Abstract
Quantitative trait loci (QTL) mapping was used to identify loci controlling various aspects of seed longevity during storage and germination. Similar locations for QTLs controlling different traits might be an indication for a common genetic control of such traits. For this analysis we used a new recombinant inbred line population derived from a cross between the accessions Landsberg erecta (Ler) and Shakdara (Sha). A set of 114 F9 recombinant inbred lines was genotyped with 65 polymerase chain reaction-based markers and the phenotypic marker erecta. The traits analyzed were dormancy, speed of germination, seed sugar content, seed germination after a controlled deterioration test, hydrogen peroxide (H2O2) treatment, and on abscisic acid. Furthermore, the effects of heat stress, salt (NaCl) stress, osmotic (mannitol) stress, and natural aging were analyzed. For all traits one or more QTLs were identified, with some QTLs for different traits colocating. The relevance of colocation for mechanisms underlying the various traits is discussed.
The long-term storage of seeds, especially under unfavorable conditions, leads to a loss of viability. The nature of this physiological damage is variable, e.g. short-term deterioration in the field is different from long-term deterioration during storage, which in turn is different from mechanical damage (McDonald, 1999). All parts of the seed, such as the seed coat, which is of maternal origin and acts as a physical and chemical barrier, and the embryo are susceptible to physiological damage. Seed longevity has been related to various seed properties such as color, weight, and membrane composition. The correlation between seed longevity and these traits is often species or in some cases even variety specific (McDonald, 1999). The ability of seeds to withstand stresses that occur while stored is one aspect of seed longevity. To some extent these stresses may resemble those that occur when imbibed seeds are exposed to unfavorable conditions during germination.
Very little is known about the genetic basis of differences in seed longevity because this trait is affected by environmental effects during seed formation, harvest, and storage, and is probably controlled by several genes. Only recently some studies on this complex trait have been initiated using quantitative trait loci (QTL) mapping in crop plants like cabbage (Brassica oleracea; Bettey et al., 2000), tomato (Lycopersicon sp.; Foolad et al., 1999), rice (Oryza sativa; Cui et al., 2002; Miura et al., 2002), barley (Hordeum vulgare; Mano and Takeda, 1997), and Sorghum bicolor (Natoli et al., 2002).
Arabidopsis can be a good model species for the identification of genes controlling seed longevity, because it is amenable to both classical and molecular genetic studies (Meinke et al., 1998; Page and Grossniklaus, 2002). This, combined with the multitude of publicly available molecular tools, including a complete genome sequence, implies that the cloning of genes can proceed relatively quickly in Arabidopsis.
To investigate the genetics of seed viability and vigor one can identify mutants that either have improved or reduced seed longevity. Among Arabidopsis mutants that have a poor seed longevity, evidenced by their rapid loss of viability upon storage, are mutants that affect seed maturation such as leafy cotyledon1 (lec1), lec2, fusca3 (fus3; Holdsworth et al., 1999; Finkelstein et al., 2002), and strong abscisic acid insensitive3 (abi3) alleles (Ooms et al., 1993). Mutants with smaller effects are weak alleles of abi3 (Koornneef et al., 1984; Bies-Ethève et al., 1999), the aberrant testa shape (ats) mutant (Léon-Kloosterziel et al., 1994), the transparent testa (tt) mutants (Debeaujon et al., 2000), and the green seed mutant (Clerkx et al., 2003). Other sources of genetic variation can be found in naturally occurring populations. Arabidopsis is widely distributed in the world, thus encountering substantial variation in growth environments. Therefore, phenotypic variation is expected to reflect the genetic variation important for adaptation to specific environments as summarized by Alonso-Blanco and Koornneef (2000). Seed longevity and seed vigor are traits of complex nature and thus interesting to study by quantitative genetic methods. The advent of efficient molecular marker technologies and specific statistical methods in the past decade has allowed that map positions of QTLs can be established with sufficient accuracy (Alonso-Blanco and Koornneef, 2000). Mapping QTLs requires a segregating population for which a genetic map has been established and an accurate phenotyping of the trait. Immortal mapping populations such as recombinant inbred lines (RILs) are very useful because each genotype can be tested repeatedly and by applying different test systems. The latter allows studying the pleiotropic effects of loci, which are suggested by colocation of QTLs for different traits.
A test commonly used to assess seed longevity is a controlled deterioration test (CDT), in which seeds are stored at high relative humidity and temperature. Tesnier et al. (2002) described such a test for Arabidopsis. Several mutants, but also different accessions, showed different responses to the applied treatments indicating the presence of genetic variation for the response to CDT (Tesnier et al., 2002). Bentsink et al. (2000) confirmed this by identifying several QTLs for survival after a CDT using a Landsberg erecta (Ler)/Cape Verde Islands (Cvi) RIL population. Quesada et al. (2002) simulated the effect of drought stress on germination by using NaCl and identified QTLs controlling this trait in the Ler/Columbia (Col) RIL population.
In dry desiccation-tolerant seeds, protection of proteins and membranes during desiccation occurs by water replacement. Water molecules are replaced by sugars at the hydrogen bonding sites to preserve the native structure of proteins and the spacing between phospholipids (Hoekstra et al., 2001). Besides sugars, heat-inducible hydrophilic late-embryogenesis abundant (LEA) proteins might also play a role in protein and membrane stabilization by acting as molecular chaperones (Hoekstra et al., 2001). Bettey and Finch-Savage (1998) investigated the role of two subclasses of LEAs, dehydrins and small heat shock proteins (sHSPs) in B. oleracea seeds. Dehydrins did not show a positive relationship with seed performance. However, the protein HSP17.6 showed a positive correlation with seed performance (Bettey and Finch-Savage, 1998). Further evidence for the role of sHSPs could come from the fact that in Arabidopsis cytosolic sHSPs appear to respond to specific developmental signals associated with the acquisition of desiccation tolerance (Wehmeyer and Vierling, 2000).
Seeds normally germinate in a wide range of temperatures. It seems that the major determinant of germination is the availability of water (Bewley and Black, 1994). Under stress conditions such as extreme temperatures, salt stress, and water deficit, germination is delayed or completely inhibited depending on the stress intensity and the genetic background (Foolad et al., 1999). To differentiate between loci involved specifically in germination under stress, Foolad et al. (1999) and Bettey et al. (2000) determined QTLs for speed of germination and argued that these loci are important for germination in general and are not specifically affected by stress.
In the present study we analyzed the genetic control of the response to various stress treatments applied during seed storage and imbibition to investigate if tolerance to such factors has a common genetic basis. This study was performed using a newly developed RIL population derived from a cross of Ler × Shakdara (Sha). Ler originates from Poland and Sha from the Shakdara mountain range in Tadjikistan at 3,400 m elevation (Khurmatov, 1982). Preliminary experiments indicated that Sha is one of the accessions most tolerant to various seed stresses. We were able to identify QTLs for various seed stresses viz, germination after a CDT, heat treatment and germination on NaCl, mannitol, H2O2, and abscisic acid (ABA). Furthermore, QTLs were identified for seed dormancy, seed sugar content, natural aging, and germination speed. Some of these QTLs colocate, which might indicate possible common genetic regulations.
