Abstract
The contribution of whole genome doubling to chromosomal instability (CIN) and tumour evolution is unclear. We use long-term culture of isogenic tetraploid cells from a stable diploid colon cancer progenitor to investigate how a genome-doubling event affects genome stability over time. Rare cells that survive genome doubling demonstrate increased tolerance to chromosome aberrations. Tetraploid cells do not exhibit increased frequencies of structural or numerical CIN per chromosome. However, the tolerant phenotype in tetraploid cells, coupled with a doubling of chromosome aberrations per cell, allows chromosome abnormalities to evolve specifically in tetraploids, recapitulating chromosomal changes in genomically complex colorectal tumours. Finally, a genome-doubling event is independently predictive of poor relapse-free survival in early stage disease in two independent cohorts in multivariate analyses (discovery data: HR=4.70, 95% CI 1.04-21.37, validation data: HR=1.59, 95% CI 1.05-2.42). These data highlight an important role for the tolerance of genome doubling in driving cancer genome evolution.
Keywords: genome doubling, tetraploidy, chromosomal instability, cancer evolution, colorectal cancer, intratumour heterogeneity
Introduction
Chromosomal instability (CIN), describing the continual loss and gain of whole and/or parts of chromosomes, is a common feature of most cancers (1). CIN represents a dynamic state that likely contributes to intratumour heterogeneity by creating a genetically diverse pool of tumour cells upon which selection can act. CIN is associated with both poor prognosis and intrinsic multi-drug resistance (2, 3). Several different cellular mechanisms are thought to contribute to CIN (1), and it has been suggested that specific ‘CIN tolerance mechanisms’ may be required for these cells to survive elevated genome instability (4, 5). While mutations in TP53 are likely to contribute to this CIN tolerance, a full mechanistic basis for this phenomenon remains incompletely understood (6).
Another common feature of tumour cells is large-scale alterations in ploidy. Polyploid cells have been observed in multiple cancer types, and, tetraploidy, resulting from a genome doubling event, has been proposed as an intermediate en route to aneuploidy (7-9). Tetraploidy has been shown to be an unstable cellular state, with polyploid yeast exhibiting increased requirements for genome stability maintenance pathways such as homologous recombination repair, as well as showing defects in sister chromatid cohesion (10). Furthermore, artificially generated mammalian tetraploid cell lines display increased segregation errors, due to supernumerary centrosomes (11). It is likely that the p53 pathway limits the proliferation of polyploid cells in order to protect genomic integrity (12). Several studies have observed tetraploid cancer cells before aneuploidy onset and in the transition from pre-malignant to malignant disease (13-15) suggesting a genome-doubling event can be driver of tumorigenesis. Consistent with this, tetraploid cells derived through multiple routes have an increased tumorigenic capacity (16-19). Tetraploid sub-clones have also been observed at later stages of tumour development (20), and whole genome-doubling events have been inferred to occur both before and after other copy number alterations across different cancer types (21, 22). However, the effect of a whole genome doubling event on CIN and how this could impact on genome evolution in human cancer has not been fully explored. Here we analyse the relationship between ploidy and genomic instability in colorectal cancer (CRC), and propose that tolerance of genome doubling might provide tolerance to CIN in this cancer type.
Results
A relationship between ploidy and genomic complexity in CRC
SNP6.0 data available for 404 Stage 1-4 CRC tumours from The Cancer Genome Atlas (TCGA) was used to explore the relationship between ploidy and CIN. To assess structural and numerical CIN, the weighted Genome Instability Index (wGII) was used, which estimates the proportion of the genome with aberrant copy number compared to the median ploidy, weighted on a per chromosome basis (23). We have previously shown that GII correlates with both numerical and structural CIN in cell lines (3). Significantly higher wGIIs were found in polyploid (ploidy ≥3) compared to diploid tumours (P<0.0001, Student’s T-test, Fig. 1A), although some stable tetraploid tumours were observed. In other tumour types, polyploidy was also associated with an increased wGII, suggesting these two genomic aberrations may be linked in a range of cancers (P<0.0001 for each cancer type, Student’s T-test, Supplementary Fig.1A).
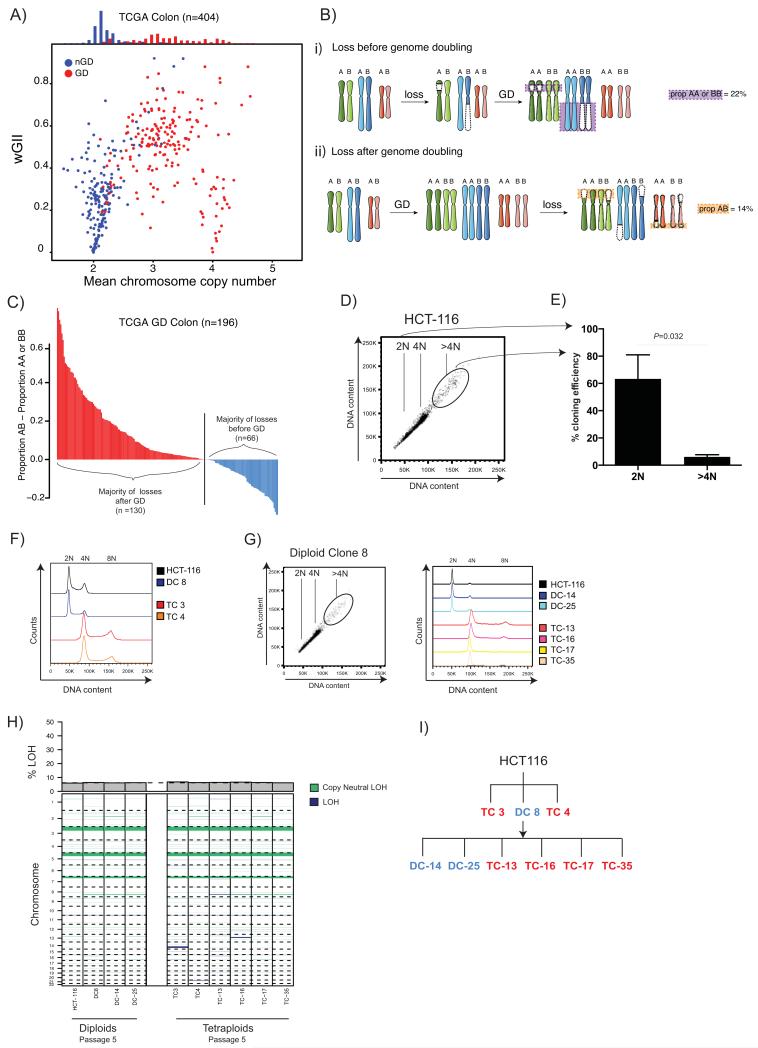
Figure 1.
