE2 (Tzfira et al., 2001, 2002). YFP-tagged VIP1 exhibited exclusively nuclear localization in roots (Fig. 6G), leaves (Fig. 6H), and stigmatic papillae of the transgenic plants (Fig. 6I). These nuclear patterns of VIP2 and VIP1 subcellular localization remained unchanged when the proteins were expressed from the 35S enhancer-containing construct (data not shown).
Cytosolic Localization
Recent data suggest that 47% of the yeast proteins are cytosolic (Kumar et al., 2002). Similarly, many plant proteins may be cytoplasmic. To illustrate the patterns expected from cytoplasmic localization, we used GSR1, the cytosolic Gln synthetase of Arabidopsis (Peterman and Goodman, 1991). Figure 7 shows the characteristic cytoplasmic pattern with transvacuolar strands that YFP- or CFP-tagged GSR1 forms in leaf (sections A and D) and root cells (section B). In this case, the use of the 35S enhancers construct did not significantly alter fluorescence levels, and it had no effect on the cytosolic YFP accumulation in roots (Fig. 7C) or leaves (data not shown).
To examine whether this tagged gene showed its known transcriptional regulation, we germinated the seedlings in light or in the dark. The YFP-tagged GSR1 exhibited the expected light-induced expression (Peterman and Goodman, 1991; Oliveira and Coruzzi, 1999) in the roots of transgenic plants (Fig. 7, E and F).
Subcellular Localization of Proteins of Unknown Function
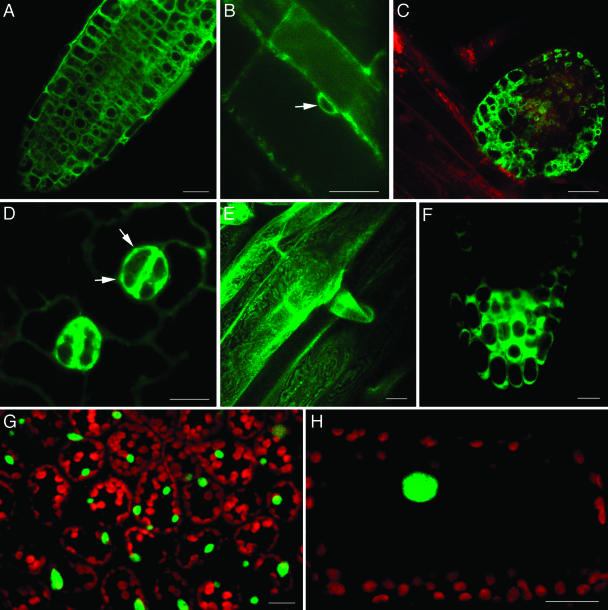
More than one-third of Arabidopsis genes have no predicted function (Wortman et al., 2003). We propose that our FTFLP approach combined with the standardized image interpretation can be used to characterize Arabidopsis proteins whose function is unknown and whose intracellular localization cannot be predicted based on their amino acid sequence. The following genes of unknown function were randomly selected for these experiments and tagged with YFP: At2g16530, At1g27090, At2g15240, At1g12170, and At1g80940. The tagged proteins were expressed in transgenic plants from their native promoters, and the subcellular localization patterns were compared to those of our reference genes.
Figure 8 shows that At2g16530, augmented with 35S enhancers, was expressed predominantly in root tissues (sections A–C), especially in lateral root primordia (section C). The tagged protein exhibited perinuclear localization (Fig. 8B, arrow), remarkably similar to that of MAF1 (see Fig. 6A). Some of the tagged protein also accumulated at the cell periphery in the cytoplasm (Fig. 8, A–C). Native expression of A1g27090 was observed mainly in guard cells, in the cytoplasm, and in unidentified subcellular structures (Fig. 8D, arrows). At2g15240 was expressed in the root and accumulated in the cytoplasm (Fig. 8E), but was not detected in other organs (data not shown). At1g12170 was expressed specifically in the root tip (Fig. 8F, compare to expression of At2g16530 throughout the root in Fig. 8A) and was cytoplasmic (Fig. 8F). Finally, At1g80940 was exclusively nuclear in the leaf (Fig. 8G) and in petioles (Fig. 8H).
