Abstract
Amelogenesis imperfecta (AI) is a genetic disease affecting tooth enamel formation. AI can be an isolated entity or a phenotype of syndromes. To date, more than 10 genes have been associated with various forms of AI. We have identified 2 unrelated Turkish families with hypoplastic AI and performed mutational analysis. Whole-exome sequencing identified 2 novel heterozygous nonsense mutations in the ENAM gene (c.454G>T p.Glu152* in family 1, c.358C>T p.Gln120* in family 2) in the probands. Affected individuals were heterozygous for the mutation in each family. Segregation analysis within each family revealed individuals with incomplete penetrance or extremely mild enamel phenotype, in spite of having the same mutation with the other affected individuals. We believe that these findings will broaden our understanding of the clinical phenotype of AI caused by ENAM mutations.
Keywords: amelogenesis imperfecta, hypoplastic, enamel, tooth, enamelin, expressivity
Introduction
The outer surface of the human tooth is covered by dental enamel, which provides a beautiful appearance with a luster and enables efficient feeding with its exceptional hardness. To achieve such fine characteristics, enamel formation should be well coordinated throughout all the stages, including the presecretion, secretion, transition, and maturation stages. This coordination includes not just components of the extracellular matrix secreted by ameloblasts but also other genetic factors influencing the function of ameloblasts themselves.
Amelogenesis imperfecta (AI) is a group of hereditary diseases affecting tooth enamel formation. AI is heterogeneous in etiology as well as in phenotypes (Witkop, 1988). Genes encoding enamel matrix proteins have been suspected as primary candidates of AI, and indeed, disease-causing mutations have been identified in AI patients. Mutations in amelogenin (AMELX; MIM *300391; Kim et al., 2004), enamelin (ENAM; MIM *606585; Rajpar et al., 2001; Mardh et al., 2002), enamelysin (MMP20; MIM *604629; Kim et al., 2005b), and kallikrein 4 (KLK4; MIM *603767; P.S. Hart et al., 2004) have been identified by candidate gene approach. Genetic analysis approaches, such as linkage analysis, autozygosity mapping, and especially whole-exome sequencing, have identified new players in AI: family with sequence similarity 83 member H (FAM83H; MIM *611927; Kim et al., 2008), WD repeat-containing protein 72 (WDR72; MIM *613214; El-Sayed et al., 2009), family with sequence similarity 20 member A (FAM20A; MIM *611062; O’Sullivan et al., 2011), chromosome 3 open reading frame 26 (C4orf26; MIM *614829; Parry et al., 2012), solute carrier family 24 member 4 (SLC24A4; MIM *609840; Parry et al., 2013), laminin beta 3 (LAMB3; MIM *150310; Poulter et al., 2014b), and integrin beta 6 (ITGB6; MIM *147558; Poulter et al., 2014a; Wang et al., 2014).
Clinically, AI can be classified as hypoplastic, hypocalcification, and hypomaturation. Hypoplastic enamel is thin but hard in most cases. Hypocalcified enamel is very soft, and hypomatured enamel has reduced mineral density and brown discoloration. However, it is not possible or extremely difficult to determine the clinical phenotype exactly in some cases. Hypocalcified enamel is easily broken down after tooth eruption due to extreme softness, with the remaining surfaces being rough and discolored. In some cases, hypoplastic enamel occurs in combination with hypomaturation AI (Sundell and Valentin, 1986).
Hypoplastic AI without other systemic conditions is caused as an X-linked pattern by mutations in the AMELX gene or as an autosomal pattern by mutations in the ENAM, LAMB3, and ITGB6 genes. In this study, we recruited 2 hypoplastic AI families and identified 2 novel nonsense ENAM mutations. Most interesting, in both families, we identified incomplete penetrance or an extremely mild clinical phenotype even though these individuals carried the same mutations as each proband.