A) Relationship between weighted mean chromosome copy number and wGII. Each circle represents one TCGA CRC tumour sample. Red depicts genome-doubled (GD) samples; blue non-genome-doubled (nGD) samples (see Methods). A histogram of weighted mean chromosome copy number for GD (red) and nGD (blue) is shown above.
B)
i) Copy number losses that occur on the background of a diploid genome prior to genome doubling will result in LOH, whereby one of the parental alleles is lost. In tumours that harboured chromosomal instability prior to genome doubling the majority of losses will be unbalanced, involving LOH. Unbalanced losses to two copies (AA or BB) are depicted with a purple box with purple dotted lines.
ii) In tumours where genome doubling was an early event, prior to the onset of chromosomal instability, the majority of losses to two copies will be balanced without LOH. Balanced losses to two copies (AB) are depicted with an orange box with orange dotted lines around them.
C) Timing of genome doubling estimated using copy number and LOH profiles. Each bar represents one genome-doubled tumour and its height corresponds to the proportion of AB – proportion AA or BB copy number states. Tumour genomes where the majority of losses to two copies are likely to have occurred after genome doubling are shown in red (n =130; proportion AB > proportion AA or BB), whereas those where the majority of losses are likely to have occurred before doubling are shown in blue (n=66; proportion AB < proportion AA or BB).
D) Flow cytometry shows a >4N population in the MIN colon cancer cell line HCT-116. 2N, 4N and <4N populations are indicated on the flow cytometry plot.
E) Cloning efficiency of 2N and >4N cells is shown with mean and standard error of mean (3 experiments). Tetraploid cloning efficiency was significantly lower than diploid cloning efficiency (P=0.0322, Student’s t-test). Diploid cloning efficiency was assessed using CellTiter-Blue (CTB) reagent (colonies were identified as wells with CTB value >1.5x mean average of blank wells) and the Poisson-corrected cloning efficiency was calculated (see Methods). Tetraploid cloning efficiency was calculated by verifying the percentage of surviving tetraploid clones using flow cytometry (flow cytometry data not shown).
F) After single-cell sorting, DNA content was assessed by flow cytometry with Hoescht staining. Two tetraploid clones (TC 3 and TC 4, at passage 3), one diploid clone (DC 8) and HCT-116 are shown.
G) Flow cytometry of the diploid clone DC 8 also shows a small >4N sub-population. Two further diploid clones (DC-14 and DC-25) and four tetraploid clone (TC-13, TC-16, TC-17 and TC-35) were isolated from DC 8, and their DNA content as assessed by flow cytometry with Hoescht staining is shown (passage 3).
H) LOH states of early (passage 5) diploid and tetraploid clones analysed by SNP6.0. Dark-blue LOH events in tetraploid clones are likely to have occurred prior to genome doubling. A barplot depicting the proportion of the genome displaying LOH is shown above, the black dotted line depicts the mean proportion LOH in diploids. The majority of LOH events are present in both diploid and tetraploid clones.
I) A family tree depicting all diploid and tetraploid clones used in this study. Tetraploid clones are shown in red; diploid clones in blue.
Next, we applied an algorithm that identifies tumours that are likely to have undergone a genome-doubling event, even if they are no longer polyploid (adapted from (7), see Methods). Significantly higher wGIIs were observed in tumours classified as genome-doubled compared to non genome-doubled (P<0.0001, Student’s T-test, Fig. 1A), suggesting a potential relationship between genome doubling and genome complexity. The majority of tumours with a triploid karyotype appeared to have undergone a genome-doubling event (105/110 tumours). However, we also observed non-genome doubled near-diploid tumours with high wGIIs, consistent with there being multiple routes to an unstable genome in CRC.
Genome doubling is an early event in the majority of CRCs
Copy number losses occurring on the background of a diploid genome will result in loss of heterozygosity (LOH). This LOH will leave a permanent footprint in the genome, persisting after a genome-doubling event (Fig. 1Bi). By contrast, losses occurring after genome doubling are less likely to exhibit LOH (Fig. 1Bii). The types of losses in a genome-doubled sample may thus shed light on the timing of genome doubling relative to copy number losses in the genome (Fig. 1B and see Methods). In the majority of genome-doubled TCGA CRC tumours, genome doubling likely occurred as a relatively early event, prior to the majority of copy number losses (Fig. 1C). In over 20% of samples where genome doubling was classified as an early event, the proportion of the genome exhibiting LOH was less than the mean proportion of LOH in chromosomally stable tumours with microsatellite instability (MIN). These data suggest that CIN can occur after genome doubling in vivo.
An isogenic cell line system to study the effects of tetraploidy
We next explored the acute effects on genome stability following a genome-doubling event. HCT-116, a diploid MIN CRC cell line, was found to have a small sub-population (<2%) with >4N DNA content (Fig. 1D). Single tetraploid and diploid cells were isolated by flow cytometry. The cloning efficiency of tetraploid cells was lower than diploid cells, suggesting tetraploidy is poorly tolerated in HCT-116 cells under standard culture conditions (2N= 63%, 4N= 6%, Fig. 1E). We expanded one diploid clone (DC 8) and two rare surviving tetraploid clones (TC 3, TC 4, Fig. 1F). It was possible to isolate a second generation of tetraploid clones (TC-13, TC-16, TC-17, TC-35) as well as two diploid clones (DC-14, DC-25) from the diploid clone DC 8 (Fig. 1G). Second-generation tetraploid clones had therefore arisen from a single diploid cell spontaneously within the time of the experiment. All tetraploid clones were found to have a seemingly functional p53 response to DNA damage (Supplementary Fig. 1B), and no mutations were found in the coding regions of TP53 or CDKN1A (p21) (data not shown), suggesting alternative tolerance routes. Although tetraploids grew slower than diploids at early passages, they grew at approximately the same rate by later passages (approximately 18 months in culture, Supplementary Fig. 2A-C).