Figure 8.
Subcellular localization and expression patterns of selected unknown genes. A–C, YFP-At2g16530 in root tissue. Arrow indicates perinuclear staining. Enhanced expression in lateral root primordia is evident in section C. D, YFP-At2g27090 in guard cells. Arrows indicate labeling of an unidentified subcellular compartment. E, YFP-At2g15240 in root. F, YFP-At1g12170 in root tip. G and H, YFP-At1g80940 in leaf and petiole cells, respectively. Sections E and G are projections of several confocal sections, the other images are single confocal sections. Bars represent 20 μm.
DISCUSSION
The complete sequence of the Arabidopsis genome has laid the foundation for functional characterization of its proteome. Elucidation of protein function often begins with homology-based predictions. However, in the absence of significant homology, determination of expression pattern and subcellular localization provide important clues to the potential function(s) of a gene. Thus, it is important to design a simple, reliable, and efficient procedure to directly assay native expression and subcellular targeting specificities of proteins in planta. To this end, we developed the FTFLP protocol, which is distinguished by three major features: (1) the fluorescent tag is inserted into the protein internally, minimizing interference with targeting signals at the N and C termini, (2) the tagged gene includes native 5′, intron, and 3′ regulatory sequences, allowing detection of the native promoter activity, and (3) the tagging procedure is simple, involving only two sequential PCR reactions followed by efficient recombination-based cloning into Agrobacterium binary vectors. These aspects make FTFLP the ideal methodology for high-throughput characterization of unknown Arabidopsis gene products.
As proof of concept, we utilized FTFLP to tag Arabidopsis gene products with known subcellular targeting specificities. Each of the selected genes was tagged with the citrine variant of YFP and the cyan variant of GFP (CFP) using TT-PCR. We could efficiently amplify gene fusions up to 8 kb, corresponding to a median length of nontranscribed region of 1.5 kb, a transcribed region of 4.0 kb, and 5′ and 3′ regulatory sequences of 3.0 kb and 1.0 kb, respectively. Our analysis of the Arabidopsis genome revealed that approximately 40% of all its genes fall within these values, indicating that the FTFLP approach is suitable for tagging of a substantial proportion of the Arabidopsis proteome. Although the PCR resulted in some amplification errors, and an amino acid substitution rate of 0 to 0.3/kb, these changes had no effect on the known subcellular localization patterns of the reference proteins we tested. Indeed, most subcellular targeting sequences are short and partially redundant, so single amino acid changes would not be expected to change protein targeting.
To stably express the tagged proteins, we made two Gateway destination binary vectors. For native expression, pBIN-GW has a Gateway cassette in its T-DNA region and no additional regulatory sequences. The second vector, pMN-GW, carries a tetramerized CaMV 35S enhancer that functions to elevate gene expression without altering tissue-specific expression patterns (this work and Weigel et al., 2000). Our analyses of transgenic Arabidopsis plants expressing various tagged genes confirmed that the 35S enhancers generated higher fluorescent signals, but did not alter tissue or cell type distribution or change the specificity of subcellular localization. Without exception, all proteins tested in this study had the expected subcellular targeting patterns, and protein targeting to diverse subcellular addresses was easily distinguished. Thus, FTFLP represents a simple and efficient approach to fluorescently tag full-length Arabidopsis genes and determine their expression patterns and subcellular localization.
