SUMMARY
Fission yeast cells utilize Arp2/3 complex and formin to assemble diverse filamentous actin (F-actin) networks within a common cytoplasm for endocytosis, division and polarization. Although these homeostatic F-actin networks are usually investigated separately, competition for a limited pool of actin monomers (G-actin) helps regulate their size and density. However, the mechanism by which G-actin is correctly distributed between rival F-actin networks is not clear. Using a combination of cell biological approaches and in vitro reconstitution of competition between actin assembly factors, we discovered that the small G-actin binding protein profilin directly inhibits Arp2/3 complex-mediated actin assembly. Profilin is therefore required for formin to compete effectively with excess Arp2/3 complex for limited G-actin, and to assemble F-actin for contractile ring formation in dividing cells.
INTRODUCTION
Within a common cytoplasm cells simultaneously assemble and maintain multiple F-actin networks of different organization and dynamics for diverse processes (Blanchoin et al., 2014; Michelot and Drubin, 2011). Fission yeast assembles three primary F-actin network structures, each of which depends upon a specific actin assembly factor (Kovar et al., 2011). Approximately 15,000 active Arp2/3 complexes, distributed between 30 to 50 endocytic actin patches, assemble short-branched F-actin networks that consume up to 50% of the actin (Sirotkin et al., 2010; Wu and Pollard, 2005). Less than 1,000 active formins use ~20% of the actin to assemble long-unbranched F-actin for either contractile rings (formin Cdc12) or polarizing actin cables (formin For3) (Kovar et al., 2011; Wu and Pollard, 2005). We recently discovered that actin patches, contractile rings and actin cables are in homeostasis, whereby their density and size are regulated in part by competition for G-actin (Burke et al., 2014). How then is actin properly distributed into different networks, and how can ~10-fold fewer formins successfully compete with an excess of Arp2/3 complex?
Despite an effective critical concentration for actin assembly of only 0.1 μM, cells maintain a reserve of tens to hundreds micromolar unassembled G-actin (Pollard et al., 2000). High concentrations of unassembled actin are maintained by a combination of G-actin binding proteins that prevent spontaneous nucleation of new filaments, and barbed end capping proteins that prevent elongation of filaments (Pollard et al., 2000). Profilin is the primary evolutionarily conserved small G-actin binding protein (Carlsson et al., 1977), which is typically present in concentrations similar to unassembled G-actin (Kaiser et al., 1999; Lu and Pollard, 2001). Profilin binds tightly (Kd=0.1 μM) to ATP-G-actin (Perelroizen et al., 1994; Vinson et al., 1998), and thereby prevents spontaneous actin nucleation and elongation of the actin filament pointed end (Kang et al., 1999; Pollard and Cooper, 1984; Pring et al., 1992).
Profilin has been regarded as a general actin cytoskeleton housekeeping factor, which is assumed to maintain a reserve of unassembled G-actin that is similarly available for incorporation into diverse actin filament networks (Blanchoin et al., 2014). However, profilin binds directly to continuous proline residue stretches in actin assembly factors such as formin and Ena/VASP (Ferron et al., 2007). Profilin thereby significantly increases the elongation rate of formin-assembled filaments (Kovar et al., 2006; Kovar et al., 2003; Romero et al., 2004), and is required for formin to assemble F-actin efficiently in vivo (Evangelista et al., 2002). Here, we utilized complementary in vivo fission yeast experiments and single molecule in vitro reconstitution approaches to test our hypothesis that profilin regulates competition for G-actin by favoring formin-mediated over Arp2/3 complex-mediated actin assembly.
RESULTS
The ratio of profilin to actin is critical for proper F-actin network homeostasis
We previously reported that specific actin expression levels are critical for proper F-actin network distribution in fission yeast (Burke et al., 2014). One possibility is that altering actin expression disrupts the appropriate ratio of profilin to actin. Actin overexpression (low profilin/actin ratio) favors Arp2/3 complex actin patches, whereas actin underexpression (high profilin/actin ratio) favors formin contractile rings (Burke et al., 2014).
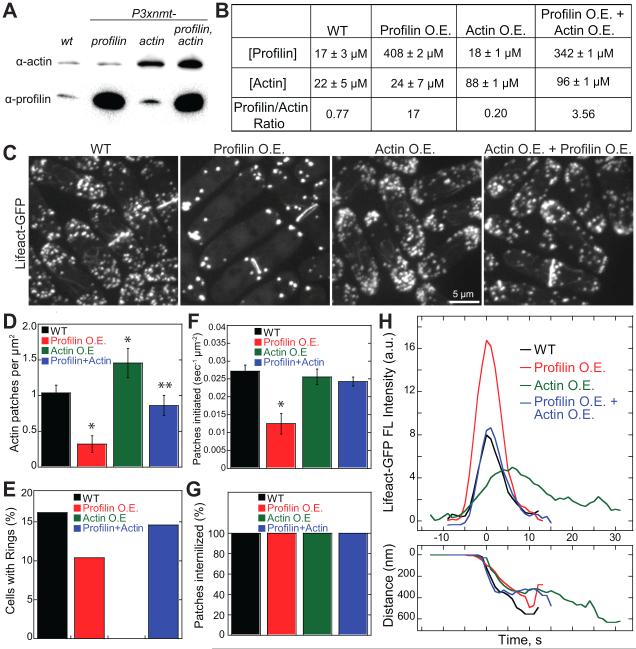
The ratio of soluble profilin to actin in wild type cells is ~0.8 (Figures 1A and 1B). We perturbed this ratio by overexpressing (O.E.) actin, profilin SpPRF (cdc3), or both in fission yeast cells by replacing their endogenous promoters with the thiamine-repressible Pnmt1 promoter (Figure 1). Growing cells in the absence of thiamine for 22 hours increases soluble profilin ~20-fold, and soluble actin ~4-fold (Figures 1A and 1B). In general O.E. profilin (profilin/actin=17) favors formin Cdc12 contractile rings, whereas O.E. actin (profilin/actin=0.2) favors Arp2/3 complex actin patches, and O.E. both (profilin/actin=~3.6) restores F-actin network homeostasis (Figures 1C). Specifically, O.E. profilin reduces the density of actin patches more than 2-fold (Figure 1D), because the actin patch initiation rate is reduced by more than half (Figure 1F), but patch internalization is not ultimately prevented (Figures 1G and 1H). These fewer actin patches have double the peak Lifeact-GFP fluorescence (Figure 1H), but the reasons are not clear. Conversely, O.E. actin increases the density of actin patches ~1.5-fold (Figure 1D) and the duration of actin patch disassembly (Figure 1H), but completely eliminates contractile rings (Figures 1E) (Burke et al., 2014). Importantly, simultaneously O.E. actin and profilin together suppresses contractile ring and actin patch defects caused by O.E. either actin, or profilin alone (Figures 1D–1H), and significantly rescues growth defects caused by O.E. actin alone (Figure S1) (Balasubramanian et al., 1994; Magdolen et al., 1993). These results emphasize a critical balance between profilin and actin that ensures the proper density and dynamics of formin Cdc12 contractile rings and Arp2/3 complex actin patches.