All clones were subject to SNP6.0 analysis, and the proportion of the genome showing LOH was determined (Fig. 1H). Tetraploid clones showed limited to no LOH beyond that harboured by the diploid clones (diploid mean: 6.17% (5.96–6.36%) of genome; tetraploid mean 6.46% (6.16-6.79%), P=0.181, Kolmogorov-Smirnov test), suggesting tetraploid clones were not CIN before genome doubling. Analysis of copy numbers gains also showed that genome doubling likely occurred prior to CIN (data not shown). All clones (Fig. 1I) were continuously cultured for over 18 months so the effects of genome doubling on genome complexity could be assessed over time.
Chromosomal instability in tetraploid clones
Clonal FISH (fluorescence in-situ hybridization) was used to assess ploidy and numerical CIN in each clone (Fig. 2A, 2B). The cell-to-cell variation in chromosome number (percentage of cells deviating from the modal chromosome number of individual colonies) provides a measure of numerical CIN emerging during colony expansion from single cells. Tetraploid colonies displayed significantly higher cell-to-cell variation than diploid colonies (Fig. 2C: passage 5 diploid mean=7% (0-23%); tetraploid mean=28% (5-57%), P<0.0001; passage 50 diploid mean=13% (3-34%); tetraploid mean=33%, (7-68%), P<0.0001, Student’s T-test). We also isolated diploid and tetraploid clones from a microsatellite competent clone of HCT-116, stably expressing a functional wild-type MLH1 gene (referred to as HCT-116_MLH1) (24) (Supplementary Fig. 2D). All four HCT-116_MLH1 tetraploid clones exhibited higher cell-to-cell variation in chromosome number than the diploid clone at passage 5 (P<0.0001, Student’s T-test, Supplementary Fig. 2E).
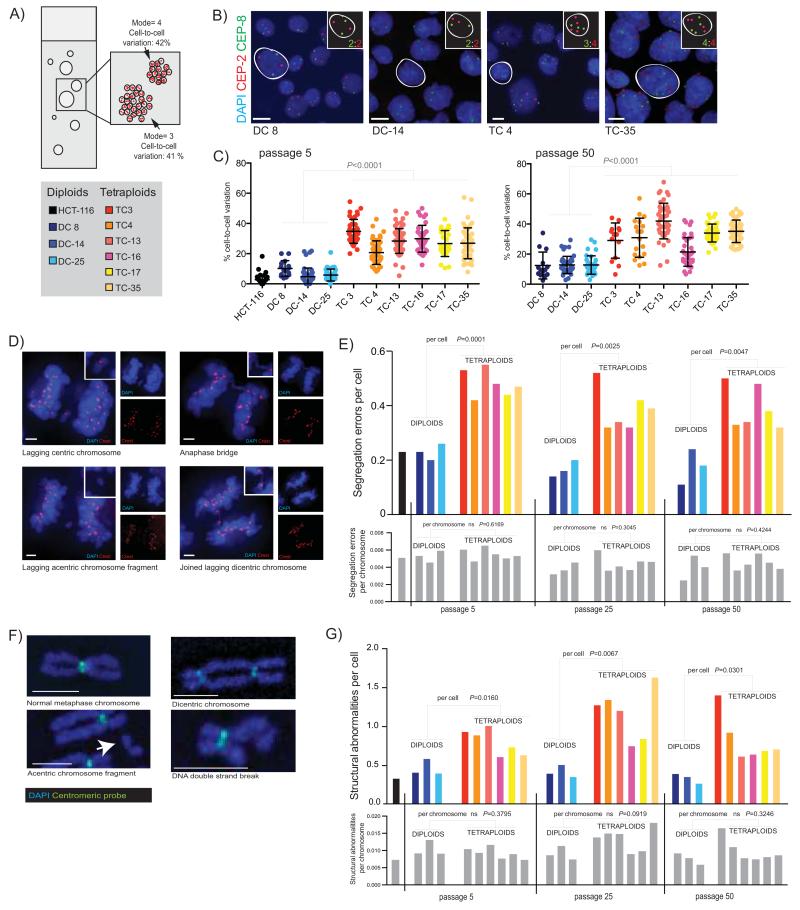
Figure 2.
A) Diagram of a clonal FISH slide, showing the two measures of chromosome number deviation that can be scored; cell-to-cell variation in chromosome number is the percentage of cells that deviate from the modal chromosome number of each individual colony. Colony-to-colony variation reflects differences in the modal chromosome copy number between colonies.
B) Example images of colonies from four clones with chromosome 2 (CEP2) shown in red, and chromosome 8 (CEP8) in green. Individual cells have been highlighted, and their copy number state for these two chromosomes is shown in the inset. Scale bar (in white) = approx.10μm.
C) Cell-to-cell variation in chromosome number. The average percentage deviation of two chromosomes, chromosome 2 and 8, is shown for all clones at both passage 5 and passage 50 (clones all scored using an Ariol automated microscope system, except DC 8, TC 3 and TC 4 at passage 50 which were scored using a DeltaVision microscope). Passage numbers shown throughout are correct to within 4 passages. Colonies with <10 cells were excluded from analysis. Each point represents one colony. Median number of cells: passage 5= 2479, passage 50= 2105.
D) Chromosome segregation errors in anaphase were visualized by immunofluorescence. Representative single z-stack images show types of segregation errors that were scored. Blue=DAPI, Red=Crest. Side panels show each channel individually, and inset shows a close-up of the segregation error. Scale bars (in white) = approx. 3μm.
E) Chromosome segregation errors on a per cell and per chromosome basis. Fifty anaphases were scored for each cell line; only data from bipolar anaphases is shown. On a per cell basis are shown on top graph (coloured bars represent individual clones, see key above). P values refer to comparisons between diploids and tetraploids at each passage (Student’s T-test). On a per chromosome basis (lower graph, all grey bars, representing the same clones as in immediately above graph) there is no significant difference in segregation errors per chromosome: P values are indicated above bars.