As any other technique, the current version of our FTFLP protocol has several limitations. First, notwithstanding our careful analysis of the predicted structure of each tagged protein for optimal tag placement, the internal YFP or CFP tag may still interfere with native targeting signals of some proteins. Furthermore, even if several tagged proteins retain their native localization patterns, e.g. peroxisomal or nuclear localization, other proteins with similar targeting specificities may be perturbed by the tag or by enhanced levels of expression from 35 enhancer-containing vectors. These considerations are especially important when tagging proteins with unknown function (see below), for which so little is known about the location of their functional domains, even using the Interpro database (Mulder et al., 2003). It is important to emphasize, however, that these possibilities are purely hypothetical because not one of the known subcellular targeting patterns of the proteins that we tested to date has been altered by the addition of the tag or by enhanced expression. Nevertheless, in the future, we plan to examine protein families (e.g. the Institute for Genomic Research's [TIGR] protein family groupings) and consider any conserved regions as potential protein domains to be preserved during tagging. Also, in specific cases where the localization of a YFP- or CFP-tagged protein looks particularly interesting or unusual, it would be prudent to confirm it by alternative and independent approaches. For example, unique restriction sites flanking the fluorescent tag may be used for its easy replacement by a smaller tag, e.g. epitopes, which is less likely to interfere with protein folding (Jarvik et al., 1996). Second, the FTFLP approach determines native expression and subcellular targeting patterns, but does not address the other functions of the protein, and thus is not an alternative to other functional characterization efforts (e.g. isolation of knockout mutants). The FTFLP data, however, represent an important resource for such efforts. For example, our subcellular and tissue specific localization data will help other researchers focus their phenotypic analyses, or may help them prioritize subsets of unknown genes for knockouts. We predict two major applications for the FTFLP approach. The first is to tag specific genes of interest. In this scenario, a single gene (or only a few genes) is tagged and its expression characterized in detail. Presently, characterization of newly-discovered genes routinely begins with tagging of the cDNA with either GUS or GFP to determine subcellular targeting, and placing the same reporters under the control of the predicted promoter to determine expression pattern. The FTFLP approach achieves both of these goals simultaneously, using fewer experimental steps and producing more reliable and consistent data based on a single full-length construct. Furthermore, any YFP-tagged full-length gene can be easily coexpressed with our CFP-tagged reference genes for colocalization studies or for interaction assays such as FRET (Tsien and Miyawaki, 1998; Pollok and Heim, 1999).
A second application for FTFLP is a high-throughput characterization of the Arabidopsis proteome. From close to 30,000 genes in the Arabidopsis genome, almost 30% could not be assigned to a functional category (Wortman et al., 2003). Since by definition these proteins have little similarity to proteins with known functions, information about their function can only be gained experimentally. Because protein function is related to localization, a useful way to shed light on the function of an unknown protein is to determine its subcellular localization and pattern of expression. As demonstrated here, the FTFLP approach is well-suited for such large-scale analysis of expression and subcellular targeting of unknown proteins.
MATERIALS AND METHODS
Conversion of Agrobacterium Binary Plasmids into Gateway Destination Vectors
The pBIN19 plasmid was digested with EcoRI and HindIII, and the ends were filled-in with Klenow. Into these blunted ends, we ligated a blunt-ended Gateway conversion cassette (reading frame C.1; Invitrogen catalog no. 11797016). The resulting Gateway destination vector, designated pBIN-GW, has the following structure in its T-DNA region: T-DNA right border-NOS terminator<-NPTII<-35S promoter-attR1->CAT->ccdB->attR2-T-DNA left border. This vector has no regulatory sequences for expression of cloned genes and, thus, is useful for producing native levels and patterns of gene expression.
Using a similar strategy, the pMN20 activation tagging plasmid (Weigel et al., 2000) was converted to a Gateway vector, following digestion with HindIII. The resulting plasmid, pMN-GW, has the following structure of its T-DNA region: T-DNA right border-(35S enhancer)4-attR1->CAT->ccdB->attR2-35S promoter-NPTII-NOS terminator-T-DNA left border. This vector has tetramerized CaMV 35S enhancers in its T-DNA region (Weigel et al., 2000) and, thus, is useful for producing elevated levels of gene expression while retaining native expression patterns. Note that pMN20-based vectors should be prepared from fresh bacterial stocks and used immediately after transferring them to Agrobacterium because they tend to lose some copies of their 35S enhancers due to recombination in Escherichia coli or Agrobacterium when stored at 4°C (Weigel et al., 2000).
Both pBIN-GW and pMN-GW vectors were propagated in the DB3.1 strain of E. coli (Invitrogen) strain carrying the gyrA462 gene which confers resistance to the toxicity of the ccdB gene (its protein product, a natural analog of the quinolone antibiotics, binds to the DNA gyrase subunit A and turns it into a poison; Bahassi et al., 1999). Following Gateway recombination, ccdB is replaced by the fluorescently-tagged gene, allowing better selection for the recombinant clones in bacterial strains that do not carry gyrA462 or F' episome (which also confers resistance to ccdB).