Figure 1. The ratio of profilin to actin facilitates F-actin network diversity in fission yeast.
(A and B) Soluble actin and profilin levels in WT cells, and cells overexpressing (O.E.) actin, profilin SpPRF or both at 22 hours without thiamine.
(A) Immunoblot analysis of equally loaded total soluble extracts.
(B) Quantification of cytoplasmic actin and profilin concentrations (mean±s.d., n=2–4), and their ratio.
(C-H) Comparison of Lifeact-GFP-labeled F-actin structures in WT cells, and cells O.E. profilin, O.E. actin, or O.E. both actin and profilin at 22 hours without thiamine.
(C) Fluorescence micrographs of actin structures labeled with Lifeact-GFP. Scale bar, 5 μm.
(D–G) Comparison of (D) actin patch density, n=6 cells per strain, (E) percentage of cells with contractile actin rings, n≥43 cells, (F) rates of patch initiation, n=3 cells per strain, and (G) percentage of patches internalized, n=5 cells. Error bars, s.d. Asterisks indicate statistical significance compared to wild type, T-test: * p<0.002, ** p<0.05.
(H) Average time courses of the fluorescence intensity (top) and the distance from origin (bottom) for actin patches labeled with Lifeact-GFP. Raw time courses for 10 patches for each strain were aligned to initiation of patch movement (time=0), and averaged at each time point.
See also Figure S1.
Profilin has separable roles in actin patch and contractile ring assembly
Because changing the ratio of profilin to actin impacts actin patches and contractile rings differently, we suspected that profilin has different roles in these processes. It is well documented that profilin is required for contractile ring assembly (see Figures 2F and 2G) (Balasubramanian et al., 1994; Lu and Pollard, 2001), presumably because profilin increases the elongation rate of filaments assembled by formin Cdc12 ~100-fold (Kovar et al., 2003). However, the role of profilin in actin patch assembly is less clear.
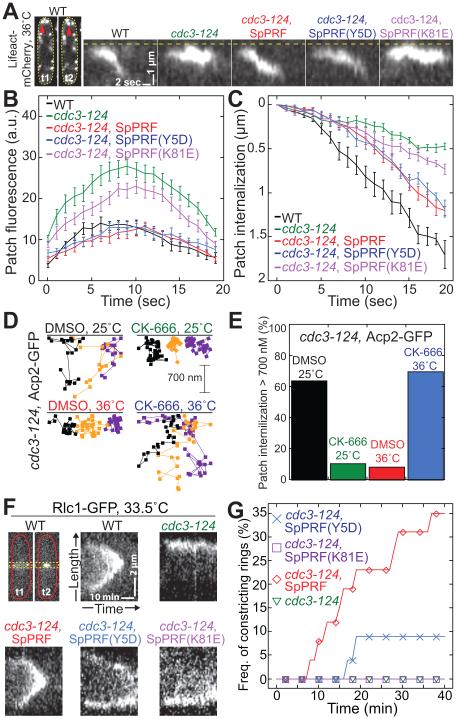
Figure 2. Profilin has roles in both endocytic actin patches and cytokinetic contractile rings.
(A–C) Profilin decreases actin incorporation into actin patches. Lifeact-mCherry labeled actin patches in WT (black), cdc3-124 (green), or cdc3-124 cells expressing WT SpPRF (red), SpPRF(Y5D) (blue), or SpPRF(K81E) (purple) at 36°C. See also Movie S1.
(A) Kymographs of representative patch internalization from the cortex (dashed line). Far-left images correspond to patches (red arrow) in a WT cell, where t2 is 1.8 sec after t1.
(B) Actin patch fluorescence intensity over time. Error bars, s.d.; n=10 patches per strain.
(C) Actin patch internalization from the cortex over time. Error bars, s.d.; n=10 patches per strain.
(D and E) Actin capping protein Acp2-GFP labeled actin patches in cdc3-124 cells at 25 or 36°C with and without 50 μM CK-666.
(D) Trajectories of three representative patches over time.
(E) ercent of patches that internalize at least 700 nm. n=50 patches per strain.
(F and G) Cytokinesis defects at 33.5°C in WT (black), cdc3-124 (green), or cdc3-124 cells expressing WT SpPRF (red), SpPRF(Y5D) (blue), or SpPRF(K81E) (purple).
(F) Kymographs of representative contractile rings labeled with Rlc1-GFP. Single Z-plane images were acquired every 1 min. Top-left images correspond to a constricting ring in a WT cell, where t1=12 min and t2=29 min after ring formation. Yellow dotted lines mark the region used for kymographs.
(G) umulative frequency of constricting contractile rings over time. n≥18 cells per strain.
We compared the behavior of actin patches, labeled with the general F-actin marker Lifeact-mCherry, in wild-type (WT) and temperature sensitive profilin mutant cdc3-124 cells. At the restrictive temperature of 36°C, patches in cdc3-124 cells incorporate ~2-fold more actin and internalize one third as far as actin patches in WT cells (Figures 2A–2C; Movie S1), indicating that profilin's role is to limit incorporation of actin into patches. Actin patch defects in cdc3-124 cells at 36°C are suppressed by inhibiting Arp2/3 complex with 50 μM CK-666 (Figures 2D and 2E), suggesting that overassembly of actin into patches in the absence of profilin inhibits patch movement, possibly by trapping an endocytic vesicle by a dense F-actin network at the plasma membrane. Conversely, inhibition of Arp2/3 complex with CK-666 at the permissive temperature of 25°C also decreases patch motility, indicating that decreased F-actin network density in the presence of CK-666 is less efficient in promoting patch internalization.