F) Representative images of normal and abnormal metaphase chromosomes are shown, stained with DAPI and probed with an all-human centromere probe. Scale bars (in white) = approx. 2.5μm.
G) Structural abnormalities on a per cell and per chromosome basis in diploid and tetraploid clones. Number of structural abnormalities per cell is shown on the top colored graph (P values refer to comparisons between diploids and tetraploid clones at each passage), and the number of structural abnormalities per chromosome is shown on the below graph with grey bars representing exactly the same clones as in above graph (P values for comparisons between diploid and tetraploids at each passage are indicated above bars). Median number of spreads scored at each passage: passage 5 = 25, passage 25 = 29, passage 50 = 27, and HCT-116 = 37.
We analysed segregation errors on a per cell and a per chromosome basis using immunofluorescence (Fig. 2D). Tetraploid cells exhibited a higher percentage of anaphases displaying segregation errors than diploids across three different passages (diploid mean=19% (11-26%); tetraploid mean=42% (32-55%), P<0.005, Student’s T-test, Fig. 2E, upper graph). However, segregation errors calculated on a per chromosome basis (dividing fraction of anaphases with errors by number of chromosomes for each clone as determined by SNP6.0 data, see Methods) were not significantly increased in tetraploid clones at any passage (Fig. 2E, lower graph). Therefore, increased segregation errors in tetraploid cells may simply result from the increased number of chromosomes. Tetraploid clones displayed a similar spectrum of segregation errors to diploid clones, including acentric and centric lagging chromosomes and anaphase bridges (Supplementary Fig. 3A, 3B). These data suggest that there may not be a tetraploid specific mechanism driving increases in any one type of segregation error in this system.
The prevalence of structural chromosome aberrations in all clones was scored from metaphase chromosome spreads hybridised to an all-centromere fluorescent probe (Fig. 2F). Tetraploid clones displayed more structural abnormalities per cell than diploid clones (Fig. 2G, diploid mean: 0.39 (0.26-0.58) abnormalities; tetraploid mean: 0.93 (0.60-1.62) abnormalities, all passage P<0.05, Student’s T test). However, no significant difference in structural abnormalities on a per chromosome basis between diploids and tetraploids was observed (chromosomes were counted directly from metaphase spreads, Fig. 2G, abnormalities per chromosome: diploid mean=0.0088 (0.0058-0.0130); tetraploid mean=0.0108 (0.0073-0.0180), P=0.1093, Student’s T-test).
Tolerance of chromosomal instability in tetraploid clones
We hypothesised that despite no increase in CIN in tetraploid clones on a per chromosome basis (Fig. 2E, 2G), increased tolerance to chromosomal segregation errors might contribute to the association between ploidy and genomic complexity (Fig. 1A). In clonal FISH experiments, the presence of colonies with modes differing from either two for diploid, or four for tetraploid clones (colony-to-colony variation) indicates tolerance of an unbalanced aneuploid genome. Individual colonies with aneuploid chromosome numbers were observed in all tetraploid clones (Fig. 3A, 3B, passage 5 mean=25% of colonies (8-44%); passage 50 mean=30% (18-43%)). In contrast, we only observed a single aneuploid colony in HCT-116 (1.7% of all colonies). One tetraploid colony was observed in a diploid clone, consistent with our findings that tetraploid cells can emerge as a rare event in diploid clones (Fig. 1G). The outcome of this assay is unlikely to be affected by differing proliferation rates, since colonies are grown from sparsely seeded single cells. These data suggest that tolerance of aneuploidy is enhanced in tetraploid clones, but a rare event in diploid cells. HCT-116_MLH1 tetraploid clones also displayed similar colony-to-colony variation in modal chromosome number (Supplementary Fig. 3C).
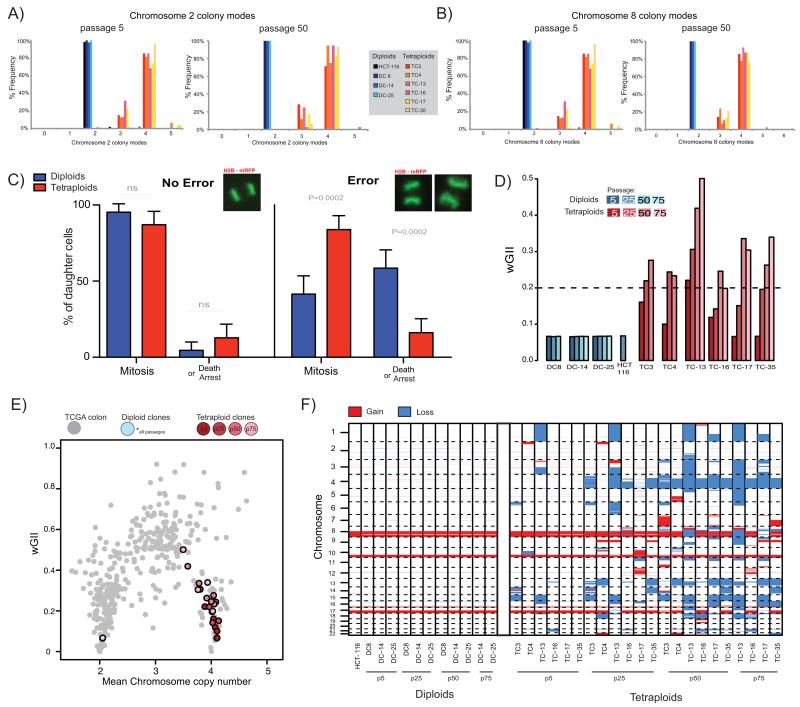
Figure 3.
A, B) Colony-to-colony variation in modal chromosome copy number for chromosome 2 (A), and chromosome 8 (B). Frequency of different colony modes from clonal FISH data is shown from all clones at passage 5 and at passage 50. Median number of colonies scored: passage 5 = 44, passage 50 = 39.