Preparation of Fluorescent Tags
YFP and CFP tags were amplified from the pRSETB-Citrine (Griesbeck et al., 2001) and pECFP-C1 (CLONTECH) plasmids as templates using the ExTaq DNA polymerase (TaKaRa, Bio, Otsu, Japan) and the following pair of primers for both genes: forward primer 5′-AAGGCCGGCCTGGAGGTGGAGGTGGAGCTGTGAGCA-3′, and reverse primer 5′-TTGGCCCCAGCGGCCGCAGCAGCACCAGCAGGATCCTTGTACAGCTCGTCCA-3′. These primers encode peptide linkers, (Gly)5Ala and AlaGly(Ala)5GlyAla, followed by recognition sites for rare-cutting endonucleases FseI and SfiI, respectively. These restriction sites, although not required for subsequent gene tagging, may be useful in future application for easy replacement of the YFP/CFP by other protein tags. The amplified fragments were subcloned into a pTOPO TA vector (Invitrogen). Using these constructs as templates, DNA fragments encoding YFP or CFP tags with linker sequences were amplified (forward primer: 5′-GGCCGGCCTGGAGGTGGAGGTGGAGCTGTGAGCA-3′, reverse primer: 5′-GGCCCCAGCGGCCGCAGCAGCACCAGCAGGATC-3′, annealing temperature 70°C) using the Pfu-Turbo DNA polymerase (Invitrogen) and gel-purified using the GFX PCR purification kit (Amersham Biosciences, Piscataway, NJ) for their subsequent use in TT-PCR.
Fluorescent Tagging of Arabidopsis Genes
For each gene, two sets of primers, P1/P2 and P3/P4 (see Fig. 1), were designed by a Perl script that takes into account the annealing temperature, the position of each primer within the genomic and protein sequence, and primer length and secondary structure in an iterative fashion until a suitable pair is detected; it uses part of the Primer3 program (available at http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). For all unknown genes, TIGR's genome release version 4.0 was used. We obtained Interpro (Mulder et al., 2003) functional domain mapping data from TIGR and secondary structure mapping from Structural Classification of Proteins (SCOP; http://scop.mrc-lmb.cam.ac.uk/scop/; J. Gough, unpublished data). The sequences of the primers are shown in Table I. Besides 18 to 25 nucleotides-long gene-specific sequences, P1 and P4 primers contained sequences 5′-GCTCGATCCACCTAGGCT-3′ and 5′-CGTAGCGAGACCACAGGA-3′, respectively, that partially overlap the Gateway primers (see below), whereas P2 and P3 primers contained sequences 5′-CACAGCTCCACCTCCACCTCCAGGCCGGCC-3′ and 5′-TGCTGGTGCTGCTGCGGCCGCTGGGGCC-3′, respectively, that partially overlap the fluorescent tag linkers (see above). For the first PCR reaction (see Fig. 1), genomic DNA was extracted from total leaf material of 6-week-old Arabidopsis ecotype Columbia plants using the DNeasy Plant Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and used as template. The gene fragments were amplified in 20 μL of the mixture containing 100 ng template DNA, 1× ExTaq reaction buffer (TaKaRa Bio), 0.2 mm dNTP, 0.2 μm of each primer, and 0.025 units/μL ExTaq. The following PCR conditions were used: 1 cycle at 95°C for 3 min; 7 touch-down cycles at 93°C for 30 s, 64°C for 30 s (this temperature is reduced by 1°C per touch-down cycle), and 68°C for 1 min/1 kb of the longest amplified sequence; 23 cycles at 93°C for 30 s, 58°C for 30 s, and 68°C for 1 min/1 kb of the longest amplified sequence; and 1 cycle at 68°C for 1 min/1 kb of the longest amplified sequence. The amplified fragments were gel-purified using a GFX PCR gel purification kit (Amersham Biosciences), and the ExTaq-generated A-overhangs were removed by incubation for 30 min at 72°C in 50 μL containing 25 μL amplified DNA, 1xPfu reaction buffer, 0.2 mm dNTP, and 0.01 units/μL Pfu polymerase (Invitrogen).