We next investigated whether it is possible to separate the roles of profilin in actin patches and contractile rings. We integrated constructs expressing WT fission yeast profilin SpPRF, and profilin mutants SpPRF(K81E) and SpPRF(Y5D) (Kovar et al., 2003; Lu and Pollard, 2001), into the leu1 locus in cdc3-124 cells. SpPRF(K81E), which has an ~100-fold lower affinity for G-actin (Lu and Pollard, 2001), does not suppress defects in either Arp2/3 complex-mediated actin patches at 36°C (Figures 2A–2C) or formin Cdc12-mediated contractile rings at the semi-permissive temperature of 33.5°C (Figures 2F and 2G). Conversely, SpPRF(Y5D), which has an ~100-fold lower affinity for proline rich motifs such as those found in formin (Chang et al., 1997; Lu and Pollard, 2001), does not suppress defects in contractile rings (Figures 2F and 2G), but does suppress the amount of actin in patches to a normal level and restores their internalization behavior (Figures 2A–2C). Therefore, contractile ring assembly requires that profilin associates with both G-actin and formin Cdc12 (Kovar et al., 2003), while profilin’s function in actin patches depends only on binding G-actin. Collectively, these in vivo experiments reveal that profilin has separate roles in actin patches and contractile rings, and that profilin specifically limits the amount of actin incorporated into patches.
Profilin inhibits Arp2/3 complex-mediated F-actin branching in vitro
Because overexpressing profilin reduces the number of actin patches, whereas actin patches in profilin loss of function mutant cdc3-124 cells incorporate more actin, we suspected that profilin inhibits Arp2/3 complex. Profilin reduces Arp2/3 complex-mediated actin assembly in ‘bulk’ pyrene actin assays (Machesky et al., 1999; Rodal et al., 2003), although the branch density was not determined. We tested whether profilin inhibits Arp2/3 complex in vitro by directly observing the assembly of Oregon Green-actin with Total Internal Reflection Fluorescence Microscopy (TIRFM). In the presence of its WASP activator SpWsp1(VCA), fission yeast SpArp2/3 complex generates a branched F-actin network with ~0.08 branches per micron after six minutes (Figures 3A and 3B; Movie S2). WT profilin SpPRF inhibits branching by at least 4-fold in a concentration dependent manner (Figures 3A and 3B). SpPRF also inhibits SpArp2/3 complex in the presence of formin Cdc12 (Figures 3A and 3B; Movie S2), as well as in ‘bulk’ pyrene actin assays (Figures S2A and S2B). Profilin also inhibits branch formation by SpArp2/3 complex and full length SpWsp1(FL), which contains a proline rich domain expected to bind profilin (Figures S2C–S2E).
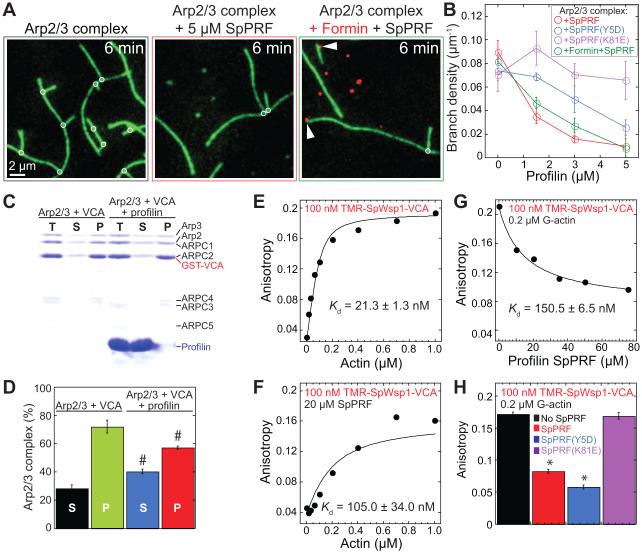
Figure 3. Profilin inhibits Arp2/3 complex branch formation.
(A–B) TIRFM visualization of 1.5 μM Mg-ATP-actin (15% Oregon Green-actin) with 40 nM SpArp2/3 complex and 80 nM SpWsp1(VCA), 10 pM formin SNAP-549(red)-SpCdc12(FH11PFH2), and 5 μM profilin SpPRF. See also Figure S2 and Movie S2.
(A) F-actin after six minutes of assembly, with marked Arp2/3 complex branches (circles) and formin-associated barbed ends (arrowheads).
(B) Dependence of Arp2/3 complex branch density on the concentration of WT SpPRF (red), SpPRF(K81E) (purple), SpPRF(Y5D) (blue), or formin and WT SpPRF (green). Error bars, s.e.; n=2 reactions.
(C–D) Pull down of 0.3 μM SpArp2/3 complex with 2 μM GST-Wsp1(VCA) by Glutathione-Sepharose in the absence or presence of 58 μM profilin SpPRF.
(C) Coomassie Blue stained SDS-PAGE gel of the total reaction before centrifugation (T), supernatant (S), and pellet (P).
(D) Percentage of Arp2/3 complex in the supernatant (S) and pellet (P). Error bars, s.e.; n=2 reactions. Pound signs indicate no statistical differences in the absence and presence of profilin, T-test: # p>0.05.
(E-H) Fluorescence anisotropy assays of 100 nM TMR-SpWsp1(VCA) equilibrium binding to (E) G-actin, (F) G-actin with 20 μM WT profilin SpPRF, (G) 0.2 μM G-actin over a range of profilin SpPRF concentrations, or (H) 0.2 μM G-actin in the absence or presence of 35 μM WT SpPRF, SpPRF(Y5D), or SpPRF(K81E). Curve fits yielded apparent equilibrium constants (Kd) for TMR-SpWsp1(VCA) binding G-actin. Anisotropy experiments were performed in duplicate. Error bars, s.e.; n=2 reactions. Asterisks indicate statistical significance compared to no profilin or SpPRF(K81E), T-test: * p<0.002.
Significantly, the ability of the ligand-specific profilin mutants to suppress actin patch defects in profilin mutant cdc3-124 cells (Figures 2A–2C) correlates with their ability to inhibit SpArp2/3 complex in vitro (Figure 3B; Figure S2B). SpPRF(Y5D) inhibits SpArp2/3 complex in vitro and largely restores normal actin patches in vivo. Conversely, SpPRF(K81E) does not inhibit SpArp2/3 complex in vitro and completely fails to suppress actin patch defects in vivo. Therefore, the ability of profilin to stimulate formin-mediated actin assembly can be separated from its ability to inhibit Arp2/3 complex, and inhibition of Arp2/3 complex by profilin is required for normal actin patch dynamics.
Profilin inhibits Arp2/3 complex by competing with WASP for G-actin
Activation of Arp2/3 complex by WASP and subsequent daughter branch formation involves multiple steps (Pollard, 2007), and profilin could inhibit one or more of these steps. For example, it is possible that by binding Arp2/3 complex (Machesky et al., 1994), profilin might prevent the association of Arp2/3 complex with WASP. Alternatively, profilin might compete with WASP for G-actin (Marchand et al., 2001; Rodal et al., 2003). We tested these possibilities with biochemical assays.