C) Live-cell imaging of H2B-mRFP expressing cell reveals different daughter cell-fates after segregation errors. The percentage frequency of each cell fate (mitosis or death or arrest [arrest = interphase >48hrs post division, see Methods]) in long-term live-cell imaging studies of all diploid and tetraploid clones either after no error or after a segregation error is shown. Example images of mitoses are shown above each panel. Data shown is an amalgamation of all clones (for individual results and n numbers see Supplementary Fig. 4G, and also see Supplementary Movies A-F).
D) wGII at different passages for diploid and tetraploid clones. Dashed line indicates wGII=0.2, a threshold separating MIN and CIN cell lines (23).
E) Weighted mean chromosome copy number versus wGII for CRC tumours from TCGA (grey), diploid clones (blue) and tetraploid clones (red) at different passages. Diploid clones at all passages overlay the same point. Lighter colours represent later passages for tetraploid clones.
F) Genome-wide copy number losses and gains for all clones at passage 5, 25, 50 (and passage 75 for DC-14, DC-25, TC13, TC-16, TC17 and TC-35). Blue sections represent loss and red sections represent gain (relative to ploidy).
We used live-cell imaging to track the fate of histone2B mRFP tagged cells following a chromosome segregation error. In diploid clones, daughter cells, derived from a parental cell that had undergone a segregation error, frequently died or underwent cell-cycle arrest (death or arrest - diploid mean: 58%, HCT-116: 43%, DC 8: 68%, DC-14: 55%, DC-25: 68%, Fig. 3C) whilst almost all cells that underwent a normal division continued through a subsequent mitosis (Fig. 3C, Supplementary movies A-F and Supplementary Fig. 4A-G). However, daughter cells arising from tetraploid clones after a segregation error (including anaphase bridges and lagging chromatin – data not shown), died or arrested less frequently, with the majority continuing through to a normal mitosis in the subsequent cell cycle (death or arrest – tetraploid mean: 16%, TC 3: 18%, TC 4: 12%, TC-13: 11%, TC-16: 10%, TC-17: 34%, TC-35: 12%, P=0.0002, Student’s T-test, Fig. 3C). These data indicate that tetraploid progeny have a greater tolerance of chromosome segregation errors relative to diploids.
We used SNP6.0 data for all clones at multiple different passages over 18 months and calculated genomic complexity using the wGII. Overall, wGII significantly increased from passage 5 to passage 75 in tetraploid clones, but remained stable in diploid clones (One way ANOVA test for passage 5, 25 and 50, tetraploids P=0.0002, diploids P=0.5907, Fig. 3D). We observed that late passage tetraploid clones had wGIIs similar to those of genomically complex polyploid CRCs in the TCGA data set (Fig. 3E). Changes in tetraploid clones over time appear to fit a model of genome doubling occurring as a precursor to a complex triploid karyotype, commonly observed in CRC (Fig. 1A). This is also supported by flow cytometry data, showing a reduction in ploidy over time (Supplementary Fig. 5).
Tetraploid clones evolve specific chromosome losses
In every clone chromosomal gains and losses relative to the cell line ploidy were assessed over long-term culture (Fig. 3F). Chromosomal aberrations present in parental HCT-116 were observed in all clones. No novel losses were common to all early passage tetraploids. However, a non-contiguous region of chromosome 4q containing 362 genes was lost to three copies in all tetraploid clones by passage 50 (approximately 1 year), consistent with selection for loss of this region during prolonged culture (Fig. 3F, and Supplementary Table 1). Copy number loss of chromosome 4q occurs after genome doubling as it did not occur before passage 25 in any clone, and does not display LOH (Fig. 3F, and Supplementary Fig. 6A). A similar pattern of chromosome losses and increasing genome complexity was found in HCT-116_MLH1 clones (Supplementary Fig. 6B). This suggests that elevated genome complexity and selection of chromosome 4q loss in HCT-116 tetraploid clones is not driven by MIN.
We assessed the correlation between loss of genes on chromosome 4q and wGII in the TCGA data set, controlling for the increased likelihood of loss in unstable tumours (see Methods). Loss of genes on chromosome 4q was significantly correlated with increasing wGII (P<0.001, Fig. 4A), and chromosome 4 was one of the chromosomes whose loss was most strongly correlated with genomic instability (Supplementary Fig. 7).
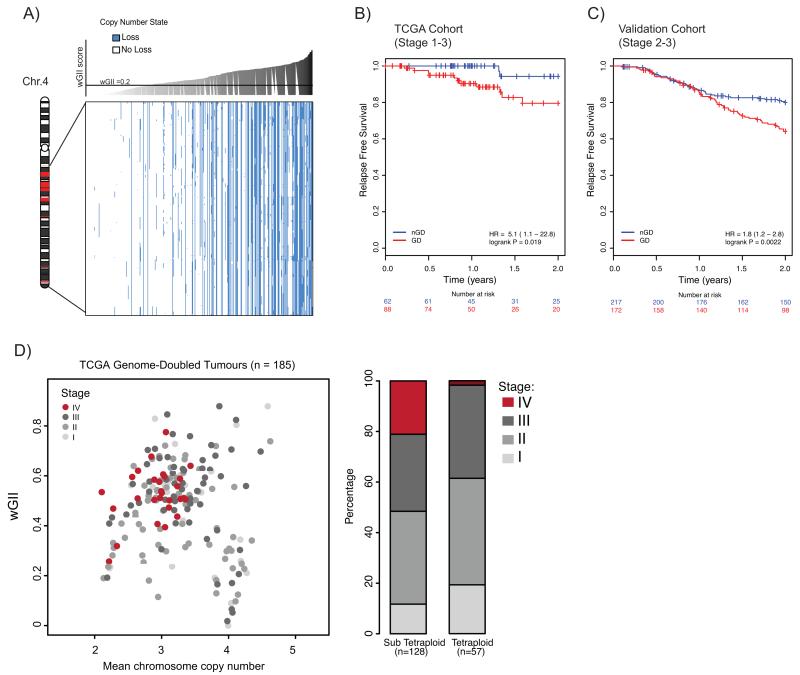
Figure 4.