Table I.
Tagged Arabidopsis genes with known targeting specificities and sequences of DNA primers used for their amplification
| Gene Name | Position on Chromosome
|
Primer Sequence (5′ => 3′)
|
|||||
|---|---|---|---|---|---|---|---|
| AGI Code | Start | End | P1 | P2 | P3 | P4 | |
| MFP2 | At3g06860 | 2161875 | 2166315 | GCTCGATCCACCTAGGCTCATTGTTATGAACGGTTGGTCTTTGG | CACAGCTCCACCTCCACCTCCAGGCCGGCCGCTCTGTAATACAAATGGAAG | TGCTGGTGCTGCTGCGGCCGCTGGGGCCGCTCCCGTGAAACAAGCC | CGTAGCGAGACCACAGGAGTTGGTCGTCGACTATCC |
| Pex5 | At5g56290 | 22803755 | 22809345 | GCTCGATCCACCTAGGCTTTCGGATCCTAGATCGGGTCTT | CACAGCTCCACCTCCACCTCCAGGCCGGCCGAGATTCCTTGACTCACAAGCTTCTAT | TGCTGGTGCTGCTGCGGCCGCTGGGGCCGATCTCTTGCAGAAAGAATTCCCGCTGTG | CGTAGCGAGACCACAGGATCCGTGAGACCCCCTTTTT |
| delta-TIP | At3g16240 | 5503301 | 5507094 | GCTCGATCCACCTAGGCTCAAGCATCTTCACAGGTTTTGG | CACAGCTCCACCTCCACCTCCAGGCCGGCCGAAATCAGCAGAAGCAAGAGGA | TGCTGGTGCTGCTGCGGCCGCTGGGGCCGTTCCTCTTGCTTCTGCTGATTTC | CGTAGCGAGACCACAGGACCATCCATTAATTTGTCATTGTTGT |
| PIP2A | At3g53420 | 19819151 (-) strand | 19814307 | GCTCGATCCACCTAGGCTTTGGTCGGATCTACGATGTTCTC | TCCACCTCCACCTCCAGGCCGGCCCGTTCTGAGAGCTTCAGGTTCTAAGTCT | TGGTGCTGCTGCGGCCGCTGGGGCCCTTGGATCATTCAGAAGTGCTG | CGTAGCGAGACCACAGGACTCGAACTTGGCTGAGGATT |
| PRP2 | At2g21140 | 9072115 | 9067216 | GCTCGATCCACCTAGGCTCACCGTCTCCGAGTCTCTTC | TCCACCTCCACCTCCAGGCCGGCCTTGTCTGCCCTCCTCTCACT | TGGTGCTGCTGCGGCCGCTGGGGCCTGATCTAAAAACATCCCCTTGAT | CGTAGCGAGACCACAGGAAATGCACATAGATTCTACAGTTCCATAG |
| Fim1 | At4g26700 | 13463674 | 13467605 | GCTCGATCCACCTAGGCTCGCCAAATTAAACAGCAACAA | CACAGCTCCACCTCCACCTCCAGGCCGGCCGATTTCAGAAACCGCATCACCAAC | TGCTGGTGCTGCTGCGGCCGCTGGGGCCACCACGGTCTCAGAGGAAGC | CGTAGCGAGACCACAGGAAAGTCATTTTTGCGCCAGATTA |
| MAF1 | At5g43070 | 17309870 | 17305500 | GCTCGATCCACCTAGGCTAACCAAATCGGTTAACATTTTCAAT | CACAGCTCCACCTCCACCTCCAGGCCGGCCATCAACAGCTGATTCAATTCCATC | TGCTGGTGCTGCTGCGGCCGCTGGGGCCTCGAAGATTGATTCAAGTGAAGCT | CGTAGCGAGACCACAGGATTCTAGAGTTTAAGAAAGCACTCCCA |
| VIP2 | At5g59710 | 24072992 | 24079768 | GCTCGATCCACCTAGGCTGCAATCGTCGGTTCGTCACT | CACAGCTCCACCTCCACCTCCAGGCCGGCCGTACTTGATAACAAAATGTTCCTGCGATA | TGCTGGTGCTGCTGCGGCCGCTGGGGCCGAGCTTATGGAAAAGAGACCAAGC | CGTAGCGAGACCACAGGAGGATGAAGAGAGAATGTTCTTCGAG |
| VIP1 | At1g43700 | 16485051 | 16488971 | GCTCGATCCACCTAGGCTGCTTGAGCGTTTATTCTGGC | CACAGCTCCACCTCCACCTCCAGGCCGGCCCTGCCCGTTTGTACTCATCTG | TGCTGGTGCTGCTGCGGCCGCTGGGGCCCCATCGCTCCCAAGCTACATG | CGTAGCGAGACCACAGGAGCACCATTTCTCATTTGTTGGTAT |
| GSR1 | At5g37600 | 14955558 | 14949978 | GCTCGATCCACCTAGGCTTGAGCATGGTTGTAGTCATTTCG | CACAGCTCCACCTCCACCTCCAGGCCGGCCAGCAATCATGGAAGTGACAATGT | TGCTGGTGCTGCTGCGGCCGCTGGGGCCGAGACTACAATCCTCTGGAATCCT | CGTAGCGAGACCACAGGAGTTACCGGAGAGTGGATTAGCG |