An excess of 58 μM WT profilin SpPRF only slightly decreases the amount of 0.3 μM SpArp2/3 complex that is pulled down by 2 μM GST-SpWsp1(VCA) with glutathione-sepharose beads (Figures 3C and 3D). On the other hand, a series of fluorescent anisotropy assays revealed that profilin decreases the apparent affinity of SpWsp1(VCA) for G-actin by ~5- to 7-fold (Figures 3E–3H). TMR-labeled SpWsp1(VCA) binds to G-actin with a dissociation constant Kd of ~21 nM (Figure 3E), which is similar to the affinity measured by FRET (Kd=15.5 nM) (Nolen and Pollard, 2008). The inclusion of near saturating amounts of SpPRF increases the apparent Kd to 105 nM (Figure 3F), and a similar increase (apparent Kd=~150 nM) was measured by adding a range of profilin concentrations in a competition-binding assay (Figure 3G). Importantly, the ability of ligand specific profilin mutants to disrupt the association of G-actin with SpWsp1(VCA) (Figure 3H) correlates with their ability to inhibit branching by SpArp2/3 complex (Figure 3B). SpPRF(Y5D) disrupts the association of G-actin with SpWsp1(VCA) and inhibits branching, whereas SpPRF(K81E) does not inhibit either. Together these results reveal that profilin directly inhibits Arp2/3 complex-mediated branching in vitro by sequestering G-actin away from WASP Wsp1(VCA), consistent with partial overlap of WH2 domain and profilin binding sites on the barbed end of an actin monomer (Chereau et al., 2005).
Reconstitution of competition between Arp2/3 complex and formin in vitro
Profilin reduces the ability of both Arp2/3 complex and formin to nucleate filaments (Figure 3 and Supplemental Figure 2) (Kovar et al., 2003; Li and Higgs, 2003; Paul and Pollard, 2008; Pring et al., 2003), but specifically increases the elongation rate of formin-assembled filaments by as much as 10-fold (Kovar et al., 2006; Romero et al., 2004). Profilin may therefore be capable of favoring formin- over Arp2/3 complex-mediated actin assembly.
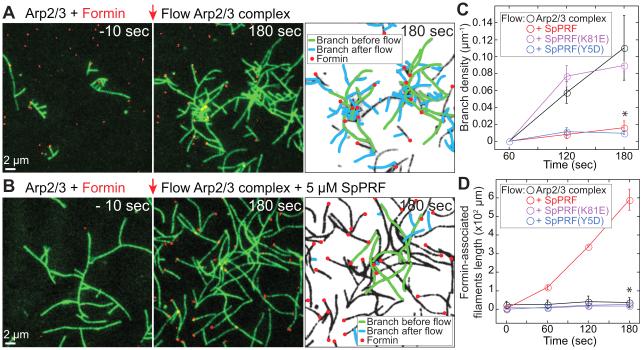
We tested this possibility by utilizing two-color TIRFM to observe the consequences of adding profilin to in vitro reactions containing a mixture of an ~400-fold excess of SpArp2/3 complex to fluorescently-labeled formin SNAP-549(red)-Cdc12(FH11PFH2) (Figures 4A–4D; Movie S3). Initially Oregon Green-actin was assembled in reactions containing SpArp2/3 complex, SpWsp1(VCA) and formin Cdc12, which produces filaments that are formed (a) spontaneously, (b) nucleated by formin Cdc12 or (c) branches nucleated by Arp2/3 complex. These initial conditions without profilin are favorable for all three types of nucleation, but not elongation by Cdc12. We then flowed in a new mix of Oregon Green-actin with SpArp2/3 complex and SpWsp1(VCA), in the absence or presence of SpPRF. Without profilin, SpArp2/3 complex is favored because the branch density increases over time, whereas Cdc12-associated filaments elongate ~20-fold slower than free barbed ends (Figures 4A, 4C and 4D; Movie S3). Conversely, addition of WT SpPRF favors formin because the rate of formation of new branches is reduced ~10-fold (Figures 4B – 4D; Movie S3), whereas the elongation rate of Cdc12-associated filaments increases 25-fold, resulting in an ~15-fold increase of Cdc12-assembled F-actin after 180 seconds (Figures 4B–4D; Movie S3) (Kovar et al., 2006; Kovar et al., 2003). Neither separation of function profilin mutant enhances formin-mediated actin filament elongation (Figure 4D; Figures S3A and S3B) (Kovar et al., 2003), and only SpPRF(Y5D) inhibits SpArp2/3 complex-mediated branch formation (Figure 4C; Figures S3A and S3B). Thus, although profilin reduces nucleation by both Arp2/3 complex and formin, profilin favors formin-mediated actin assembly by specifically increasing the elongation rate of formin-associated filaments.
Figure 4. Profilin favors formin-over Arp2/3 complex-mediated actin assembly in vitro.
(A–D) TIRFM visualization of 1.5 μM Mg-ATP-actin (15% Oregon Green-actin) with 40 nM SpArp2/3 complex and 80 nM SpWsp1(VCA), 100 pM formin SNAP-549(red)-SpCdc12(FH11PFH2), and 5 μM profilin SpPRF. See also Figure S3 and Movie S3.
(A and B) Addition of profilin into mixtures of Arp2/3 complex and formin. Initial reactions contained Mg-ATP-actin, Arp2/3 complex, SpWsp1(VCA) and formin. At t=0 sec (red arrow) additional Mg-ATP-actin, Arp2/3 complex and SpWsp1(VCA) were flowed into the chamber in the (A) absence or (B) presence of WT SpPRF. Inverted micrographs indicate formin-associated filaments (red dots), and Arp2/3 complex branches initiated before (green) and after (blue) flow.
(C) The branch density and (D) length of total formin-associated F-actin over time are plotted in the absence and presence of WT and mutant profilin. Error bars, s.e.; n=2 reactions. Asterisks indicate statistical significance compared to (C) SpPRF(K81E) and (D) SpPRF, T-test: * p<0.02.