A) Relationship between copy number loss and wGII in TCGA cohort for genes identified as recurrently lost in tetraploid clones (see Supplementary Table 1). Blue represents loss and white represents no loss (relative to ploidy). CRC tumours (columns) are ordered according to increasing wGII score, from left to right. Every gene (rows) shows a significant correlation between copy number loss and wGII, even when taking into account an increased likelihood of loss in high wGII tumours (P<0.001; see Methods). Chromosome schematic shows where genes reside on chromosome 4; genes within regions shown in red are not depicted in the plot as they are not recurrently lost in all tetraploid clones.
B) Kaplan Meier relapse free survival curves, censored at 2 years for genome-doubled (GD; red) and non-genome doubled (nGD; blue) TCGA cohort CRC tumours (n=150). P=0.019, log-rank test (for full survival curves see Supplementary Fig. 8A).
C) Kaplan Meier relapse-free survival curves, censored at 2 years for genome-doubled (GD; red) and non-genome doubled (nGD; blue) validation cohort CRC tumours (n=389). P=0.0022, log-rank test (for full survival curves see Supplementary Fig.8B).
D) Relationship between wGII, ploidy and tumour stage in genome-doubled tumours. Each circle represents one genome-doubled tumour. The barplot shows the proportion of different tumour stages for tetraploid and sub-tetraploid samples. P=0.0062, Cochrane-Armitage test for trend.
Genome doubling is associated with poor prognosis in CRC tumours
Given the established relationship between genomic instability and poor clinical outcome, we reasoned that genome doubling could be a useful prognostic marker in early stage CRC. Survival data was obtained for 150 stage 1-3 CRC TCGA patients and an Australian validation cohort, comprising 389 stage 2-3 CRC patients. Relapse-free survival was used as the endpoint for both datasets, censored at 2 years, given that 80% of recurrences occur within this period (25), and also due to the paucity of survival data available in the TCGA cohort beyond this point. A genome-doubling event was significantly associated with relapse in both the TCGA (Fig. 4B, P=0.019, hazard ratio (HR)=5.1, 95% CI 1.1-22.8) and the validation cohort (Fig. 4C, P=0.0022, HR=1.80, 95% CI 1.2-2.8). When extending outcome to beyond 2 years, genome doubling remained significant in the larger validation cohort, but not the TCGA cohort (Supplementary Fig. 8). Genome doubling remained significant in both cohorts in multivariate analysis when tumour stage, age and MSI status were included (TCGA data: P=0.045, HR=4.70, 95% CI 1.04-21.37, validation data: P=0.028, HR=1.59, 95% CI 1.05-2.42, Supplementary Table 2).
By contrast, wGII was only significant in univariate analysis for one of the cohorts (TCGA data: P=0.1296, HR=6.09, 95% CI 0.57-64.93; validation data: P=0.00649, HR=3.81, 95% CI 1.44-10.03), and was not significant in multivariate analysis when including tumour stage, age and MSI status in either cohort (Supplementary Table 2). In the larger validation cohort, genome doubling was significant when restricting to just diploid tumours (P=0.001, Supplementary Fig. 8C), and also when including polyploidy (ploidy ≥ 3) in a multivariate analysis (P=0.0209, Supplementary Table 2). A genome-doubling event may therefore provide prognostic relevance with a greater sensitivity than aneuploidy to detect high-risk tumours. Furthermore genome doubling was significant in predicting overall 5-year survival when restricting to just early stage 1-2 tumours (available for the TCGA cohort only, Supplementary Table 2).
Within genome-doubled tumours, sub-tetraploid (ploidy <4) samples were genomically more complex than tetraploid samples, and enriched for higher tumour stage (Fig. 4D, P=0.0062, Cochran-Armitage test). This supports a model where genome doubling is an early event in some CRCs, permitting the evolution of more genomically complex, sub-tetraploid, higher stage tumours.
Discussion
Through long-term culture of naturally occurring, rare surviving tetraploid clones, we observed the evolution of genomic complexity specifically in tetraploid genomes over time. A year post genome doubling, the HCT-116 genome was markedly altered relative to the genomically stable diploid progenitors, suggesting that tolerance of genome doubling permits rapid genome evolution (Fig. 3F).
Compton and colleagues have shown that increasing the segregation error rates in diploid MIN CRC cells does not result in the propagation of chromosomally unstable progeny, suggesting that CIN is a complex phenotype requiring both initiation of segregation errors and tolerance of the ensuing altered genomic content in daughter cells (4). Although tetraploid cells displayed elevated structural and numerical CIN relative to diploid cells, diploids and tetraploids had similar segregation errors and structural abnormalities on a per chromosome basis (Fig 2E, 2G). These data suggest that there is no additional instability initiated by tetraploidisation on a per chromosome basis in our system, in which segregation defects are already observed in diploid cells. However, long-term live-cell imaging revealed increased tolerance to chromosome missegregation events in tetraploids (Fig 3C), and we observed many aneuploid colonies in tetraploid clones (Fig. 3A). Thus, in rare tetraploid cells that survive cloning, the same MIN cell line that cannot propagate artificially induced (4) or endogenous segregation errors is now able to propagate an unstable genome.
Within the limits of SNP6.0 based analyses we conclude that tetraploid clones were unlikely to have been CIN before genome doubling (Fig. 1H), however it is unlikely that a genome-doubling event alone is sufficient for cells to tolerate CIN. It is possible that chromosome segregation error tolerance mechanisms can be activated in diploid cells that may or may not undergo genome doubling. Whole genome doubling may therefore represent a consequence of a prior somatic event permitting the tolerance of genome instability, which can then exacerbate genomic evolution due to elevated chromosomal aberrations occurring on a per cell basis. Defining the mechanistic basis for such tolerance mechanisms in the presence of wild-type p53 is clearly a high priority.
Strikingly, all tetraploid clones displayed convergent loss of regions of chromosome 4q, which we also found to be commonly lost in CRC tumours with elevated genomic complexity (Fig. 4A). Interestingly chromosome 4q loss has previously been suggested as a predictor of outcome in early stage CRC (26). Conceivably, tetraploidy can provide a permissive genetic background for selection of high-risk genomic copy number aberrations over time.