For the second PCR reaction (TT-PCR, see Fig. 1), a pair of standard, gene-nonspecific Gateway primers were designed. The forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTCGATCCACCTAGGCT-3′, contained the attB1 sequence, and the reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGTAGCGAGACCACAGGA-3′, contained the attB2 sequence. These primers were combined with three templates, i.e. the fluorescent tag and two gene fragments, in 20 μL of a mixture containing 100-ng long fragment (P1-P2), 50-ng short fragment (P3-P4), 50-ng YFP or CFP fragment, 1× ExTaq reaction buffer, 0.2 mm dNTP, 0.2 μm of each Gateway primer, and 0.02 units/μL ExTaq. TT-PCR was performed under the following conditions: 1 cycle at 95°C for 3 min; 20 cycles at 93°C for 30 s, 54°C for 30 s, and 68°C for 1 min/1 kb of the longest amplified sequence; and 1 cycle at 68°C for 1 min/1 kb of the longest amplified sequence.
The TT-PCR product was gel-purified as described above and recombined into the Gateway donor vector pDONR207 (Invitrogen) in 10 μL BP Clonase (Invitrogen) reaction containing 300-ng TT-PCR fragment, 150-ng pDONR207, 2 μL BP Clonase, and 2 μL BP Clonase buffer. Following overnight incubation at 25°C, 1 μL Proteinase K (2 μg/μL) was added, and the incubation continued for 10 min at 37°C, after which 1 to 2 μL of the reaction mixture was transformed into the DH5α or DH10B strain of E. coli, and the recombinants were selected on a gentamycin-containing medium. Clones with the correct inserts were identified by colony PCR using a pair of vector-specific attL primers, i.e. forward attL1 primer: 5′-TCGCGTTAACGCTAGCATGGATCTC-3′, and reverse attL2 primer: 5′-GTAACATCAGAGATTTTGAGACAC-3′.
Finally, the tagged genes were transferred from their donor constructs to both of the binary destination vectors pBIN-GW and pMN-GW in 10 μL of a single LR Clonase reaction containing 150 to 200 ng donor construct, 200 ng pBIN-GW and pMN-GW (1:1 w/w mixture), 5 units topoisomerase (Invitrogen), 2 μL LR Clonase (Invitrogen), and 2 μL LR Clonase buffer. After overnight incubation at 25°C, 1 μL of Proteinase K (2 μg/μL) was added, and the incubation continued for 10 min at 37°C, following which 2 μL of the reaction mixture was transformed into the DH5α or DH10B strain of E. coli, and the pBIN-GW and pMN-GW recombinants were selected on a kanamycin- or spectinomycin-containing medium, respectively; note that different antibiotic resistance markers of pBIN-GW and pMN-GW allow simultaneous recombination of the tagged genes into both of these binary vectors in the same reaction mixture. The correct clones were identified by colony PCR using P1 and P4 primers. All gene amplification reactions were performed using ExTaq (TaKaRa Bio) DNA polymerase (which, under our conditions, exhibited the highest fidelity while still capable of efficiently amplifying 8–11 kb-long DNA fragments), and the amplification products were verified by DNA sequencing.