Materials & Methods
Enrollment of Human Subjects
Two unrelated Turkish families having hypoplastic AI were recruited for the genetic studies. The study protocol was independently reviewed and approved by the Institution Review Board at Seoul National University Dental Hospital and University of Istanbul. Clinical examinations were performed, and blood samples were collected with the understanding and written consent of each participant according to the Declaration of Helsinki.
Whole-exome Sequencing
Whole-exome sequencing was performed with DNA samples from the probands of the 2 AI families after exome capturing with NimblGen exome capture reagent (family 1) or Illumina TruSeq DNA sample prep kit (family 2); 75-bp (family 1) and 90-bp paired-end sequencing reads were obtained with Illumina HiSeq 2000 (Yale Center for Mendelian Genomics, West Haven, CT, USA; or Macrogen, Seoul, Korea). Sequencing reads were aligned to the NCBI human reference genome (NCBI build 37.2, hg19), and the sequence variations were annotated with dbSNP build 137.
Polymerase Chain Reaction and Sequencing
Identified variation in the ENAM gene was confirmed with Sanger sequencing, and segregation within each family was confirmed. DNA sequences of the primers and polymerase chain reaction (PCR) protocol were previously described (Kim et al., 2005a). PCR amplifications were done with the HiPi DNA polymerase premix (Elpis Biotech, Taejeon, Korea), and PCR amplification products were purified with a PCR Purification Kit and protocol (Elpis Biotech). DNA sequencing was performed at a DNA sequencing center (Macrogen).
Results
Family 1
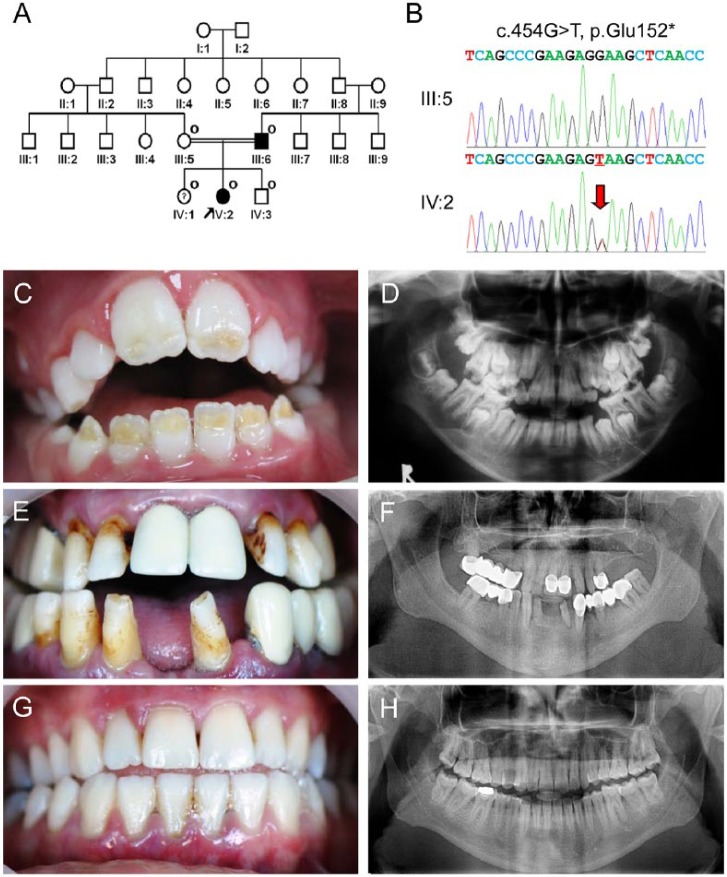
The proband was a ten-year-old girl presenting a localized form of hypoplastic AI from a consanguineous family (Fig. 1A). Whole-exome sequencing identified a novel guanine to thymine transversion in exon 7 of the ENAM gene (c.454G>T; Fig. 1B). There was no other rare or novel variant in other AI candidate genes, including AMELX, AMBN, MMP20, KLK4, FAM83H, C4orf26, WDR72, SLC24A4, CNNM4, and FAM20A. This mutation changes glutamic acid (GAA) to the ochre termination codon (TAA) at codon position 152 (p.Glu152*). Because this mutation introduces an early termination codon that is not located in the last exon, the mutant mRNA would be degraded by the nonsense-mediated decay system so that no truncated protein could be generated. Sanger sequencing of the participating family members confirmed the existence of this mutation in the father (III:6), proband (IV:2), and sister of the proband (IV:1). All participating family members had uneventful pregnancy and delivery, and there was no systemic condition that could influence amelogenesis. Clinical examination revealed that the proband had hypoplastic regions located on the incisal half