Consistent with the effect of tetraploidisation upon emerging genome instability, a genome-doubling event is an independent predictor of poorer relapse-free survival in CRC from the TCGA and in a larger validation cohort in both univariate and multivariate analyses (Fig. 4B, 4C, Supplementary Table 2). These data support studies that have linked aneuploidy with disease outcome in CRC (27), and genome doubling with poor prognosis in ovarian cancers (7). Genome doubling may forecast the onset and tolerance of elevated CIN, which has previously been shown to be associated with both poor prognosis and intrinsic drug resistance (2, 3).
A genome-doubling event could represent a macro-evolutionary leap in tumours, analogous to saltation in ecology, which both precipitates and sustains extensive chromosomal rearrangements. Other examples of punctuated evolutionary events occurring in tumours have been proposed, such as chromothripsis and chromoplexy, both involving complex chromosomal rearrangement events (28, 29). Whole genome-doubling events can drastically alter the evolution of whole organisms, for example by facilitating sub-functionalisation of duplicated genes (30), so there may be additional benefits to a genome-doubling event beyond the propagation of CIN. It will be important to investigate mechanisms leading to the emergence of tetraploid cells in tumours, such as cellular stress, cytokinesis failure or telomere shortening (8).
On the basis of these data we suggest that the tolerance of genome doubling combined with an elevated chromosome segregation error rate on a per cell basis provides the fuel for rapid genomic change, accelerating evolution of tumours from a karyotypically stable to a more complex state. Deciphering the cause of CIN tolerance is likely to have important therapeutic implications in guiding efforts to limit tumour diversity, evolution and adaptation.
Materials and Methods
Cell culture
HCT-116 was obtained from ECACC by CRUK cell services, and STR fingerprinted on 10/10. HCT-116_MLH1.3 clone was a gift of Françoise Praz, and was STR fingerprinted on 02/10, with similar results to HCT-116 except at vWA. Cells were maintained at 37°C in 5% CO2 in DMEM High Glucose with L-Glutamine (Invitrogen), with 10% FBS and 1×PenStrep (Sigma). Clones were passaged approx. once a week and were split at the same dilution. Passage numbers represented in figures are within <4 passages of passage used. CellTiter-Blue® (Promega) assays were performed following the manufacturers instructions. Cloning efficiency was estimated using the Poisson distribution: efficiency = (−100) * ln(#of wells with no colony / total #of wells)% (as in (31)).
FACS
Cells were stained with 10μg/ml Hoescht 33342 (Sigma) for 1hr at 37°C. A MofLo (Beckman Coulter) cell sorter was used to sort single cells into 96 well plates with 20% FBS media. Assessment of DNA content was carried out by flow cytometry using Propidium Iodide (Sigma) with RNase (Life Technologies), or Hoescht 33342, after fixation in 70% Ethanol.
Clonal FISH, metaphase spreads & immunofluorescence
FISH probing for chromosomes 2 (CEP2 D271, SO), and 8 (CEP8, D872, SGn, both Abbott Molecular probes) was performed as described (23). Slides were scored semi-automatically using the Ariol system (Leica Microsystems). Colonies were scanned at 40× magnification with z-stacks of 9×0.7μm, and analysed with the automated SPOT assay before manual curation. Three slides were scored using an Olympus DeltaVision RT microscope (Applied Precision, LLC) equipped with a Coolsnap HQ camera with an Olympus ×40 1.3 numerical aperture UPlanSApo oil immersion objective. Metaphase spreads were prepared and probed with an all-human centromere probe (Posiedon) as described (23). Immunofluorescence and segregation error classification were performed as described (23).
H2B-mRFP transfection and live-cell imaging analysis
Cells were transfected with pH2B-mRFP (gift from A. Straube) using Fugene 6.0 (Promega), and selected in 1 mg ml−1 G418 (Life technologies) before flow-sorting for mRFP expression. Cells were maintained in 500μg ml−1 G418 and imaged in an 8-well chamber (LabTek) using the same DeltaVision microscope in 5% CO2 atmosphere at 37°C. 14μm z-stacks were taken every 3mins for 6hrs and every 15mins thereafter, for approx. 60 hours, and analysed using Softworx Explorer (Applied Precision, LLC). Cells were scored as arrested if they failed to divide within 48 hours of the parental division. Multipolar divisions were excluded from analysis.
SNP array processing
Cell-lines were analyzed with Affymetrix SNP6.0 arrays. PICNIC (32) was used for normalization and integer copy number estimation.
TCGA Affymetrix SNP6.0 data were downloaded for 422 CRC, 898 Breast, 391 Lung (Ad), 407 Lung (SC), 506 Ovarian and 503 Renal cancers (http://tcga.data.nci.nih.gov/tcga/). Samples that failed Affymetrix Genotyping Console QC were excluded. LogR and BAFs were obtained using the aroma R package (33). Integer copy numbers were estimated using OncoSNP (34).
Validation cohort analysis was performed on Illumina 610 Quad arrays. LogRs and BAFs were obtained using GenomeStudio V2011.1 and Genotyping Module V1.9.4. For QC, samples with moving standard deviation >0.28 were discarded. Integer copy numbers were estimated using OncoSNP (34).
Ploidy was estimated for each sample by summing the weighted median integer copy for each chromosome and dividing by number of chromosomes analyzed (n=22). The number of chromosomes in each sample was estimated by summing the modal copy numbers from the segmented copy number profile of each chromosome. Each segment was weighted according to the number of base pairs it covered. Copy number segments of loss and gain were defined relative to ploidy. wGII was calculated as in (23).
Validation cohort
Validation cohort CRC patients were recruited from the Royal Melbourne Hospital, Western Hospital Footscray and St Vincent’s Hospital Sydney in Australia. The study was ethics approved, and patients gave informed consent.
Genome Doubling Algorithm
A modified version of a published algorithm (7) was used. Each sample, s, was represented as an aberration profile of major and minor allele copy numbers at chromosome arm resolution. From this profile, we calculated Ns, the total number of aberrations (relative to diploid) and Ps, the probabilities of loss/gain for each allele at each chromosome arm. 10,000 simulations were run for each sample s. In each simulation, Ns sequential aberrations, based on Ps, were applied to a diploid profile. A p-value for genome doubling was obtained by counting the percentage of simulations where the proportion of chromosome arms with a major allele copy number ≥2 was higher than that observed in the sample. For samples with ploidy ≤3, a p-value threshold of 0.001 was used. To avoid underestimating genome doubling in high-ploidy samples, p-value ≤ 0.05 was used if ploidy=4, and all samples where ploidy≥5 were classified as genome-doubled.