Agrobacterium-Mediated Transformation of Arabidopsis
The binary constructs were introduced into the A. tumefaciens strain GV3101 and the resulting bacterial cultures were used to transform Arabidopsis ecotype Columbia by the standard flower dip method (Clough and Bent, 1998) or its modified version (Kim et al., 2003). Seed stocks for all transgenic plants expressing the tagged proteins described in this study are available from the Arabidopsis Biological Resource Center (ABRC).
Specimen Preparation and Imaging
Live seedlings and plant tissue samples were mounted in water between number 1 1/2 coverglasses, using silicon vacuum grease to create spacers between the glass surfaces. Images were collected with one of three laser scanning confocal microscope systems, a Leica TCS SP2/UV (Wetlzer, Germany), a Zeiss LSM 5 Pascal (Jena, Germany), or a Bio-Rad MRC 1024 (Hercules, CA). In all cases, a high numerical aperture (1.2–1.3) water immersion objective (60–63×) was employed. A 488-nm or a 514-nm laser line from an argon ion was used to excited YFP and a 442-nm line from a HeCd laser or a 457-nm line from an argon ion laser was used to excite CFP.
Quantitative RT-PCR
Total RNA was extracted from 2.0 g of roots, leaves, stems, flower buds, and mature flowers of wild-type Arabidopsis using TRI Reagent (Molecular Research Center, Cincinnati), and reverse-transcribed with M-MuLV reverse transcriptase using the dT23VN primer (BioLabs). The resulting first strand cDNAs were PCR-amplified as described (Kang et al., 1995; Ni et al., 1998), using a mixture of forward and reverse primers derived from VIP2 (AGI code AT5G59710) and Arabidopsis ACT2/ACT8 actin (AGI code AT1G49240, An et al., 1996). RT-PCR products were then detected by ethidium bromide staining of agarose gels. VIP2 forward and reverse primers, 5′-AAACATGGTTGGTGGAGGTAACTT-3′ and 5′-GGAGACGCAAATGTCTTGTAGAGA-3′, respectively, generated a 948-bp product while actin forward and reverse primers, 5′-ACCTTGCTGGTCGTGACCTT-3′ and 5′-GATCCCGTCATGGAAACGAT-3′, respectively, generated a 632-bp product.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AF295433.
Acknowledgments
We thank Dr. Tsien for his generous gift of the pRSETB-Citrine plasmid. This is Carnegie publication number 1673.
This work was supported by a grant from the 2010 Project of the National Science Foundation to V.C., D.E., D.J., N.R., and S.R.