Biomimetic reconstitution of competition between Arp2/3 complex and formin in vitro
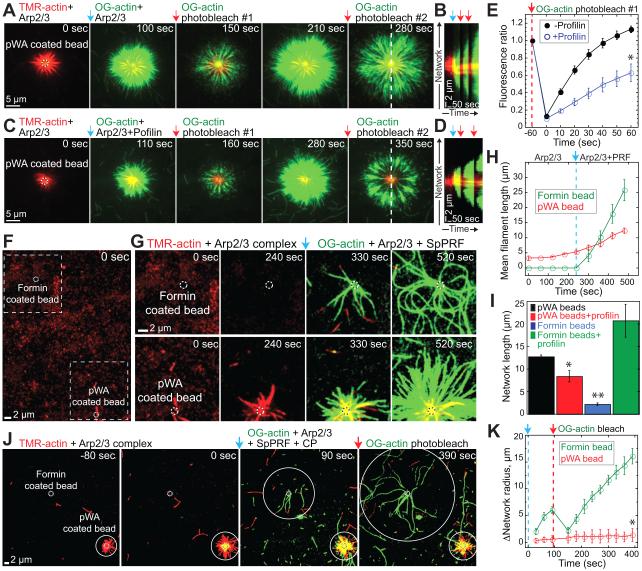
Within cells Arp2/3 complex nucleation promoting factors like WASP are clustered, and different F-actin networks are assembled at distinct locations (Blanchoin et al., 2014). We used two-color TIRFM biomimetic assays with WASP and formin clustered on different polystyrene beads, which spatially separates the assembling F-actin networks. Because the activity of fission yeast formin Cdc12 is low on polystyrene beads, these reactions contained vertebrate formin mDia2, mammalian Arp2/3 (mArp2/3) complex, a pWA fragment of its WASP activator that includes a central proline-rich domain, and human profilin HPRO1 or fission yeast profilin SpPRF.
Profilin HPRO1 potently inhibits the nucleation of branched networks from beads by mArp2/3 complex, but has little effect on their elongation. Figures 5A–5E and Movie S4 show F-actin networks assembled by mArp2/3 complex from TMR(red)-actin on the surface of beads coated with GST-pWA. Addition (time=0 sec, blue arrows) of Oregon Green-actin with mArp2/3 complex labels new actin incorporation into the networks. Two successive rounds of fluorescence recovery after photobleaching Oregon Green-actin (red arrows) reveal that in the absence of profilin, new mArp2/3 complex branched filaments are continuously generated from the bead surface (Figure 5E). Conversely, the inclusion of 5 μM profilin HPRO1 significantly reduces Oregon Green-actin fluorescence recovery at the bead surface, indicating that mArp2/3 complex branch formation is inhibited (Figure 5E).
Figure 5. Biomimetic reconstitution of the effect of profilin on competition between formin and Arp2/3 complex.
(A–E) TIRFM visualization of the assembly of 1.5 μM Mg-ATP-actin monomers from beads coated with mammalian Arp2/3 complex activator GST-pWA. Initial reactions contained 10% TMR(red)-actin and (A-D) 10 nM or (E) 50 nM mArp2/3 complex. At t=0 sec (blue arrow) 15% Oregon green-actin was flowed into the reaction chamber with mArp2/3 complex in the (A,B,E) absence and (C,D,E) presence of 5 μM profilin HPRO1. Oregon green was globally bleached twice (red arrows) to visualize incorporation of new G-actin. See also Figure S4 and Movie S4.
(A, C) Fluorescent micrographs of time series.
(B, D) Kymographs of merged TMR and Oregon green F-actin fluorescence (dashed white line in (A, C)) over the course of two rounds of photobleaching in (B) the absence or (D) the presence of 5 μM profilin HPRO1.
(E) Oregon-green-actin fluorescence recovery after photobleach #1. Error bars, s.e.; n=4 beads. Asterisk indicates statistical significance, T-test: * p<0.004.
(F–K) TIRFM visualization of 1.5 μM Mg-ATP-actin assembly from beads coated with either mArp2/3 complex activator GST-pWA or formin SNAP-mDia2(FH1FH2). See also Movie S5.
(F–I) Initial reactions contained 10% TMR(red)-actin with 2 nM mArp2/3 complex. After 240 seconds (blue arrow) 15% Oregon green-actin, 2 nM mArp2/3 complex, and 5 μM profilin were flowed into the chamber.
(F) Full field containing GST-pWA-and formin-associated beads (dashed circles).
(G) Time-lapse of magnified boxed regions in (F).
(H) Mean filament length from the GST-pWA-(red circle) and formin-coated (green circle) beads shown in (F) and (G) over time. Profilin addition is marked by a blue arrow and dashed line. Error bars, s.d.; n=4 filaments.
(I) Mean length of F-actin networks assembled from GST-pWA-or formin-coated beads after >300 seconds. Error bars, s.e.; n=20 formin beads and 26 pWA beads from 3 different experiments. Asterisks indicate statistical significance, T-test: * p<0.03 (pWA beads) ** p<0.007 (formin beads).
(J–K) Initial reactions contained 10% TMR(red)-actin with 5 nM mArp2/3 complex. At t=0 second (blue arrow) 15% Oregon green-actin, 5 nM mArp2/3 complex, 5 μM profilin, and 10 nM capping protein (CP) were flowed into the chamber. Oregon green was bleached (red arrow) to visualize incorporation of new G-actin.
(J) Fluorescent micrographs of time series. White solid circles mark expanding networks generated from beads (dashed circles).
(K) Increase in network radius over time from formin-and pWA-coated beads. Error bars, s.e.; n=3 beads. Asterisk indicates statistical significance, T-test: * p<0.002.
On the other hand, both profilin HPRO1 and SpPRF enhance the elongation of formin mDia2-mediated networks from beads. Supplemental Figure S4 shows F-actin networks assembled on formin SNAP-mDia2(FH1FH2)-coated beads. Formin mDia2-assembled filaments elongate 4-fold slower than control filaments in the absence of profilin (Figures S4A and S4B). Conversely, mDia2-assembled filaments elongate ~4- and ~2-fold faster than control filaments in the presence of SpPRF (Figures S4C and S4D) or HPRO1 (Figures S4E and S4F) (Kovar, 2006).
When GST-pWA- and formin SNAP-mDia2(FH1FH2)-coated beads are mixed in the same reaction chamber (Figures 5F–5I; Movie S5), F-actin networks rapidly expand on GST-pWA-coated beads in the absence of profilin whereas filaments elongate slowly on SNAP-mDia2(FH1FH2)-coated beads. However, the addition of profilin SpPRF completely changes the system dynamics, facilitating the rapid elongation of F-actin networks mediated by mDia2 (Figures 5F–5H; Movie S5). On average ~35 to 40% of both GST-pWA- and formin SNAP-mDia2(FH1FH2)-coated beads are active and grow filaments. After six minutes, the average filament network length from multiple GST-pWA-coated beads is half as long with profilin, whereas the average filament length from mDia2-coated beads is ~10-fold longer with profilin (Figure 5I).