of the anterior permanent teeth with anterior open bite occlusion (Fig. 1C). A panoramic radiograph showed that developing molars had hypoplastic enamel (Fig. 1D). The affected father had many crowns and bridges, but the remaining teeth showed horizontal hypoplastic enamel regions, which are considered as a characteristic feature of ENAM mutations (Fig. 1E, 1F). However, clinical examination could not detect any enamel defect related to the mutation in individual IV:1, who had the same ENAM mutation, even though she had several initial dental caries (both maxillary first molars and left mandibular first molar) and a restoration (right mandibular first molar; Fig. 1G, 1H).
Figure 1.
Clinical and mutational analysis of family 1. (A) Pedigree of family 1. The “O” symbol indicates members recruited for this study. The proband is indicated with a black arrow. The question symbol (?) indicates that this individual is clinically normal but has the mutation. (B) Comparison of the ENAM exon 7 sequencing chromatograms for the unaffected family member (III:5) with the wild-type (top) sequence, and the mutated allele in the proband (IV:2) reveals a G-to-T transversion: c.454G>T, p.Glu152*. A red arrow indicates the mutated nucleotide. (C) Frontal clinical photo of the proband. Yellow dentin color can be seen through the hypoplastic enamel located on the incisal half of the permanent anterior teeth. (D) Panoramic radiograph of the proband reveals unusual crown of the developing second permanent molars due to hypoplastic enamel. (E) Frontal clinical photo of the affected father shows the characteristic horizontal hypoplastic grooves. (F) Panoramic radiograph of the affected father shows multiple prosthetics and thin enamel in remaining teeth. (G) Frontal clinical photo of a sister of the proband. Despite having the mutation, her dentition has no enamel defects. (H) Panoramic radiograph of a sister of the proband reveals normal-looking tooth structures.
Family 2
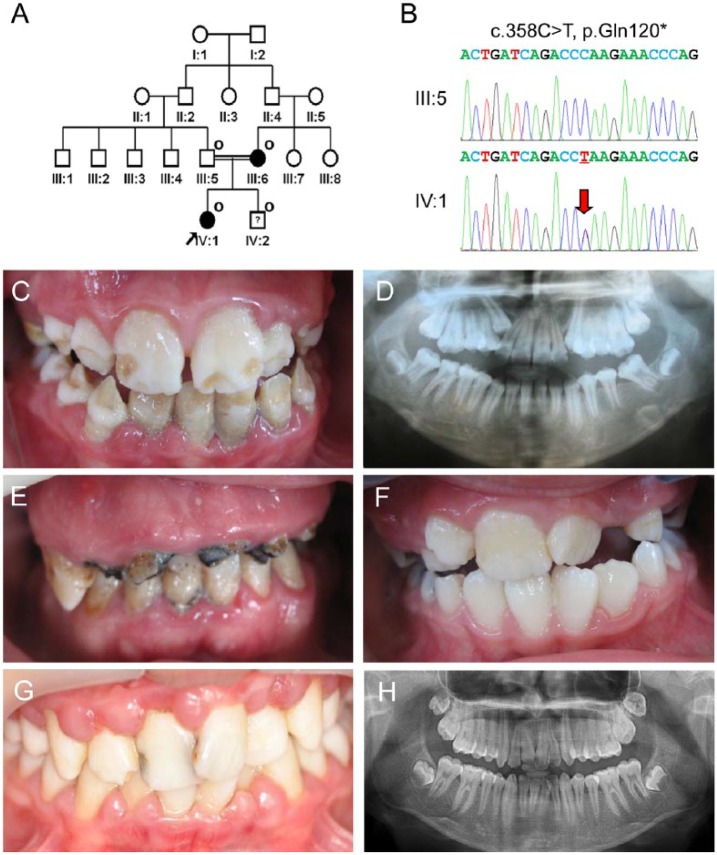
The proband was an 11-year-old girl from a consanguineous marriage, who presented a localized form of hypoplastic AI (Fig. 2A). All participating family members had uneventful pregnancy and delivery, and there was no systemic condition. Whole-exome sequencing identified a novel cytosine-to-thymine transition in exon 7 of the ENAM gene (c.358C>T; Fig. 2B). There was no other rare or novel variant in other AI candidate genes, including AMELX, AMBN, MMP20, KLK4, FAM83H, C4orf26, WDR72, SLC24A4, CNNM4, and FAM20A. This mutation changes glutamine (CAA) to the ochre termination codon (TAA) at codon position 120 (p.Gln120*). This nonsense mutation is also predicted to cause degradation of the mutant transcript, as in the case of family 1. The mutation was identified in the mother (III:6) as well as the brother of the proband (IV:2). The proband had a generalized thin enamel with some thick enamel in the middle of the mandibular permanent teeth and localized hypoplastic regions on the incisal half of the maxillary permanent teeth (Fig. 2C, 2D). The affected mother had only several remaining teeth with gross destruction of the tooth structures (Fig. 2E). The brother of the proband had completely normal-looking mandibular permanent teeth; therefore, he was considered as a normal individual at the initial screening. The only abnormal enamel found was the regions on the incisal corners of the maxillary right lateral incisor, which resembled enamel fractures or dental caries (Fig. 2F). At age 14 yr, poor oral hygiene resulted in dental caries in the maxillary central incisors and gingivitis (Fig. 2G), but panoramic radiograph revealed normal-looking tooth structures (Fig. 2H).
Figure 2.
Clinical and mutational analysis of family 2. (A) Pedigree of family 2. The “O” symbol indicates members recruited for this study. The proband is indicated with a black arrow. The question symbol (?) indicates that this individual is clinically normal but has the mutation. (B) Comparison of the ENAM exon 7 sequencing chromatograms for the unaffected family member (III:5) with the wild-type (top) sequence, and the mutated allele in the proband (IV:1) reveals a C-to-T transition: c.358C>T, p.Gln120*. (C) Frontal clinical photo of the proband shows a localized form of enamel regions with some normal-looking areas. (D) Panoramic radiograph of the proband shows an irregular form of crowns. (E) Frontal clinical photo of the affected mother. She has several remaining teeth with gross destruction. (F) Frontal clinical photo of a brother of the proband at age 8 yr. He has normal-looking teeth, even though he also has the mutation. However, he has minor enamel defects, resembling an enamel fracture or dental caries, on the incisal corners of the maxillary right lateral incisor. (G) Frontal clinical photo of a brother of the proband at age 14 yr. Dental caries of maxillary central incisors and gingivitis can be seen. (H) Panoramic radiograph of a brother of the proband at age 14 yr.
Discussion
Enamelin, one of the major proteins in the enamel matrix, is essential for proper enamel formation. A knockout mouse model demonstrated deficient enamel mineralization, resulting in no true enamel but an irregularly mineralized thin layer covering normal dentin (Hu et al., 2008). The dosage effect of enamelin was noted with the recessive ENAM mutations (T.C. Hart et al., 2003) and was further demonstrated by enamelin overexpressing transgenic mouse on the knockout background, indicating that an adequate quantity of enamelin is essential for normal enamel formation (Hu et al., 2014).