RESULTS
The Ler/Sha RIL Population
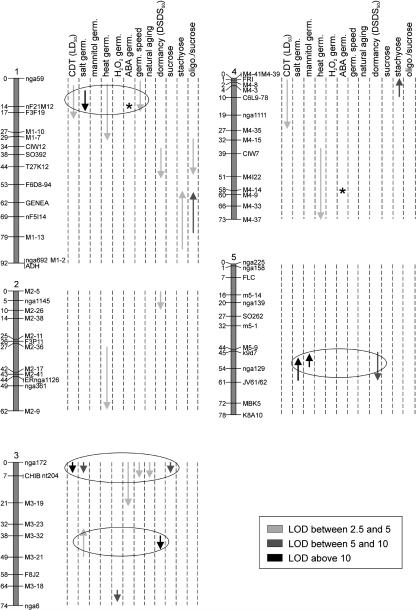
Using 114 F9 lines a genetic map (Fig. 1) was generated using 66 markers distributed over the genome, which has a total length of 378 centimorgans (cM). There was an average distance between markers of 5.7 cM, and there were 14 segments having a genetic distance between 10 and 14 cM (Table I). Of all possible marker data points 1.2% were not available, usually due to missing amplification or uncertainty in scoring. The average residual heterozygosity per locus was 0.7%, which was slightly higher than the expected 0.4% for an F9 generation. On average, Ler alleles represented 54% of the alleles; certain regions showed a significantly distorted segregation ratio. These included the top of chromosome 1 (between nga59 and F3F19), chromosome 2 (between msat2-5 and msat2-38; msat in Fig. 1 is M), chromosome 3 (around markers CHIB and nt204), chromosome 4 (between marker C6L9-78 and CIW7), and chromosome 5 (around marker K8A10). Except for the region on chromosome 3, where Sha alleles were in excess, the bias was always in favor of Ler alleles.
Figure 1.
The Ler/Sha linkage map showing the genetic locations affecting various seed-related traits (indicated above each lane). Gray scales of the arrows represent the LOD score. Arrows indicate the direction of the phenotypic effects: up, Ler increasing, Sha decreasing; down, Ler decreasing and Sha increasing. The lengths of the arrows depict the 2-LOD support intervals. Circles indicate discussed colocations. *, a significant interaction for germination on ABA with a marker colocating with a heat germination QTL.
Table I.
Map features per chromosome and entire genome
| Chromosome | Total cM | kb/cM | Avr. Dist./Marker | Gaps >10 cM |
|---|---|---|---|---|
| 1 | 92.3 | 308 | 6.2 | 4 |
| 2 | 61.7 | 293 | 4.7 | 3 |
| 3 | 74.1 | 311 | 7.4 | 4 |
| 4 | 72.6 | 239 | 4.8 | 1 |
| 5 | 77.7 | 339 | 6.0 | 2 |
| Total | 378.5 | 299 | 5.7 | 14 |
Map distances were calculated with the JoinMap program. Physical distances were calculated using the center of the BAC in which the primer is situated.
QTL Mapping
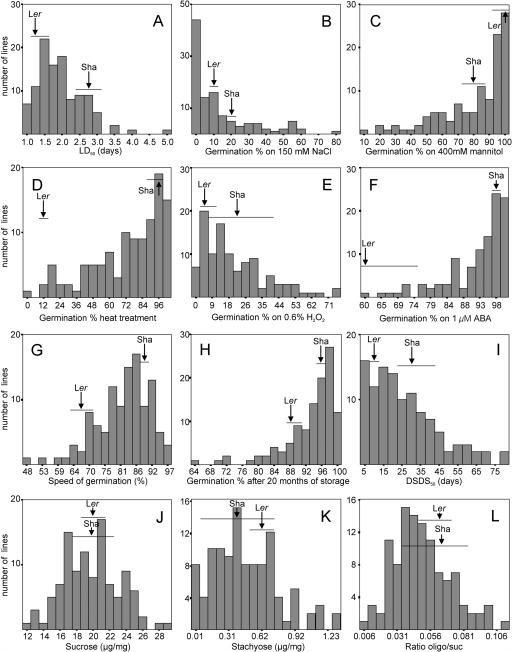
For all traits analyzed in germination experiments, broad sense heritabilities were estimated as the proportion of variance explained by between-line differences. These heritabilities ranged from 0.60 for germination on ABA up to 0.88 for seed dormancy (Table II). Table II also shows the parental values and values obtained from F2 seeds harvested from the two reciprocal F1 hybrids indicating some reciprocal differences and suggesting some maternal effects, for instance, for dormancy (expressed as days of dry seed storage to reach 50% germination, DSDS50; Alonso-Blanco et al., 2003). Frequency distributions of the various traits analyzed and the parental values are shown in the histograms in Figure 2. It was also clear from these histograms that some traits were not normally distributed. These were LD50 (the number of days CDT required to reach 50% germination); salt-, mannitol-, heat treatment-, H2O2-, and ABA-germination (respectively, Fig. 2, A–F); and speed of germination and germination after natural aging (respectively, Fig. 2, G and H). Improved normality was obtained by log-transformation of the LD50 data. A probit transformation, to improve normality, was performed on the germination data for speed of germination, salt, mannitol, heat treatment, H2O2, ABA, and natural aging. Because transformations improved normality (data not shown), the subsequent QTL analysis was performed with the transformed data.
Table II.
Trait values for Ler, Sha, and the reciprocal F2 of Ler × Sha and Sha × Ler along with the range of trait values for individual RILs and calculated heritabilities
| Trait | Ler | Sha | F2 (Ler × Sha) | F2 (Sha × Ler) | Range | Heritability |
|---|---|---|---|---|---|---|
| LD50 (d) | 1.3 ± 0.1 | 2.8 ± 0.2 | 1.3 ± 0.1 | 1.6 ± 0.0 | 0.5–5.1 | 0.82 |
| Salt germ. (%) | 8.7 ± 1.2 | 21.2 ± 0.4 | 13.7 ± 2.1 | 12.8 ± 6.3 | 0–79 | 0.82 |
| Mannitol germ. (%) | 97 ± 1.6 | 79 ± 6.6 | 83 ± 3.4 | 82 ± 8.2 | 10–100 | 0.79 |
| Heat germ. (%) | 12 ± 2.4 | 94 ± 2.1 | 82 ± 4.6 | nd | 1.3–100 | 0.93 |
| H2O2 germ. (%) | 4.9 ± 2.1 | 24 ± 17 | 9.7 ± 7.7 | 16 ± 6.4 | 0–74 | 0.78 |
| ABA germ. (%) | 57 ± 18 | 97 ± 0.8 | 82 ± 17 | 93 ± 2.4 | 59–100 | 0.60 |
| Germ. speed (%) | 67 ± 3.7 | 89 ± 1.3 | 73 ± 5.4 | 77 ± 4.9 | 48–97 | 0.68 |
| Natural aging (%) | 89 ± 2.0 | 96 ± 1.4 | 86 ± 8.2 | 89 ± 3.9 | 64–100 | 0.69 |
| DSDS50 (d) | 9.6 ± 1.0 | 31.8 ± 7.4 | 19.0 ± 3.5 | 29.4 ± 4.0 | 3.4–80.5 | 0.88 |
| Suc (μg/mg) | 19.7 ± 1.1 | 20.2 ± 2.1 | 22.9 ± 0.5 | 19.0 ± 1.1 | 12.3–28.7 | nd |
| Stachyose (μg/mg) | 0.6 ± 0.1 | 0.4 ± 0.3 | 0.2 ± 0.0 | 0.5 ± 0.1 | 0–1.3 | nd |
| RSO/Suc | 0.06 ± 0.01 | 0.06 ± 0.02 | 0.03 ± 0.00 | 0.05 ± 0.01 | 0.01–0.11 | nd |
nd, Not determined.