The addition of both profilin SpPRF and barbed end capping protein further differentiates F-actin networks assembled from formin- and GST-pWA-coated beads (Figures 5J and 5K). Formin mDia2-mediated filaments continue to elongate rapidly because formin inhibits capping protein by remaining processively associated with the elongating barbed end (Zigmond et al., 2003). On the other hand, the combination of profilin and capping protein severely blocks F-actin network assembly from GST-pWA-coated beads because profilin reduces the number of branches and capping protein blocks their elongation.
Profilin antagonizes Arp2/3 complex to facilitate formin-mediated actin assembly in fission yeast
By simultaneously inhibiting Arp2/3 complex branching and facilitating rapid formin-mediated actin elongation in vitro (Figures 4 and 5), profilin could be a key regulator of F-actin network homeostasis in cells. We therefore investigated cytokinesis in profilin mutant cdc3-124 fission yeast cells, in both the presence and absence of functional Arp2/3 complex, to test whether inhibition of Arp2/3 complex by profilin is required for formin-mediated actin assembly (Figure 6).
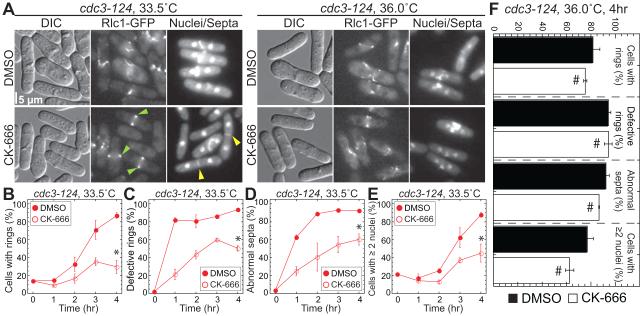
Figure 6. Profilin antagonizes Arp2/3 complex in fission yeast.
(A–F) Profilin mutant cdc3-124 fission yeast expressing the contractile ring marker Rlc1-GFP at semi-restrictive 33.5°C or fully restrictive 36°C, in the absence and presence of 50 μM Arp2/3 complex inhibitor CK-666. See also Table S1.
(A) Representative micrographs. Normal Rlc1-GFP labeled contractile rings and Calcofluor stained septa are marked with green and yellow arrowheads.
(B-E) Quantification of cytokinesis defects over time at 33.5°C in the absence (filled symbols) and presence (open symbols) of CK-666. Error bars, s.e.; n=3 experiments with ≥200 cells per strain. Asterisks indicate statistical significance, T-test: * p<0.02.
(B) ercentage of cells with rings.
(C) Percent of rings that are defective.
(D) ercent of abnormal (broad, misplaced, misoriented, and/or partial) septa.
(E) ercent of cells with ≥2 nuclei.
(F) Quantification of cytokinesis defects described in (B-E) after 4 hr at 36.0°C. Error bars, s.e.; n≥50 cells per strain. Pound signs indicate no statistical difference, T-test: # p≥0.1.
At permissive temperature, asynchronous wild-type (WT) and cdc3-124 cultures contain only ~20% of dividing cells, without cytokinesis defects. Conversely, after 4 hours at the semi-permissive temperature of 33.5°C, ~80% of cdc3-124 cells develop significant cytokinesis defects with abnormal contractile rings and septa, and ≥2 nuclei (Figures 6A–6E) (Balasubramanian et al., 1994; Lu and Pollard, 2001). Cytokinesis defects in cdc3-124 cells are significantly suppressed by inhibition of Arp2/3 complex with intermediate concentration of the Arp2/3 complex inhibitor CK-666 (Figures 6A–6E), or by mutations in Arp2/3 complex (Balasubramanian et al., 1996; McCollum et al., 1996). Thus, formin function is restored by inhibiting formin’s competitor for actin monomers, most likely by increasing the pool of G-actin available to formin. Importantly, a minimal level of profilin is required for formin Cdc12-mediated contractile ring assembly, as inhibition of Arp2/3 complex does not suppress cdc3-124 cytokinesis defects at the fully restrictive temperature of 36°C (Figures 6A and 6F) (Balasubramanian et al., 1996). Furthermore, suppression is specific to profilin mutant cdc3-124 cells, as other contractile ring mutants including formin cdc12-112, myosin light chain cdc4-8 and tropomyosin cdc8-27 are not suppressed by inhibiting Arp2/3 complex at 33.5°C (Table S1) (Balasubramanian et al., 1996).
DISCUSSION
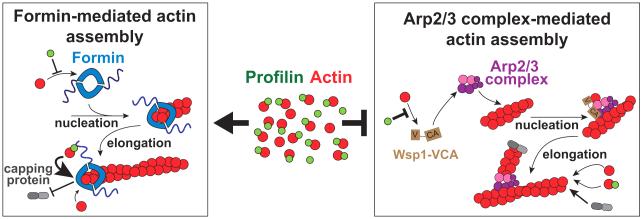
Here, and in the accompanying paper using animal fibroblast cells (Rotty et al, submitted to Developmental Cell), we discovered that profilin is not simply a housekeeping G-actin binding protein that similarly facilitates actin assembly by diverse actin assembly factors. In addition to preventing unwanted spontaneous actin assembly (Pollard and Cooper, 1984), we found that by favoring formin over Arp2/3 complex profilin is required for proper distribution of actin assembly to different homeostatic F-actin networks in fission yeast (Figure 7).
Figure 7. Cartoon model for profilin’s role in regulating F-actin network homeostasis.
The majority of G-actin in cells is bound to profilin, which helps distribute actin assembly between diverse F-actin networks. Profilin inhibits nucleation by formin but dramatically increases the elongation rate of formin-associated filaments. Profilin inhibits Arp2/3 complex-mediated daughter branch formation by disrupting the association of its activator WASP VCA with actin, but has little effect on their elongation rate. Furthermore, formin-associated filaments continue to elongate in the presence of capping protein, whereas Arp2/3 complex-branched filaments are rapidly capped. Therefore, profilin is necessary for formin to rapidly assemble unbranched actin filaments in the presence of excess Arp2/3 complex.