Enamelin gene has been associated with dental caries susceptibility and recently identified as one of the genes contributing molar-incisor hypomineralization (Jeremias et al., 2013; Chaussain et al., 2014). Even though specific structural change(s) or genetic element(s) has not been identified yet, it seems obvious that sequence variations and allelic predilection are related to caries susceptibility or molar-incisor hypomineralization.
Unlike other genes involved in AI, ENAM mutations frequently exhibit wide variable expressivity (even within a family), from minor enamel pits to severe enamel loss, where virtually no enamel is remaining (Kida et al., 2002; Chan et al., 2011). It has been also noted that the same ENAM mutation results in different clinical phenotypes among families (P.S. Hart et al., 2003). Phenotypic variation within family and between families is also shown in this study in spite of the similar nature of the mutations. A characteristic clinical feature frequently associated with ENAM mutations is horizontal hypoplastic grooves on the labial surface of the crown. The affected father of family 1 and the proband of family 2 also exhibit horizontal hypoplastic grooves.
Through the identification of recessive ENAM mutations, localized small enamel pits have been recognized as a milder phenotype of AI in heterozygous individuals (Table). Individuals with ENAM mutations in both alleles exhibited severe generalized hypoplastic AI with open bite, indicating a dosage-dependent effect of the mutation (T.C. Hart et al., 2003). Interestingly, the same mutation has been identified in other autosomal-dominant AI families with characteristic horizontal hypoplastic grooves, confirming the variable expressivity of ENAM mutations (Pavlic et al., 2007).
Table.
Disease-causing Mutations in ENAM Gene
| Location | cDNA | Protein | Inheritance Mode | References |
|---|---|---|---|---|
| Exon 4 | c.107delA | p.Asn36Ilefs*22 | AD | Simmer et al., 2013 |
| Exon 5 | c.157A>T | p.Lys53* | AD | Mardh et al., 2002; Kim et al., 2006 |
| Intron 6 | c.211-2A>C | p.Met71-Gln157del | AD | Kim et al., 2005a |
| Exon 7 | c.358C>T | p.Gln120* | AD | This report |
| Exon 7 | c.454G>T | p.Glu152* | AD | This report |
| Intron 8 | c.534+1G>A | p.Ala158-Gln178del | AD | Rajpar et al., 2001; Song et al., 2012 |
| Exon 9 | c.536G>T | p.Arg179Met | AD | Gutierrez et al., 2007 |
| Intron 9 | c.588+1delG | p.Asn197Ilefs*81 | AD | Kida et al., 2002; P.S. Hart et al., 2003; Kim et al., 2005a; Pavlic et al., 2007 |
| Exon 10 | c.647C>T | p.Ser216Leu | AR/AD | Chan et al., 2010 |
| Exon 10 | c.737C>A | p.Ser246* | AD | Ozdemir et al., 2005 |
| Exon 10 | c.1020-1021insAGTCAGTACCAGTACTGTGTC | p.Val340-Met341insSer GlnTyrGlnTyrCysVal | AR/AD | Ozdemir et al., 2005 |
| Exon 10 | c.1259-1260insAG | p.Pro422Valfs*27 | AR/AD | T.C. Hart et al., 2003; Ozdemir et al., 2005; Pavlic et al., 2007; Kang et al., 2009; Chan et al., 2010; Lindemeyer et al., 2010 |
| Exon 10 | c.2991delT | p.Leu998Trpfs*65 | AD | Kang et al., 2009 |
Sequences based on the reference sequence for mRNA (NM_031889.2) and protein (NP_114095.2), where the A of the ATG translation initiation codon is nucleotide 1.