Figure 2.
Frequency distributions of nonnormalized data of all traits in the Ler/Sha RIL population. Calculated number of days CD-treatment to reach 50% germination, LD50 (A). Germination percentages on 150 mm NaCl (B), 400 mm mannitol (C), after an 8-h 50°C treatment (D), on 0.6% H2O2 (E), and on 1 μm ABA (F). The speed of germination (G) and natural aging as the germination percentage after 20 months of storage (H). The calculated number of days of dry seed storage needed to reach 50% germination, DSDS50 (I). Measurements of Suc (J), stachyose (K) quantities, and RSO to Suc ratio (L). The average parental value is indicated with an arrow for both parents, and the horizontal bar represents the se for these parental values.
Mapping QTLs for Nonstressed Germination
To distinguish between loci specific for regulation of germination under stress versus nonstress conditions, the latter was determined using the germination speed under optimal conditions. QTL analysis revealed two loci, viz, one on chromosome 1 and one on chromosome 3, that influenced germination speed (Fig. 1). In total these two loci explained 28.6% of the variance, and in both cases Sha alleles conferred a faster germination (Table III).
Table III.
Characteristics of QTLs detected in Ler/Sha RIL population
| Trait | Chromosome | QTLa Position | LOD | Percentage Variance Explained | Confidence Interval (cM)b | QTL Effectc |
|---|---|---|---|---|---|---|
| d | ||||||
| CDT (LD50) | 1 | nF21M12 | 3.9 | 8.3 | 3–20 | −0.41 |
| 3 | nga172 | 12.5 | 35.4 | 0–5 | −0.81 | |
| 4 | nga1111 | 4.3 | 10.1 | 1–26 | −0.46 | |
| total | 48.9 | |||||
| % germ. | ||||||
| Salt germ. | 1 | nF21M12 | 12.1 | 27.4 | 6–16 | −17 |
| 3 | nga172 | 5.2 | 10.1 | 0–5 | −10 | |
| 3 | msat3-32 | 2.8 | 5.1 | 34–49 | 7 | |
| 5 | nga129 | 10.5 | 23.1 | 48–60 | 17 | |
| total | 58.6 | |||||
| % germ. | ||||||
| Mannitol germ. | 5 | K9D7 | 16.8 | 49.4 | 46–53 | 28 |
| total | 49.4 | |||||
| % germ. | ||||||
| Heat germ. | 1 | nF21M12 | 3.7 | 11.5 | 3–31 | −15 |
| 2 | msat2-41 | 2.6 | 7.9 | 28–60 | −14 | |
| 4 | msat4-14 | 2.9 | 8.8 | 36–73 | −15 | |
| total | 30.1 | |||||
| % germ. | ||||||
| H2O2 germ. | 3 | nga6 | 8.7 | 29.4 | 69–74 | −19 |
| total | 29.4 | |||||
| % germ. | ||||||
| ABA germ. | 3 | nga172 | 2.8 | 11.2 | 0–23 | −6 |
| total | 11.2 | |||||
| % germ. | ||||||
| Germ. speed | 1 | nF21M12 | 4.3 | 13.9 | 0.5–17 | −7 |
| 3 | nga172 | 5 | 16.5 | 0–6 | −7 | |
| total | 28.6 | |||||
| % germ. | ||||||
| Natural aging | 3 | nga172 | 4.6 | 17.4 | 0–6 | −6 |
| total | 17.4 | |||||
| d | ||||||
| DSDS50 | 1 | T27K12 | 4 | 8 | 35–50 | −11 |
| 2 | msat2-5 | 4 | 8.1 | 0–10 | −10 | |
| 3 | msat3-32 | 14.4 | 36.4 | 39–46 | −21 | |
| 5 | nga129 | 6.3 | 13.4 | 45–60 | −14 | |
| total | 54.6 | |||||
| μg/mg | ||||||
| Suc | 3 | nga172 | 6.9 | 25.3 | 10–4 | −3.44 |
| total | 25.3 | |||||
| μg/mg | ||||||
| Stachyose | 1 | GENEA | 3.9 | 11.7 | 56–85 | 0.21 |
| 4 | FRI | 6.6 | 20.9 | 0–10 | 0.28 | |
| Interactiond | GENEA × FRI | 4.8 | ||||
| total | 35.1 | |||||
| ratio | ||||||
| RSO/Suc | 1 | CIW12 | 3 | 9.8 | 30–48 | −0.013 |
| 1 | GENEA | 6.6 | 22.9 | 57–77 | 0.02 | |
| Interactiond | CIW12 × GENEA | 3.3 | ||||
| total | 26.1 |
Cofactor nearest to highest LOD score: also cofactor fixed.
Positions given correspond to 2-LOD confidence intervals.
QTL effect of untransformed data, negative value means that the Ler allele at this QTL reduces the effect of this parameter.
Interaction between QTLs calculated as indicated in “Materials and Methods.”
Mapping QTLs for Seed Longevity
Bentsink et al. (2000) and Tesnier et al. (2002) had shown that a CDT can be used to identify genetic differences for seed storability. QTL analysis was performed using the LD50 value. Ler was more sensitive (reduced longevity) to the CDT conditions than Sha, and most values for the RILs were in between the parental values (Fig. 2A). Three QTLs were identified (Fig. 1) affecting viability after controlled deterioration viz, on chromosomes 1, 3, and 4, respectively (Table III). In total these loci explained 48.9% of the variance. In all cases the Sha allele increased the number of days of treatment that the seeds could endure, which was in agreement with the limited transgression observed in the population (Fig. 2A).
To test whether CDT mimics natural aging, seeds that had been stored for 20 months at ambient conditions were germinated. The QTL mapping revealed only one QTL viz, on chromosome 3 (Fig. 1), which explained 17.4% of the total variance (Table III). The Sha allele conferred a higher longevity to prolonged storage, similar to the CDT QTL found at the same position.
Mapping QTLs for Stressed Germination on NaCl, Mannitol, H2O2, and after Heat Treatment
The ability of seeds to cope with water stress can be tested by germination on media containing NaCl or mannitol. For germination on NaCl, four QTLs, one on chromosome 1, two on 3, and one on 5 (Fig. 1), were identified; in total they explained 59.4% of the variance (Table III). For two of these QTLs (chromosomes 3 and 5) the Ler alleles increased the germination percentage and for the other two, the Sha alleles (chromosomes 1 and 3) showed higher germination, which could explain the observed transgression (Fig. 2B). Germination on NaCl might also be explained as toxicity to sodium rather than the ability to cope with water stress. To distinguish between these two effects germination on mannitol was tested. Only one QTL for this trait was detected viz, on chromosome 5 (Fig. 1), which explained 49.4% of the variation (Table III). The Sha allele decreased the germination percentage.
A factor commonly suggested in seed deterioration is reactive oxygen species (ROS). To mimic the effect of ROS we applied H2O2 during imbibition and subsequent germination. QTL analysis revealed only one QTL viz, on chromosome 3, which explained 29.4% of the variance (Table III), the Sha allele increasing the tolerance to H2O2.