Although profilin slows nucleation by formins (Kovar et al., 2003; Li and Higgs, 2003; Paul and Pollard, 2008; Pring et al., 2003), once filaments are formed profilin increases their elongation rate as much as 10-fold faster than the rate of free barbed ends assembled by Arp2/3 complex (Kovar et al., 2006; Romero et al., 2004). This results in fewer filaments that elongate quickly. Additionally, formins remain continuously associated on growing barbed ends (Kovar and Pollard, 2004), allowing rapid elongation in the presence of capping protein (Zigmond et al., 2003). Conversely, profilin significantly reduces Arp2/3 complex-mediated branch formation by inhibiting the association of its activator WASP VCA with actin (see Figure 3) (Egile et al., 1999; Higgs et al., 1999; Machesky et al., 1999; Rodal et al., 2003), and profilin does not increase the elongation rate of Arp2/3 complex-nucleated filaments. Furthermore, filaments with free barbed ends produced by Arp2/3 complex are rapidly capped by capping protein (Blanchoin et al., 2000). Therefore, the combination of profilin and capping protein dramatically favors formin over Arp2/3 complex (Figures 5J and 5K).
These opposing effects of profilin allow formin to assemble F-actin networks by efficiently competing with Arp2/3 complex for G-actin. Significantly, fission yeast cells expressing the profilin separation of function mutant SpPRF(Y5D), which binds to G-actin and inhibits Arp2/3 complex but cannot bind to formin, fail cytokinesis. Conversely, cytokinesis defects in profilin mutant cdc3-124 cells are suppressed by inhibition of Arp2/3 complex. Therefore, both enhancing formin and suppressing Arp2/3 complex are important functions of profilin.
How does profilin inhibit Arp2/3 complex? Activation of Arp2/3 complex by WASP and subsequent branch formation requires multiple steps (Pollard, 2007): (i) Arp2/3 complex binds two WASP molecules, each associated with an actin monomer (Padrick et al., 2011; Ti et al., 2011) (ii) the WASP-actin-Arp2/3 complex associates with a mother filament (Smith et al., 2013), (iii) Arp2/3 complex adopts an active conformation resembling a short-pitch actin dimer (Goley et al., 2010; Rouiller et al., 2008), (iv) WASP delivers and helps position the first and second actin subunits of the daughter filament (Machesky et al., 1999; Marchand et al., 2001; Padrick et al., 2011), (v) WASP dissociates from a nascent actin branch (Smith et al., 2013), and (vi) new actin monomers are added to the barbed end of the Arp2/3 complex nucleus. Profilin might theoretically modify several steps in actin branch formation. However, based on experimental evidence we favor a simple hypothesis that profilin and WASP compete for actin monomers (Machesky et al., 1999; Rodal et al., 2003). First, both human and fission yeast WASP and profilin cannot bind actin monomers simultaneously (Figures 3E–3H) (Marchand et al., 2001). Second, the profilin SpPRF(K81E) mutant does not inhibit the association of WASP with G-actin (Figure 3H) and does not inhibit Arp2/3 complex (Figure 3B). By competing with VCA for binding actin monomers, profilin prevents the delivery of the first actin subunit to the daughter filament, thereby blocking branch formation. We also found that profilin slightly disrupts the association of WASP with Arp2/3 complex (Figures 3C and 3D), so we cannot fully rule out that profilin might also disrupt the association of the WASP-G-actin complex with Arp2/3 complex or sterically hinder Arp2/3 complex activation by binding Arp2 (Mullins et al., 1998). On the other hand, we can rule out the importance of profilin binding directly to WASP because (1) profilin similarly inhibits Arp2/3 complex activation by WASP fragments with or without the proline rich domain (Figures S4C–S4E), and (2) the profilin SpPRF(Y5D) mutant inhibits Arp2/3 complex as well as WT SpPRF. Interestingly, profilin also inhibits formin-mediated nucleation (Kovar et al., 2003; Li and Higgs, 2003; Paul and Pollard, 2008; Pring et al., 2003), suggesting that a general role of profilin is to limit the nucleation step of actin assembly, even by nucleation factors.
The role of profilin as a regulator of homeostatic F-actin networks may be evolutionarily conserved. Modifying profilin levels alters specific F-actin networks in diverse cell types. Reducing profilin levels often leads to defects specifically in formin-mediated processes such as cytokinesis in diverse cells including fission yeast, worms and mice (Balasubramanian et al., 1994; Severson et al., 2002; Witke et al., 2001). Conversely, increased profilin levels often lead to the disappearance of Arp2/3 complex-mediated networks such as lamellipodia in animal cells (Cao et al., 1992), Listeria actin comet tails (Sanger et al., 1995), or actin patches in yeast (Balasubramanian et al., 1994). Similarly, Rotty et al. (submitted to Developmental Cell) found that profilin antagonizes Arp2/3 complex in animal fibroblasts. We suspect that the spatial and temporal regulation of profilin may be a key molecular switch that allows assembly of diverse F-actin networks via different actin assembly factors. For example, profilin may be concentrated where formin-mediated actin assembly is required, such as the middle of dividing fission yeast cells (Balasubramanian et al., 1994), and the projection tip of mating fission yeast cells (Petersen et al., 1998). Higher eukaryotic cells with multiple profilin isoforms might have particular isoforms expressed at a particular time and place, and/or tailored to facilitate actin assembly for different F-actin networks (Mouneimne et al., 2012). Furthermore, phosphorylation of profilin (Fan et al., 2012), and/or release of profilin’s inhibitory association with membrane phosphoinositides (Goldschmidt-Clermont et al., 1991), might also impact the spatial and temporal regulation of actin assembly by formins and Arp2/3 complex.
EXPERIMENTAL PROCEDURES
Fission yeast strain construction and assay conditions
Fission yeast strains used in this study are listed in Table S2. Cells were maintained on standard YE5S complete and EMM5S minimal growth media. All strains combining two or more markers and/or mutants were constructed by genetic crosses and tetrad dissection. To overexpress profilin, the endogenous Pcdc3 promoter was replaced with the thiamine-repressible 3xPnmt1 promoter by integrating pFA6a-kanMX6-P3nmt1 cassette (Bahler et al., 1998) in place of -1 to -140 nucleotide region in the 5’UTR of the endogenous cdc3 locus. For data in Figure S1, profilin was overexpressed from the high expression vector pREP3x-cdc3, which was transformed by lithium acetate into WT cells or cells O.E. actin (Burke et al., 2014).
Stock solutions of 10 mM CK-666 (Sigma, St. Louis, MO) were prepared in DMSO (Nolen et al., 2009). 700 μL of exponentially growing cells were treated with DMSO or 50 μM CK-666, incubated at 25°C, 33.5°C, or 36°C and then visualized each hour by DIC and epifluorescence microscopy. Aliquots were fixed in MeOH and labeled with DAPI and Calcofluor to visualize nuclei and septa. For actin patch tracking in cdc3-124 cells, cells were grown for four hours at 25°C or 36°C in the presence or absence of CK-666, and then imaged. Strains expressing WT or mutant profilins were generated by integration of pJK148-SpPRF constructs into leu1-32 locus of cdc3-124 cells. In these strains profilin expression is under control of the 81xPnmt1 promoter that was induced by growing cells for 26 hours in EMM5S without thiamine. Cells were grown for an additional two hours at 36°C for actin patch tracking, and for an additional two hours at 33.5°C for contractile ring tracking. Actin and profilin overexpression strains were grown in YE5S at 25°C for 24 hours, then grown in EMM5S without thiamine at 25°C for 22 hours before imaging.