Most striking, a sibling had normal-looking enamel despite having the same mutation (c.211-2A>C) with other affected family members (Kim et al., 2005a). In this study, we identified additional cases of incomplete penetrance or extremely mild clinical phenotype with 2 different nonsense mutations. Recently, LAMB3 mutations have been identified to cause nonsyndromic AI by a dominant negative effect (Kim et al., 2013; Poulter et al., 2014b), and a case with very mild expressivity has been noted in a truncating mutation with a prediction of a minimal dominant negative effect (Lee et al., 2014).
The possibility of the hypoplastic AI with incomplete penetrance was noted in an epidemiologic study of Swedish population (Sundell and Valentin, 1986). It is highly possible that among them, there were individuals having less harmful mutations in genes such as LAMB3, ENAM, ITGB6, or other yet unidentified genes; therefore, the phenotypes were variable and very mild in some cases. However, ENAM mutations with incomplete penetrance in a previous study and this study caused severe hypoplastic AI in other affected individuals.
Generally, mutations in the ENAM gene cause hypoplastic enamel, but the severity of the enamel defects is sometimes highly variable, even among individuals with the same mutation. At this moment, there is no direct explanation or mechanism to support the lack of penetrance or extremely mild clinical phenotype in an individual. Other genetic factors, such as yet unidentified cis- or trans-acting elements, could regulate and influence the expression of enamelin or ENAM protein function. Further genetic and functional studies are needed to understand the dynamic and variable nature of enamel formation.
Acknowledgments
We thank the participants in this study for their cooperation.
Footnotes
This work was supported by grants from the Bio & Medical Technology Development Program (2013037491) and by the Science Research Center grant to the Bone Metabolism Research Center (2008-0062614) by the Korea Research Foundation.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Chan HC, Mai L, Oikonomopoulou A, Chan HL, Richardson AS, Wang SK, et al. (2010). Altered enamelin phosphorylation site causes amelogenesis imperfecta. J Dent Res 89:695-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC. (2011). Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci 119(Suppl 1):311-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussain C, Bouazza N, Gasse B, Laffont AG, Opsahl Vital S, Davit-Beal T, et al. (2014). Dental caries and enamelin haplotype. J Dent Res 93:360-365. [DOI] [PubMed] [Google Scholar]
- El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, et al. (2009). Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet 85:699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez SJ, Chaves M, Torres DM, Briceno I. (2007). Identification of a novel mutation in the enamalin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch Oral Biol 52:503-506. [DOI] [PubMed] [Google Scholar]
- Hart PS, Michalec MD, Seow WK, Hart TC, Wright JT. (2003). Identification of the enamelin (g.8344delG) mutation in a new kindred and presentation of a standardized ENAM nomenclature. Arch Oral Biol 48:589-596. [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. (2004). Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet 41:545-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Hart PS, Gorry MC, Michalec MD, Ryu OH, Uygur C, et al. (2003). Novel ENAM mutation responsible for autosomal recessive amelogenesis imperfecta and localised enamel defects. J Med Genet 40:900-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, et al. (2008). Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem 283:10858-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Hu Y, Lu Y, Smith CE, Lertlam R, Wright JT, et al. (2014). Enamelin is critical for ameloblast integrity and enamel ultrastructure formation. PLoS One 9:e89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeremias F, Koruyucu M, Kuchler EC, Bayram M, Tuna EB, Deeley K, et al. (2013). Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol 58:1434-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HY, Seymen F, Lee SK, Yildirim M, Tuna EB, Patir A, et al. (2009). Candidate gene strategy reveals ENAM mutations. J Dent Res 88:266-269. [DOI] [PubMed] [Google Scholar]