Imbibed Sha seeds showed a higher tolerance when exposed to high temperatures than Ler seeds. Therefore, imbibed seeds of all RILs were exposed to 50°C for 8 h and germination was scored after 7 d. Germination frequencies are shown in Figure 2D. Three QTLs were identified for this trait, viz, on chromosomes 1, 2, and 4 (Fig. 1), which explained 30.1% of the total variance observed. In all cases the Sha alleles conferred a higher tolerance to the treatment (Table III).
Mapping QTLs Controlling Dormancy and Tolerance to ABA
Dormancy QTLs were mapped using the DSDS50 value (Alonso-Blanco et al., 2003) for which in total 54.9% of the variance could be explained by four QTLs located on chromosomes 1, 2, 3, and 5, respectively (Fig. 1 and Table III). In all cases the Sha alleles increased seed dormancy. The frequency distribution (Fig. 2I) showed that there was some transgression beyond the Sha parental value, indicating that loci with minor effect, where Ler alleles increase dormancy, might have remained undetected.
To test the sensitivity of the RIL lines to ABA we applied 1 μm ABA during germination. This revealed only one QTL, viz, on chromosome 3 (Fig. 1), which explained 11.2% of the variation. The Sha allele conferred a higher tolerance to applied ABA (Table III).
Mapping QTLs Controlling Seed Soluble Oligosaccharide Content
QTL mapping was performed on the quantity of the three major soluble sugars in Arabidopsis seeds—Suc and the raffinose series oligosaccharides (RSO; raffinose and stachyose)—and the ratio of RSO to Suc (Fig. 2, J–L). Raffinose levels were low (data not shown) and no QTL for the content of this oligosaccharide could be detected. In four genomic regions QTLs were detected for Suc and stachyose content. For Suc a major QTL explaining 25.3% of the variation was found on the top of chromosome 3 (Fig. 1 and Table III), the Sha allele increasing the Suc content. Two QTLs, for which Ler alleles increased the stachyose levels, were detected on chromosome 1 and 4, respectively (Fig. 1 and Table III). Interaction between these two loci was detected, explaining 4.8% of the variation observed. Sha alleles at the FRI marker had a synergistic effect when Ler alleles at the GENEA marker were present. For the ratio RSO to Suc two QTLs with opposite allelic effects were detected; one of these is apparently due to the higher stachyose content at the GENEA marker on chromosome 1 (Fig. 1). The QTL for the ratio RSO/Suc at the position of marker CIW12 might have been influenced by a minor QTL for stachyose at the same position (log of the odds [LOD] 2.2; data not shown). The interaction between the two loci, CIW12 and GENEA, explained 3.3% of the variance found for this trait. Ler alleles at the GENEA marker had a synergistic effect when Sha alleles at the CIW12 marker were present (Table III).
Epistatic Interactions
We hypothesized that similar physiological processes, like germination under different stresses, might have a common genetic basis. However, QTL mapping might not be able to detect a significant QTL due to statistical inaccuracy resulting from a low heritability and/or a relatively small population size. The latter effect is enforced by epistasis, a situation where variation for one locus is only observed in the background of a specific allele at another locus, thereby reducing the effective population size. Interactions between loci could be present and significant, despite the fact that the effects of the individual loci were not found to be significant. Therefore, we tested interactions at those positions where at least one of the traits showed a significant QTL using ANOVA. We only found one significant interaction for the germination on ABA between two markers that colocate with heat germination QTLs (msat4-14 and nF21M12) indicated as asterisks in Figure 1. It appears that the Ler allele of msat4-14 is epistatic over nF21M12 adding a QTL for ABA germination to the cluster of seed stress QTL at the top of chromosome 1, where also the Sha allele increases germination.
DISCUSSION
The Ler/Sha RIL Population
A linkage map has been generated for a novel Arabidopsis Ler/Sha RIL population. These two accessions were used since preliminary tests (data not shown) had indicated that the accession Sha was very resistant to applied stress during germination. The map was made with 65 microsatellite, cleaved-amplified polymorphic sequence (CAPS), and indel markers, the latter being based on the Cereon database (Jander et al., 2002), and the erecta mutation. The possibility of genotyping a population using only microsatellites was already shown by Loudet et al. (2002) and allows perfect anchoring to the physical map of Arabidopsis. The order of the markers in the Ler/Sha RIL population was in agreement with the published Col sequence. The relative length of the various chromosomes is similar to what has been published for maps based on other Arabidopsis RIL populations (Lister and Dean, 1993; Alonso-Blanco et al., 1998; Loudet et al., 2002). The average distance per marker in kb per cM showed that only chromosome 4 was very different from the mean relative ratio of 320 kb per cM (Table I), which was also seen in the Bayreuth (Bay-0)/Sha map described by Loudet et al. (2002). Large segregation distortion could compromise the QTL analysis. However, since the segregation ratios that we observed never exceeded a 1:1.8 ratio, we did not expect that this affected the present analysis. Similar biases in the segregation ratios were observed in other populations like Ler/Col, Ler/Cvi, Col/Kashmir, and Bay-0/Sha (Lister and Dean, 1993; Alonso-Blanco et al., 1998; Wilson et al., 2001; Loudet et al., 2002).
Comparison of QTL Positions with QTLs in Previously Analyzed Arabidopsis Populations
The finding of QTLs at a similar position for the same trait in different mapping gives an indication that allelic variation in different accessions might result from similar loci. However, the relative inaccuracy of the map positions obtained never excludes that in different crosses, distinct but closely linked genes segregate. Comparing the QTLs for storability found in the Ler/Sha population to those found in the Ler/Cvi population (Bentsink et al., 2000) revealed that the QTLs on chromosomes 1 and 2 could be in the same regions. In both cases the Ler alleles decreased longevity, while the Sha and Cvi alleles increased longevity.
Comparison of the QTLs found for germination on NaCl in the Ler/Sha and Ler/Col populations (Quesada et al., 2002) showed that two QTLs could be in the same region. In both populations the Ler allele decreased germination for the QTL found on chromosome 1. In the region where Quesada et al. (2002) located a QTL for days needed to reach 50% germination on salt-supplemented medium (QT50), a QTL was also found in the Ler/Sha RILs, although these had opposite effects. In the Ler/Sha population, Ler increases the tolerance and in the Ler/Col, Col increases the tolerance. Thus, the relative effects of the alleles on salt tolerance could be Col > Ler > Sha, if this is due to the same locus.
Despite the low level of explained variance observed for seed sugar content, a comparison of the present data with those published by Bentsink et al. (2000) revealed that the QTL for stachyose on the lower arm of chromosome 1 in the Ler/Sha population and in the Ler/Cvi (Bentsink et al., 2000) population could be at a similar position. For Suc content only one QTL could be detected, on top of chromosome 3, which was also detected in the Ler/Cvi population (Bentsink et al., 2000).
The four dormancy QTLs detected in the Ler/Sha RIL population colocated with QTLs previously identified in the Ler/Col RIL population (van der Schaar et al., 1997) and all except one (chromosome 2) were also found in the Ler/Cvi population (Alonso-Blanco et al., 2003).