References
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M (1999) Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem 274: 10936–10944 [DOI] [PubMed] [Google Scholar]
- Barton MK (2001) Giving meaning to movement. Cell 107: 129–132 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Casey PJ (1995) Protein lipidation in cell signaling. Science 268: 221–225 [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263: 802–805 [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 25: 333–336 [DOI] [PubMed] [Google Scholar]
- Citovsky V (1993) Probing plasmodesmal transport with plant viruses. Plant Physiol 102: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Zambryski PC (2000) Systemic transport of RNA in plants. Trends Plant Sci 5: 52–54 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (2001) Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol 125: 1802–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20: 448–455 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences: a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Doyle T, Botstein D (1996) Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA 93: 3886–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler TJ, Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol 121: 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Benevolenskaya EV, Birchler JA (1998) Regena (Rga), a Drosophila homolog of the global negative transcriptional regulator CDC36 (NOT2) from yeast, modifies gene expression and suppresses position effect variegation. Genetics 148: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J, Heitman J, Hall MN (1991) Nuclear protein localization. Biochim Biophys Acta 1071: 83–101 [DOI] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis F, Peffer NJ, Meier I (1999) MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Ozkan M, Carter D, Raikhel NV (2003) Towards a modeling infrastructure for studying plant cells. Plant Physiol 132: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276: 29188–29194 [DOI] [PubMed] [Google Scholar]
- Hanson MR, Kohler RH (2001) GFP imaging: methodology and application to investigate cellular compartmentation in plants. J Exp Bot 52: 529–539 [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Jackson D (2000) Opening up the communication channels: recent insights into plasmodesmal function. Curr Opin Plant Biol 3: 394–399 [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Adler SA, Telmer CA, Subramaniam V, Lopez AJ (1996) CD-tagging: a new approach to gene and protein discovery and analysis. Biotechniques 20: 896–904 [DOI] [PubMed] [Google Scholar]
- Jauh GY, Fischer AM, Grimes HD, Ryan CA Jr, Rogers JC (1998) delta-Tonoplast intrinsic protein defines unique plant vacuole functions. Proc Natl Acad Sci USA 95: 12995–12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh GY, Phillips TE, Rogers JC (1999) Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11: 1867–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jeong SY, Rose A, Meier I (2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Kuhn JE, Schafer P, Immelmann A, Henco K (1995) Quantification of DNA and RNA by PCR. In MJ McPherson, BD Hames, GR Taylor, eds, PCR 2, A Practical Approach. IRL Press, Oxford
- Kertbundit S, Linacero R, Rouze P, Galis I, Macas J, Deboeck F, Renckens S, Hernalsteens JP, De Greve H (1998) Analysis of T-DNA-mediated translational beta-glucuronidase gene fusions. Plant Mol Biol 36: 205–217 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Kotlizky G, Katz A, van der Laak J, Boyko V, Lapidot M, Beachy RN, Heinlein M, Epel BL (2001) A dysfunctional movement protein of Tobacco mosaic virus interferes with targeting of wild-type movement protein to microtubules. Mol Plant Microbe Interact 14: 895–904 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Gibbon BC, McCurdy DW, Staiger CJ (2001) Fluorescently-labeled fimbrin decorates a dynamic actin filament network in live plant cells. Planta 213: 390–395 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Staiger CJ, Weaver EA, McCurdy DW (2000) AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al. (2002) Subcellular localization of the yeast proteome. Genes Dev 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A (1989) Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem 58: 913–949 [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG, Beachy RN (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN (1997) Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN (2001) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6: 256–261 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12: 773–786 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M (2003) CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Coruzzi GM (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol 121: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Prior DAM, Santa-Cruz S, Padgett HS, Beachy RN (1997) Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J 12: 781–789 [DOI] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM (1991) The glutamine synthetase gene family of Arabidopsis thaliana: light regulation and differential expression in leaves, roots, and seeds. Mol Gen Genet 230: 145–154 [DOI] [PubMed] [Google Scholar]
- Pollok BA, Heim R (1999) Using GFP in FRET-based applications. Trends Cell Biol 9: 57–60 [DOI] [PubMed] [Google Scholar]
- Roberts IM, Boevink P, Roberts AG, Sauer N, Reichel C, Oparka KJ (2001) Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218: 31–44 [DOI] [PubMed] [Google Scholar]
- Rolls MM, Stein PA, Taylor SS, Ha E, McKeon F, Rapoport TA (1999) A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J Cell Biol 146: 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, Furuya M, Nakano A (2002) A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A (1983) Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219: 493–495 [DOI] [PubMed] [Google Scholar]
- Taylor CB (1997) Promoter fusion analysis: an insufficient measure of gene expression. Plant Cell 9: 273–275 [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Tsien RY, Miyawaki A (1998) Seeing the machinery of live cells. Science 280: 1954–1955 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Rhee Y, Chen M-H, Citovsky V (2000) Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu Rev Microbiol 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20: 3596–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2002) Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA 99: 10435–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12: 259–265 [DOI] [PubMed] [Google Scholar]
- Walhout A, Temple G, Brasch M, Hartley J, Lorson M, van den Heuvel S, Vidal M (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328: 575–592 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al. (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Piston DW, Johnson CH (1999) A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci USA 96: 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65: 241–269 [DOI] [PubMed] [Google Scholar]