- Kida M, Ariga T, Shirakawa T, Oguchi H, Sakiyama Y. (2002). Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary. J Dent Res 81:738-742. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hu YY, Lin BP, Boyd C, Wright JT, et al. (2004). Amelogenin p.M1T and p.W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J Dent Res 83:378-383. [DOI] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, et al. (2005a). ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res 84:278-282. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, et al. (2005b). MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet 42:271-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Lin BP, Seymen F, Bartlett JD, Hu JC. (2006). Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci 114:(Suppl 1):3-12; discussion 39-41, 379. [DOI] [PubMed] [Google Scholar]
- Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, et al. (2008). FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet 82:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lee KE, Ko J, Yildirim M, Tuna EB, et al. (2013). LAMB3 Mutations Causing Autosomal-dominant Amelogenesis Imperfecta. J Dent Res 92:899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KE, Ko J, Tran Le CG, Shin TJ, Hyun HK, Lee SH, et al. (2014). Novel LAMB3 mutations cause non-syndromic amelogenesis imperfecta with variable expressivity. Clin Genet [Epub ahead of print 2/4/2014]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemeyer RG, Gibson CW, Wright TJ. (2010). Amelogenesis imperfecta due to a mutation of the enamelin gene: clinical case with genotype-phenotype correlations. Pediatr Dent 32:56-60. [PMC free article] [PubMed] [Google Scholar]
- Mardh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. (2002). A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2). Hum Mol Genet 11:1069-1074. [DOI] [PubMed] [Google Scholar]
- O’Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, et al. (2011). Whole-exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet 88:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. (2005). Phenotype of ENAM mutations is dosage-dependent. J Dent Res 84:1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Brookes SJ, Logan CV, Poulter JA, El-Sayed W, Al-Bahlani S, et al. (2012). Mutations in C4orf26, encoding a peptide with in vitro hydroxyapatite crystal nucleation and growth activity, cause amelogenesis imperfecta. Am J Hum Genet 91:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Poulter JA, Logan CV, Brookes SJ, Jafri H, Ferguson CH, et al. (2013). Identification of mutations in SLC24A4, encoding a potassium-dependent sodium/calcium exchanger, as a cause of amelogenesis imperfecta. Am J Hum Genet 92:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlic A, Petelin M, Battelino T. (2007). Phenotype and enamel ultrastructure characteristics in patients with ENAM gene mutations g.13185-13186insAG and 8344delG. Arch Oral Biol 52:209-217. [DOI] [PubMed] [Google Scholar]
- Poulter JA, Brookes SJ, Shore RC, Smith CE, Abi Farraj L, Kirkham J, et al. (2014a). A missense mutation in ITGB6 causes pitted hypomineralized amelogenesis imperfecta. Hum Mol Genet 23:2189-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter JA, El-Sayed W, Shore RC, Kirkham J, Inglehearn CF, Mighell AJ. (2014b). Whole-exome sequencing, without prior linkage, identifies a mutation in LAMB3 as a cause of dominant hypoplastic amelogenesis imperfecta. Eur J Hum Genet 22:132-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. (2001). Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet 10:1673-1677. [DOI] [PubMed] [Google Scholar]
- Simmer SG, Estrella NM, Milkovich RN, Hu JC. (2013). Autosomal dominant amelogenesis imperfecta associated with ENAM frameshift mutation p.Asn36Ilefs56. Clin Genet 83:195-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YL, Wang CN, Zhang CZ, Yang K, Bian Z. (2012). Molecular characterization of amelogenesis imperfecta in Chinese patients. Cells Tissues Organs 196:271-279. [DOI] [PubMed] [Google Scholar]
- Sundell S, Valentin J. (1986). Hereditary aspects and classification of hereditary amelogenesis imperfecta. Community Dent Oral Epidemiol 14:211-216. [DOI] [PubMed] [Google Scholar]
- Wang SK, Choi M, Richardson AS, Reid BM, Lin BP, Wang SJ, et al. (2014). ITGB6 loss-of-function mutations cause autosomal recessive amelogenesis imperfecta. Hum Mol Genet 23:2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkop CJ., Jr (1988). Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol 17:547-553. [DOI] [PubMed] [Google Scholar]