The Significance of Colocation and Candidate Genes
Colocation of QTLs for different traits might be a first indication that the locus has pleiotropic effects on these traits, due to a common mechanistic basis. In some cases these pleiotropic effects were unexpected as found for the circadian period length (Swarup et al., 1999) and water use efficiency (McKay et al., 2003). These effects were associated with the FLC flowering time locus and could be confirmed by studying mutants at this locus. Since it is assumed that seed longevity is determined by an overall tolerance to various seed stresses we expected, and found evidence for, colocations for CDT QTLs and QTLs for tolerance to other seed stresses. However, in some cases colocations were not found, indicating that tolerance to each specific stress has its own genetic basis. Because the germination percentage is the only parameter used to determine germination-related stress traits it should be taken into account that dormancy, which is also assayed by germination, might affect the data. However, no colocation between stress and dormancy QTLs was observed except on chromosomes 3 and 5 where salt tolerance and dormancy QTLs colocate in opposite directions. The ability of QTL approaches to uncover novel loci, which determine stress tolerance traits, is also demonstrated in the case of heat tolerance. No QTL for this trait mapped to the bottom of chromosome 1, at the location of the HSP101 gene, which when mutated leads to severe heat-sensitivity of imbibed seeds (Hong and Vierling, 2000).
ROS as a Common Factor
Colocation of QTLs related to germination under stressful conditions was observed on top of chromosome 1, where the Sha allele confers a higher tolerance to CDT survival, germination under saline conditions, and germination after heat treatment. A common factor in all these stresses could be the release of ROS. For CDT it is known that seed deterioration can occur through the generation of oxidative stress (Khan et al., 1996). Saline conditions are also known to generate ROS (for review, see Xiong and Zhu, 2002). Stressing seeds might also induce the expression of LEA proteins and after heat shock, sHSPs are specifically expressed (Wehmeyer and Vierling, 2000). Besides acting as protector in preventing irreversible protein denaturation, sHSPs may also modulate cellular redox state (Grene, 2002). This and the localization of several known ROS scavenging enzymes in this region, viz, catalase (Frugoli et al., 1996) and a superoxide dismutase (Kliebenstein et al., 1998) might point toward the production of active oxygen species as a mechanism in seed deterioration. Several other stress-related genes were mapped in this region, e.g. genes related to freezing tolerance, possibly also related to oxidative stress (Thorlby et al., 1999), and genes involved in drought stress, one of them having homology to a glutathione S-transferase, an enzyme involved in reactive oxygen scavenging (Taji et al., 1999). However, germination on H2O2 could not confirm the relationship between ROS and germination. It might be expected that seeds with a higher tolerance to endogenously produced ROS might be more tolerant to exogenously applied H2O2. Therefore a QTL for H2O2 tolerance might be expected at this position on chromosome 1, but was not detected. However, the transgression toward higher values indicates that there are more loci involved in H2O2 resistance than the one detected on chromosome 3. Alternatively, applying H2O2 might not reveal all types of ROS tolerance and hence not reveal all loci involved in ROS scavenging. The presence of a QTL for speed of germination, on the top of chromosome 1, might indicate that this locus is involved in germination in general and is not specific for germination under stress conditions, as was shown in tomato (Foolad et al., 1999) and B. oleracea (Bettey et al., 2000). A specific control of ROS during germination by this locus cannot be excluded since in barley (Bethke and Jones, 2001; Fath et al., 2001) and radish (Raphanus sativus cv Eterna; Schopfer et al., 2001) it was shown that germination is accompanied by an increase of ROS, although our data did not provide arguments for this. The absence of colocation of a QTL for germination on H2O2 with any of the other stresses might be due to the fact that this locus is only involved in the metabolism of applied H2O2 and not involved in scavenging of endogenous H2O2.
Relationship between Inhibition of Germination and Dormancy
QTLs related to seed dormancy and germination on NaCl colocate at chromosomes 3 and 5. Colocation of QTLs found for germination under salt stress and for dormancy was also observed in a barley (H. vulgare) mapping population. In the Steptoe/Morex barley lines, dormancy on chromosome 7(5H) is conferred by the Steptoe allele (Gao et al., 2003), and the salt tolerance is conferred by the Morex allele (Mano and Takeda, 1997). This is similar in the Ler/Sha population; the more dormant alleles have increased sensitivity to NaCl (Sha/Steptoe), while the less dormant alleles are more resistant to NaCl (Ler/Morex). Since at these loci salt germination correlates with increased dormancy, these pleiotropic effects might be a consequence of the fact that seeds with a high germination potential (less dormant) might allow germination at higher osmotic values. A common factor in these processes might be the plant hormone ABA, which is involved in dormancy regulation. High salt conditions induce high levels of ABA and inhibit germination (Xiong and Zhu, 2002). The observation that nondormant, ABA-deficient mutants (Léon-Kloosterziel et al., 1996) and ABA-insensitive mutants (Quesada et al., 2000) can germinate at higher salt concentrations compared to wild type also suggests a role for ABA. We did not obtain indications that ABA sensitivity per se was involved in this population since no QTLs for ABA sensitivity colocated at these loci. However, there are mutants showing an altered response to applied ABA, abi4 and abi5, without an altered dormancy (Finkelstein, 1994) and not all reduced dormancy (rdo) mutants have an altered ABA sensitivity (Peeters et al., 2002). Mano and Takeda (1997) suggested a role for ABA in germination under saline conditions in barley doubled haploid lines. They were only able to find colocation of QTLs for ABA and salt response in a population of Harrington/TR306 barley doubled haploid lines, while in the Steptoe/Morex doubled haploid lines no colocation could be found. We could not confirm this relationship between salt and dormancy by locating QTLs for ABA response at similar positions.
The only significant QTL for ABA sensitivity at the top of chromosome 3 colocates with the pleiotropic seed stress locus (LD50), and better germination on ABA correlates with better germination in other stress conditions. This suggests that this locus affects germination in many conditions, which might be illustrated by a QTL for germination speed. A similar colocation of germination under various stresses and germination speed is observed on top of chromosome 1.
Regulation of Germination, Sugars, and Longevity
On top of chromosome 3, QTLs for germination under stress and nonstress conditions colocate. Mapping QTLs for germination speed could indicate a locus involved in the regulation of germination as such, as suggested by Foolad et al. (1999) and Bettey et al. (2000). The presence of a QTL for Suc could also point in this direction since Suc is probably the first metabolite involved in the initial germination process (Bewley and Black, 1994). Therefore, high endogenous Suc levels may enhance germination even under stress conditions. Breakdown of lipids to sugars is not essential for germination as was shown by germination of seeds lacking the glyoxylate cycle but is essential for proper seedling establishment (Eastmond et al., 2000). However, the presence of a QTL for both CDT and natural aging might point toward an essential role for Suc in seed longevity. Suc and stachyose are often associated with desiccation tolerance and seed longevity (Obendorf, 1997; Sinniah et al., 1998; Bailly et al., 2001). In a comparison of the QTLs obtained for seed longevity, by CDT survival, and for seed oligosaccharide (OS) content it was already shown by Bentsink et al. (2000) that the variation observed for OS content does not clearly affect seed storability. The Ler/Sha population does not rule out the possibility of a protective role for OS in seed longevity, although only a very low amount of the total variance for seed sugar content could be explained. Sinniah et al. (1998) found that acquiring desiccation tolerance and seed longevity are separate processes; Suc content could not be correlated to desiccation tolerance, but it could be correlated with potential seed longevity. In the Ler/Sha RIL the latter correlation can be seen on top of chromosome 3 where a higher Suc content from Sha alleles colocates with a higher CDT survival and better germination under salt stress. In both cases Sha alleles conferred to a higher tolerance. This colocation of QTLs for CDT survival and higher Suc content on chromosome 3 was also seen in the Ler/Cvi population (Bentsink et al., 2000). A possible explanation might be the protection of membranes by Suc in the desiccated seeds due to water replacement (Hoekstra et al., 2001).