Estimating Timing of Genome Doubling
Each genome-doubled sample was represented as an array of genotype proportions reflecting copy numbers ranging from zero to eight. Sixteen possible genotypes were discriminated: zero copies, A|B (1 copy); AA|BB and AB (2 copies); AAA|BBB and AAB|ABB (3 copies); etc. (where A and B represent the two parental alleles). Only losses to two copies (AA, BB, AB) were used as these can either reflect losses before (AA or BB) or after genome doubling (AB). Samples with a higher proportion AB compared to AA/BB were classified as having genome doubled before the majority of losses, whereas those where AA/BB > AB were classified as having genome doubled after the majority of losses.
Significance of correlation between wGII and copy number loss
Based on the observed probability for loss, given by the percentage of genome that is lost in that sample, we generated an aberration state (loss or no loss) for each sample separately. A point-biserial correlation between aberration state and wGII was then calculated across samples. This process was repeated 10,000 times and a p-value was obtained for each gene by counting the percentage of simulations showing a greater correlation coefficient than that observed for that gene.
Statistical analyses
Survival curves were plotted according to the Kaplan Meier method. Log-rank test statistics were used to assess significance for univariate analysis. Cox proportional hazards regression models were conducted for multivariate survival analysis (R package survival). Survival times were censored at 2 years unless otherwise stated. All statistical analyses were carried out using the R statistical environment or GraphPad Prism.
Supplementary Material
Significance.
Our work sheds light on the importance of whole genome doubling events in colorectal cancer evolution. We show that tetraploid cells undergo rapid genomic changes and recapitulate the genetic alterations seen in unstable tumours. Furthermore we demonstrate that a genome doubling event is prognostic of poor relapse free survival in this disease type.
Acknowledgements
We thank the lab of Tim Hunt for the p21 antibody, Anne Straube for the H2BmRFP plasmid, Françoise Praz for the HCT-116_MLH1 cell line and we are very grateful to the cell services facility at CRUK.
We acknowledge the Victorian Cancer Biobank for the provision of patient specimens for the validation cohort and BioGrid Australia for providing de-identified clinical data.
The results published here are in part based upon data generated by the Cancer Genome Atlas pilot project established by the NCI and NHGRI (information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at hhtp://cancergenome.nih.gov/). The data were retrieved through dbGaP authorization (Accession No. phs000178.v5.p5).
Financial support
This project was funded by grants from Cancer Research UK (S.M.D, N.M, R.A.B, C.S), the Medical Research Council (A.J.R, C.S, ID:G0701935/2), EU Framework 7 (E.G, C.S, T.J, Z.S, projects PREDICT and RESPONSIFY), The Prostate Cancer Foundation (C.S), The Rosetree Trust (C.S), The Breast Cancer Foundation (C.S) and the NHMRC through a Project Grant (Application ID 489418; to O.M.S, P.G, R.L.W), and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Abbreviations
- CIN
Chromosomal instability
- MIN
Microsatellite Instability
- CRC
Colorectal cancer
- TCGA
The Cancer Genome Atlas
- FISH
Fluorescence in-situ hybridization
Footnotes
The authors declare no competing conflicts of interest.
References
- 1.Janssen A, Medema RH. Genetic instability: tipping the balance. Oncogene. 2012 doi: 10.1038/onc.2012.576. [DOI] [PubMed] [Google Scholar]
- 2.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–38. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–6. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 6.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davoli T, de Lange T. The Causes and Consequences of Polyploidy in Normal Development and Cancer. Annual Review of Cell and Developmental Biology. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 9.Shackney SE, Smith CA, Miller BW, Burholt DR, Murtha K, Giles HR, et al. Model for the genetic evolution of human solid tumors. Cancer Res. 1989;49:3344–54. [PubMed] [Google Scholar]
- 10.Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–7. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 11.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–66. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 13.Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27:337–43. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- 15.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nature genetics. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 16.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–76. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 18.Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A Virus Causes Cancer by Inducing Massive Chromosomal Instability through Cell Fusion. Current Biology. 2007;17:431–7. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen HG, Makitalo M, Yang D, Chinnappan D, St. Hilaire C, Ravid K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. The FASEB Journal. 2009;23:2741–8. doi: 10.1096/fj.09-130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob S, Aguado M, Fallik D, Praz F. The role of the DNA mismatch repair system in the cytotoxicity of the topoisomerase inhibitors camptothecin and etoposide to human colorectal cancer cells. Cancer Res. 2001;61:6555–62. [PubMed] [Google Scholar]
- 25.Hellinger MD, Santiago CA. Reoperation for recurrent colorectal cancer. Clinics in colon and rectal surgery. 2006;19:228–36. doi: 10.1055/s-2006-956445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosens RP, Belt EJ, Haan JC, Buffart TE, Carvalho B, Grabsch H, et al. Deletion of chromosome 4q predicts outcome in stage II colon cancer patients. Cellular oncology. 2011;34:215–23. doi: 10.1007/s13402-011-0042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araujo SE, Bernardo WM, Habr-Gama A, Kiss DR, Cecconello I. DNA ploidy status and prognosis in colorectal cancer: a meta-analysis of published data. Diseases of the colon and rectum. 2007;50:1800–10. doi: 10.1007/s10350-007-9013-6. [DOI] [PubMed] [Google Scholar]
- 28.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huminiecki L, Conant GC. Polyploidy and the evolution of complex traits. International journal of evolutionary biology. 2012;2012:292068. doi: 10.1155/2012/292068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leight ER, Sugden B. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Molecular and cellular biology. 2001;21:4149–61. doi: 10.1128/MCB.21.13.4149-4161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–75. doi: 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bengtsson H, Ray A, Spellman P, Speed TP. A single-sample method for normalizing and combining full-resolution copy numbers from multiple platforms, labs and analysis methods. Bioinformatics. 2009;25:861–7. doi: 10.1093/bioinformatics/btp074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yau C, Mouradov D, Jorissen RN, Colella S, Mirza G, Steers G, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome biology. 2010;11:R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.