Cell imaging
Confocal images were collected with a Cascade 512BT camera (Photometrics, Tucson, AZ) on a Zeiss Axioimager M1 equipped with 63x, 1.4 numerical aperture oil-immersion lens and a Yokogawa CSU-X1 spinning-disk unit (Solamere, Salt Lake City, UT) with a Tempcontrol-37 objective heater (Zeiss, Thornwood, NY). Z-stacks were acquired with a 100x, 1.4 NA objective on a Zeiss Axiovert 200M equipped with a Yokogawa CSU-10 spinning-disk unit (McBain, Simi Valley, CA) illuminated with a 50-milliwatt 473-nm DPSS laser, and a Cascade 512B EM-CCD camera (Photometrics, Tucson, AZ) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Differential interference contrast (DIC) and epifluorescence images were collected using an Orca-ER camera (Hamamatsu, Bridgewater, NJ) on an IX-81 microscope (Olympus, Tokyo, Japan) equipped with a Plan Apo 60x/N.A. 1.4 N.A. objective. Wild type, actin and profilin overexpressing cells in Figure 1 were imaged on pads of 25% gelatin in EMM5S using UltraView VoX (PerkinElmer, Waltham, MA) spinning disk confocal system equipped with C9100-50 EMCCD camera (Hamamatsu), installed on a Nikon Ti-E microscope with a 100×/1.4 N.A. Plan Apo lens, and controlled by Volocity software. Actin patch dynamics were analyzed using ImageJ and the plugin TrackMate from the program Fiji (Schindelin et al., 2012). At least ten patches for each strain were measured for fluorescence intensity and distance traveled from the cell cortex over time. Time courses for individual patches were aligned to the initiation of patch movement (time zero in Figures 1H and 2C) or peak intensity (in Figure 2B) and averaged at each time point.
TIRF Microscopy
Microscope slides (#1.5, Fisher Scientific) were coated with mPEG-Silane (Winkelman et al., 2014). TIRF microscopy images of Oregon Green-labeled actin (488 nm), TMR-labeled actin (561 nm), and SNAP-549(red)-SpCdc12(FH11PFH2) (561 nm) were collected at 10-s intervals with an iXon EMCCD camera (Andor Technology) using an Olympus IX-71 microscope equipped with through-the-objective TIRFM illumination. Mg-ATP-actin (1.5 μM, 15% Oregon Green-actin or 15% TMR-actin) was mixed with other proteins in a polymerization mix containing 10 mM Imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, and 0.5% (wt/vol) methylcellulose 400 centipoise to induce assembly, and transferred to a flow cell (Winkelman et al., 2014). For biomimetic assays, carboxylated 2 μm microspheres (Polysciences, Eppelheim, Germany) were coated with either 2 μM of GST-pWA or formin SNAP-mDia2(FH1FH2) (Loisel et al., 1999), and introduced into the polymerization mix.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brad Nolen (University of Oregon) for Rhodamine-labeled TMR-SpWsp1-VCA, Dennis Zimmermann for SNAP-mDia2, Jonathan Winkelman for helpful discussions, Colleen Skau for pilot experiments, and Ben Glick for the use of SnapGene for plasmid construction. This work was supported by National Institutes of Health (NIH) Grants RO1 GM079265 and the Human Frontier Science Program Grant RGY0071/2011 (to D.R.K.), NIH Molecular and Cellular Biology Training Grant T32 GM007183 (to T.A.B., J.R.C. and A.J.B.), NSF Graduate Student Fellowship DGE-1144082 (to J.R.C.) and AHA 11SDG5470024 (to V.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interests.
REFERENCES
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. Embo J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic Actin Cytoskeleton Networks Are Regulated by Assembly Factor Competition for Monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LG, Babcock GG, Rubenstein PA, Wang YL. Effects of profilin and profilactin on actin structure and function in living cells. J Cell Biol. 1992;117:1023–1029. doi: 10.1083/jcb.117.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Fan Y, Arif A, Gong Y, Jia J, Eswarappa SM, Willard B, Horowitz A, Graham LM, Penn MS, Fox PL. Stimulus-dependent phosphorylation of profilin-1 in angiogenesis. Nat Cell Biol. 2012;14:1046–1056. doi: 10.1038/ncb2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recrutiment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Kim JW, Machesky LM, Rhee SG, Pollard TD. Regulation of phospholipase C-gamma 1 by profilin and tyrosine phosphorylation. Science. 1991;251:1231–1233. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- Goley ED, Rammohan A, Znameroski EA, Firat-Karalar EN, Sept D, Welch MD. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci U S A. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Vinson VK, Murphy DB, Pollard TD. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J Cell Sci. 1999;112 ( Pt 21):3779–3790. doi: 10.1242/jcs.112.21.3779. [DOI] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Sirotkin V, Lord M. Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21:177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vanderkerckhove J, Pollard TD. Purificaiton of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen V, Drubin DG, Mages G, Bandlow W. High levels of profilin suppress the lethality caused by overproduction of actin in yeast cells. FEBS Lett. 1993;316:41–47. doi: 10.1016/0014-5793(93)81733-g. [DOI] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard T.D., and Higgs, H.N. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. Embo J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–630. doi: 10.1016/j.ccr.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Pollard TD. Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J Biol Chem. 2008;283:26490–26498. doi: 10.1074/jbc.M802607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1035. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci U S A. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J Cell Sci. 1998;111:867–876. doi: 10.1242/jcs.111.7.867. ( Pt 7) [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Pring M, Weber A, Bubb MR. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry. 1992;31:1827–1836. doi: 10.1021/bi00121a035. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JM, Mittal B, Southwick FS, Sanger JW. Listeria monocytogenes intracellular migration: inhibition by profilin, vitamin D-binding protein and DNase I. Cell Motil Cytoskeleton. 1995;30:38–49. doi: 10.1002/cm.970300106. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A Formin Homology Protein and a Profilin Are Required for Cytokinesis and Arp2/3-Independent Assembly of Cortical Microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr. Xu, M.Q., Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.