According to Sinniah et al. (1998), stachyose is possibly involved in both tolerance to desiccation and potential longevity. However, this relationship was not observed in the population described here, since a colocation on top of chromosome 4 is in opposite direction and the QTL for stachyose on chromosome 1 does not colocate with a QTL for seed germination under stress conditions. The ratio of OS to Suc could not be associated with a QTL for seed storability as was suggested by Obendorf (1997), although variation for this trait is limited in this population.
CDT simulates aging of seeds under controlled, but artificial, conditions and can be used to predict seed storage potential (Hampton and TeKrony, 1995). Bentsink et al. (2000) showed the usefulness of the CDT to predict longevity since one QTL for CDT survival colocated with a QTL for naturally aged Ler/Cvi seeds. This was confirmed in the Ler/Sha RIL population by a similar colocation on top of chromosome 3. CDT reveals four loci in Ler/Cvi (Bentsink et al., 2000) and three in Ler/Sha, while natural aging only shows one in both populations. However, in both populations the range of germination percentages, after natural aging, found in the RILs is still above 60%, indicating that finding QTLs for natural aging probably requires storage periods longer than 20 months for this batch of Ler/Sha RIL seeds and longer than 4 years in Ler/Cvi seeds used. The colocation of the CDT QTL on the top of chromosome 1 with QTLs for germination on salt and after a heat treatment indicates that one aspect of seed longevity is a general tolerance to stress. However, seed longevity might also be affected by genes that control other aspects like the amount of Suc (top of chromosome 3), which was found to colocate with QTLs for germination on NaCl and CDT survival. The colocation of CDT survival and Suc was also observed in the Ler/Cvi population (Bentsink et al., 2000).
The QTL mapping approach appears to be a valuable method in elucidating the genetics but also the physiological background of traits involved in seed germination and seed longevity. Further analysis such as fine mapping and the study of mutants of candidate genes will be needed to prove the pleiotropic effects that are now only suggested by colocations of QTLs.
MATERIALS AND METHODS
Genotypes and Culture Conditions
A new RIL population was obtained from a cross between the accessions Landsberg erecta (Ler - NW20) and Shakdara (Sha - N929; pollen parent), previously also described as Shahdara (Loudet et al., 2002). The population of 114 RILs was obtained via selfing and single seed descent from individual F2 plants. F9 seeds were sown in petri dishes on water-saturated filter paper and incubated at 4°C for 4 d and then transferred to a growth chamber at 25°C. After 2 d of incubation, germinated seeds were transferred to soil and cultivated in an air-conditioned greenhouse (18°–23°C) in a 16-h photoperiod. Two randomized plots containing six plants per line were grown, and seeds were bulked from five plants per line. The sixth plant was used to isolate DNA and seeds were harvested separately. Seeds were stored under ambient conditions until further use. To reduce the developmental and environmental effects on seed characteristics, the onset of flowering was synchronized. For that the late flowering RILs were planted 2 weeks before the entire set of 114 lines was planted again. The lines and marker data will be made available through the Arabidopsis stock centers.
DNA Isolation and Genotyping
DNA was isolated from greenhouse-grown plants, one plant per line per plot. The Bernatzky and Tanksley (1986) protocol was adapted for rapid extraction of small quantities. Flower buds were harvested in liquid nitrogen and ground in 330 μL of a preheated (65°C) extraction solution (125 μL extraction buffer (0.35 m sorbitol, 100 mm Tris, 5 mm EDTA, pH 7.5 (HCl) together with 175 μL lysis buffer (200 mm Tris, 50 mm EDTA, 2 m NaCl, 2% (w/v) cetyl-trimethyl-ammonium bromide) to which 30 μL sarkosyl (10% w/v)) was added. The mixture of crude plant material and extraction solution was incubated for 30 min at 65°C; during this period occasional shaking was applied. Hereafter a solution of 400 μL chloroform/isoamyl alcohol (24:1) was added and vortexed. After centrifuging for 5 min at maximum speed in an Eppendorf centrifuge the water phase was transferred to a new tube. An equal amount of cold isopropanol was added to precipitate the DNA by carefully inverting the tube several times. After 10 min centrifugation at maximum speed in an Eppendorf centrifuge the water-alcohol mixture was discarded and the pellet washed with 70% cold ethanol. The pellet was left to dry and dissolved in water containing RNAse A and incubated 30 min at 37°C. Thereafter it was stored at 4°C. If the genotyping required more DNA, this was isolated from seedlings grown on half-strength Murashige and Skoog media containing 10% (w/v) Suc from seeds that had been harvested separately. DNA was now isolated using the Wizard magnetic 96 (Promega, Madison, WI; FF3760) DNA isolation kit.
Genotyping was done on F9 plants using microsatellite and CAPS markers. The CAPS and microsatellite markers were either found in the TAIR database (http://www.arabidopsis.org) or taken from http://www.inra.fr/qtlat/msat. Primers for markers that cannot be found in these two databases are summarized in Table IV. The ADH CAPS marker polymorphism was detected with the enzyme XbaI. All markers were first checked for polymorphism between Ler and Sha; thereafter, the polymorphic markers were used to genotype all individual RIL lines. For the microsatellite markers a standard protocol of 30 s 94°C, 30 s 50°C, and 30 s 72°C (35 cycles) was used except for FRI (54°C, 1-min extension) and FLC (52°C annealing, 2-min extension).
Table IV.
New markers used in genotyping the Ler/Sha RIL population
| Chromosome | Marker name | BAC | Primer 1 (5′ to 3′) | Primer 2 (5′ to 3′) | Origin |
|---|---|---|---|---|---|
| 1 | F3F19 | F3F19 | CGTCCAGACACTGACATTGGTTTTAGG | CCACTCACGTTCAGTGGGGTTAAACT | Genetics |
| 1 | F6D8-94 | F6D8 | GTCATTGGTTGCAATACGAGAGC | GCTGCCTCTTCCTTGTAAAGCC | Indel |
| 2 | F3P11-6b | F3P11 | TTCAATCTTCTCTACTGTCTTCG | AGCAGGAAGTAGTAAGTGGAATA | Alonso Blanco |
| 3 | F8J2 | F8J2 | GTGACCCAAGTGGGATCTCTC | ACGTGTGGGCAATCTTTTAAT | Genetics |
| 4 | FRI | F6N23 | CATGTCGTAATCATGCAACC | GAAGATCATCGAATTGGC | Tony Gendall |
| 4 | C6L9-78 | C6L9 | TGCTTTGTGAAAGTCTCTCATGCC | CCCTTTGATTGCTCAGTGATATCG | Indel |
| 4 | M4I22-22 | M4I22 | CGCTTTTAGGGGTAATATCGTCAC | CTGTGTGATCAGGCAAAACCAGT | Indel |
| 5 | FLC | T31P16 | CATTGGATAACTAATCTTTGAGC | CAGGCTGGAGAGATGACAAAA | Tony Gendall |
| 5 | K9D7 | K9D7 | GCTGTTGTAATTTGTGATAGGG | CATATGCCACGTTTCTTAATAG | Genetics |
| 5 | K8A10 | K8A10 | AATGCCAAGGATCAAAAGTGTT | GATGATCGGAGGAAAATGAAAA | Genetics |
Genetics: Primers developed by co-workers in genetics department.
Indel: Primers developed based on Ler and Col indel as found in the Cereon database (Jander et al., 2002).
Alonso Blanco: Primers were a gift from Dr. Alonso Blanco.
Tony Gendall: Primers were a gift from Dr. Tony Gendall (Gazzani et al., 2003).
Map Construction and QTL Analysis
A set of 66 markers covering most of the Arabidopsis genetic map at intervals of 1 to 15 cM was constructed with the JoinMap program (version 3.0, Plant Research International, Wageningen, The Netherlands). The computer program MapQTL (version 4.0, Plant Research International) was used to identify and locate QTL linked to molecular markers using both interval mapping and multiple-QTL model mapping (MQM) methods as described in its reference manual (http://www.plant.wageningen-ur.nl/products/). The estimated additive effect and the percentage of variance explained by each QTL as well as the total variance explained by all of the QTLs affecting a trait were obtained with MapQTL in the final MQM model. For this, different cofactor markers were tested around a putative QTL position (van Ooijen and Maliepaard, 1996) selecting as final cofactors the closest marker to each QTL, i.e. those maximizing the LOD score. A LOD threshold of 2.6 was applied to declare the presence of a QTL. We verified this threshold for interval mapping by applying the permutation test to each data set (10,000 repetitions) and found a P = 0.05 LOD varying between 2.4 and 2.6. Two-LOD support intervals were established as 95% confidence intervals (van Ooijen, 1992). For every trait, two-way QTL interactions were analyzed at a significance level of P < 0.05, using the general linear model module of the statistical package SPSS version 11.0.1 (SPSS, Chicago). For each analysis, the closest linked markers to the corresponding detected QTL were used as random factors in the ANOVA (the same markers used as cofactors in the MQM mapping MapQTL). A Bonferoni correction to the 0.05 threshold of significance (P value) was applied if multiple tests were performed on the same data set. Heritability as the proportion of the variance explained by the between-line effects was calculated using the general linear model module of the statistical package of SPSS version 11.0.1 (SPSS). The QTL effect was calculated using the untransformed data; the value was obtained by fixing the same cofactors as with the transformed data used to determine the positions of the QTLs.
Sugar Content Measurement
One hundred seeds from bulks of five plants of one replicate were weighed. Sugars were extracted from these 100 seeds by heating for 15 min at 76°C in 80% (v/v) methanol with the addition of 25 μg melezitose as internal standard. After heating the homogenate was centrifuged for 5 min at 10,000g. The supernatant was vacuum-evaporated and its residue resuspended in 0.5 mL pure water and injected into a Dionex HPLC system (Dionex Corporation, Sunnyvale, CA). Sugar content was determined with a high-pH-anion-exchange HPLC, using a gradient pump module (model GP40) and an ED40 pulsed electrochemical detector (Dionex Corporation, Sunnyvale, CA). Sugars were chromatographed on a CarboPac PA100 4- × 250-mm column (Dionex) preceded by a guard column (CarboPac PA100, 4 × 50 mm). Mono-, di-, and trisaccharides were separated by elution in increasing concentration of NaOH (50–200 mm), with a flow rate of 1 mL per min. Peaks were identified by coelution of standards. Sugar quantity was corrected by means of the internal standard and transformed to micrograms of sugar per milligram of seed.
Germination Assays
To assess seed dormancy, two replicas of 50 to 100 seeds were sown on water-saturated filter paper in petri dishes. Germination was scored as seeds with radicle protusion after 7 d incubation in a growth chamber (25°C, 16-h light period). All the lines were sown at several intervals after harvest until the germination was between 95% and 100%; meanwhile seeds were stored under dry conditions. Seed dormancy of a genotype was estimated in a single parameter as the number of days of seed dry storage required to reach 50% germination (DSDS50). To estimate the DSDS50 value of each genotype, all the measurements of germination proportions at various times during seed storage were used for probit regression on a logarithm time scale applying the regression module of the statistical package SPSS version 11.0.1 (SPSS; Alonso-Blanco et al., 2003). The DSDS50 values were individually assessed for all RILs and each replicate, thereafter the DSDS50 values were averaged and QTL analysis was performed with this average value.
Germination assays were performed, in duplicate, with seeds bulked from five plants. These bulks were harvested from two greenhouse-grown replicates. Prior to the transfer to the growth chamber (25°C, 16-h light period) seeds were stored for 4 d at 4°C unless stated differently.
Sodium and mannitol tolerance was estimated by germinating 50 to 80 vapor-sterilized seeds on water agar containing 150 mm of NaCl or 400 mm of mannitol. Plates were stored for 3 d at 4°C. Germination was counted after 13 d (NaCl) or 7 d (mannitol). The germination data were corrected for germination on medium containing no salt and then probit transformed for each line and both replicates. The average value was used for QTL analysis.
Vapor sterilization of seeds was done by placing seeds in opened Eppendorf tubes in a desiccator jar. Then a 250-mL beaker containing 100 mL commercial bleach was placed inside and 3 mL concentrated HCl was added. The desiccator jar was closed and the seeds were sterilized by chlorine gas. After 2 to 3 h the jar was opened and the Eppendorf tubes were closed until use.
Tolerance to hydrogen peroxide or ABA was estimated by germinating 50 to 80 seeds on filter paper either saturated with 200 mm H2O2 or 1 μm ABA. Plates were stored for 7 d (only ABA) at 4°C and final germination was counted after 7 d. The germination percentages per duplicate were first averaged, then corrected for germination at day 7 on water and thereafter probit transformed for each line and both replicates. These values were then averaged and used for QTL analysis.
Natural aging was determined by germinating 50 to 80 seeds of seed lots that had been stored for 20 months at ambient conditions on water-saturated filter paper. Final germination was counted after 7 d. The germination percentages per duplicate were first averaged; thereafter both replicates were probit transformed and averaged to perform the QTL analysis.
Speed of germination was determined by germinating 50 to 80 seeds on water-saturated filter paper. Germination was first determined at 36 h after transfer to the light, a second germination percentage was determined after 60 h, and final germination was determined after 7 d. Both duplicates for each replicate were first corrected for germination at day 7 on water; thereafter these corrected germination percentages for 36 and 60 h were averaged per duplicate. This average germination percentage was then probit transformed for each duplicate; these were averaged within the greenhouse-grown replicate, and thereafter both replicates were averaged to perform the QTL analysis.
CDT
CDT was performed according to Tesnier et al. (2002). Briefly, seeds were equilibrated at 85% relative humidity (15°C), and day 0 controls were immediately dried back at 32% relative humidity. Treatment was done by storing the seeds (at 85% relative humidity) for a number of days at 40°C (2, 4, and 6 d). Then these seeds were also dried back at 32% relative humidity (20°C) and stored at 4°C until the germination assay was performed. Two replicates of 50 seeds were tested for each day of treatment. Seed deterioration was estimated as a single parameter as the number of days of treatment required to reach 50% germination (LD50). Germination proportions of all treatments were used for probit regression on a time scale, in days, applying the regression module of the statistical package SPSS version 11.0.1 (SPSS). The LD50 points of both replicates were then log transformed, to improve normality, averaged, and used in QTL mapping procedures.
Heat Assay
Seeds, harvested from plants grown in a climatized greenhouse in Tucson, Arizona were sown on water-saturated filter paper. Thereafter they were left to imbibe for 18 h at room temperature, transferred to 50°C, and left for 8 h as heat treatment. The germination percentage was determined after 7 d. This was done in three replicates of each line; to normalize the data all germination percentages were probit transformed, and QTL mapping was with the average probit.
Acknowledgments
We thank Elena Matteucci, Sohaee Negar, and Corrie Hanhart for performing the salt germination assay; Shannon Parrington for the heat-stress germination assay; Olivier Loudet and Carlos Alonso-Blanco for providing marker information before publication; Leónie Bentsink for instructions about the MapQTL program; and Steve Tonsor and the STW Supervision Committee for useful suggestions and discussions.
This work was supported by the Technology Foundation STW (Stichting Toegepaste Wetenschappen), Applied Science Division of the Netherlands Organization for Scientific Research (project no. WBI4737 to E.J.M.C., G.J.R., H.B.-D.V.), the NATURAL program of the European Union (contract no. QLG2–CT–2001–01097), and by a fellowship from the Government of Egypt to M.E.E.-L. E.V. was supported by the National Science Foundation POWRE Grant, the Guggenheim Foundation, and the U.S. Department of Agriculture National Research Initiative Competitive Grants Program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036814.
References
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankenstijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an under exploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pots J, Kuiper MTR (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Bailly C, Audigier C, Ladonne F, Wagner M-H, Coste F, Corbineau F, Côme D (2001) Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. J Exp Bot 52: 701–708 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124: 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD (1986) Genetics of actin-related sequences in tomato. Theor Appl Genet 72: 314–324 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Jones RL (2001) Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J 25: 19–29 [DOI] [PubMed] [Google Scholar]
- Bettey M, Finch-Savage WE (1998) Stress protein content of mature Brassica seeds and their germination performance. Seed Sci Res 8: 347–355 [Google Scholar]
- Bettey M, Finch-Savage WE, King GJ, Lynn JR (2000) Quantitative genetic analysis of seed vigor and pre-emergence seedling growth traits in Brassica oleracea. New Phytol 148: 227–286 [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination. Plenum Press, New York
- Bies-Ethève N, da Silva Conceicao A, Giraudat J, Koornneef M, Léon-Kloosterziel KM, Valon C, Delseny M (1999) Importance of the B2 domain of the Arabidopsis ABI3 protein and 2S gene regulation. Plant Mol Biol 40: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, Blankenstijn-de Vries H, Ruys GJ, Groot SPC, Koornneef M (2003) Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol 132: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui KH, Peng SB, Xing YZ, Xu CG, Yu SB, Zhang Q (2002) Molecular dissection of seedling-vigour and associated physiological traits in rice. Theor Appl Genet 105: 745–753 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol 122: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath A, Bethke PC, Jones RL (2001) Enzymes that scavenge reactive oxygen species are down regulated prior to gibberellic acid induced programmed cell death in barley aleurone. Plant Physiol 126: 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad MR, Lin GY, Chen FQ (1999) Comparison of QTLs for seed germination under non-stress, cold stress and salt stress in tomato. Plant Breed 118: 167–173 [Google Scholar]
- Frugoli JA, Hong Zhong H, Nuccio ML, McCourt P, McPeek MA, Thomas TL, McClung CR (1996) Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol 112: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Clancy JA, Han F, Prada D, Kleinhofs A, Ullrich SE (2003) Molecular dissection of a dormancy QTL region near the chromosome 7 (5H) L telomere in barley. Theor Appl Genet 107: 552–559 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grene R (2002) Oxidative stress and acclimation mechanism in plants. In CR Somerville, EM Meyerowitz eds, The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD. doi/10.1199/tab.0036, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Hampton JG, TeKrony DM (1995) Handbook of Vigour Test Methods. The International Seed Testing Association, Zurich, pp 117
- Hoekstra FA, Golovina EA, Buitink J (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6: 431–438 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJK, Kurup S, McKibbin R (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4: 275–280 [Google Scholar]
- Hong S-W, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, Hendry GF, Atherton NM, Vertucci-Walters CW (1996) Free radical accumulation and lipid peroxidation in testas of rapidly aged soybean seeds: a light-promoted process. Seed Sci Res 6: 101–107 [Google Scholar]
- Khurmatov KK (1982) Heterogeneity of natural populations of the Arabidopsis thaliana (Pamiro-Alay) in the flowering time. Arabidopsis Inf Serv 19: 62–66 [Google Scholar]
- Kliebenstein D, Monde R, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Léon-Kloosterziel KM, Alvarez Gil M, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Keijzer CJ, Koornneef M (1994) A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 X Shahdara recombinant inbred line population: a powerful tool for genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94: 263–272 [Google Scholar]
- McDonald MB (1999) Seed deterioration: physiology, repair and assessment. Seed Sci Technol 27: 177–237 [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T (2003) Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol Ecol 12: 1137–1151 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M (1998) Arabidopsis thaliana: a model plant for genome analysis. Science 282: 662–682 [DOI] [PubMed] [Google Scholar]
- Miura K, Lin SY, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104: 981–986 [DOI] [PubMed] [Google Scholar]
- Natoli A, Gorni C, Chegdani F, Ajmone Marsan P, Colombi C, Lorenzoni C, Marocco A (2002) Identification of QTLs associated with sweet sorghum quality. Maydica 47: 311–322 [Google Scholar]
- Obendorf RL (1997) Oligosaccharides and galactosyl cyclitols in seed desiccation tolerance. Seed Sci Res 7: 63–74 [Google Scholar]
- Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana - a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet 3: 124–136 [DOI] [PubMed] [Google Scholar]
- Peeters AJM, Blankestijn-de Vries H, Hanhart CJ, Léon-Kloosterziel KM, Zeevaart JAD, Koornneef M (2002) Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol Plant 115: 604–612 [DOI] [PubMed] [Google Scholar]
- Quesada A, Garcia-Martinez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Plachy C, Frahry G (2001) Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol 125: 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinniah UR, Ellis RH, John P (1998) Irrigation and seed quality development in rapid-cycling Brassica; seed germination and longevity. Ann Bot (Lond) 82: 309–314 [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77 [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Yamaguchi Shinozaki K, Kamada H, Giraudat J, Shinozaki K (1999) Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol 40: 119–123 [DOI] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, van Pijlen JG, van der Geest AHM, Bino RJ, Groot SPC (2002) A controlled deterioration test of Arabidopsis thaliana reveals genetic variation in seed quality. Seed Sci Technol 30: 149–165 [Google Scholar]
- Thorlby G, Veale E, Butcher K, Warren G (1999) Map positions of SFR genes in relation to other freezing related genes in Arabidopsis thaliana. Plant J 17: 445–452 [DOI] [PubMed] [Google Scholar]
- Van der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, Van Ooijen JW, Koornneef M (1997) QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 79: 190–200 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84: 803–811 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Maliepaard C (1996). MapQTL™ version 4.0: Software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen, The Netherlands
- Wehmeyer N, Vierling E (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IW, Schiff CL, Hughes DE, Somerville SC (2001) Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accessions Kashmir-1. Genetics 158: 1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu J-K (2002) Salt tolerance. In CR Somerville, EM Meyerowitz eds, The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD. doi/10.1199/tab.0048, http://www.aspb.org/publications/arabidopsis/