Abstract
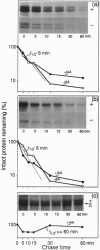
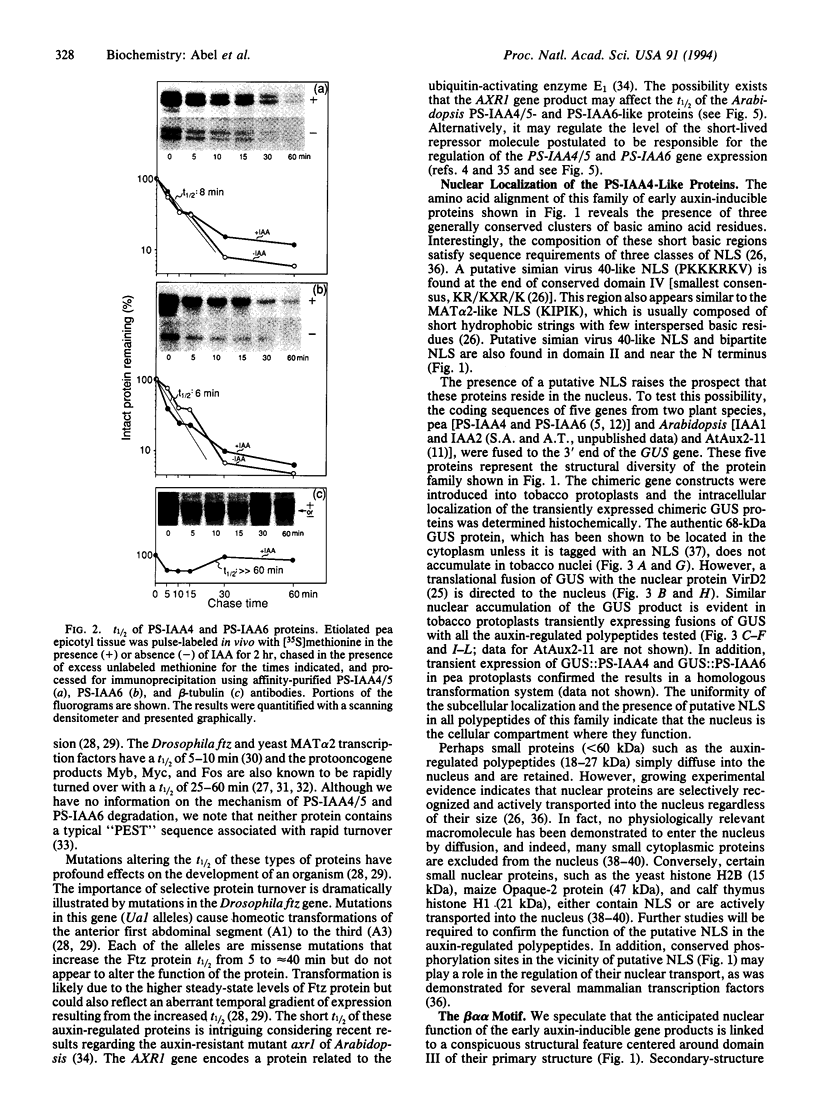
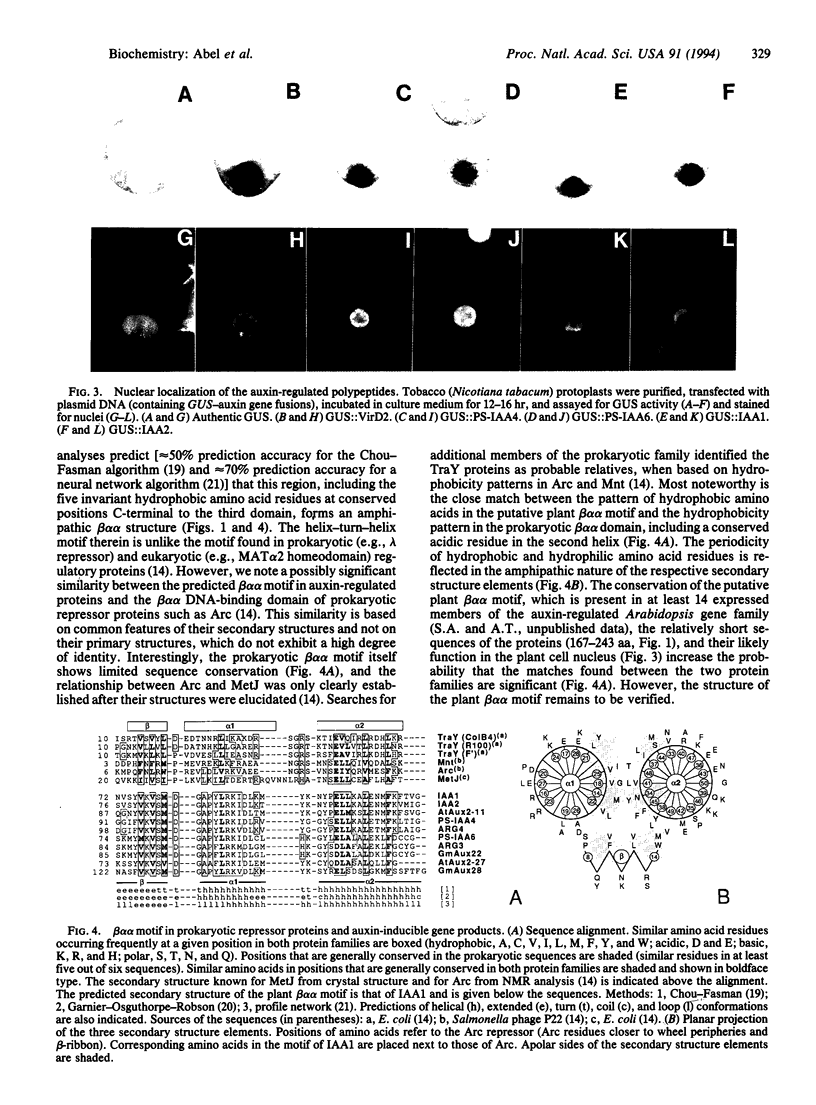
The plant growth hormone indoleacetic acid (IAA) transcriptionally activates gene expression in plants. Some of the genes whose expression is induced by IAA encode a family of proteins in pea (PS-IAA4 and PS-IAA6) and Arabidopsis (IAA1 and IAA2) that contain putative nuclear localization signals that direct a beta-glucuronidase reporter protein into the nucleus. Pulse-chase and immunoprecipitation experiments have defined the t1/2 of the PS-IAA4 and PS-IAA6 proteins to be 8 and 6 min, respectively. Their most prominent feature is the presence of a beta alpha alpha motif similar to the beta-sheet DNA-binding domain found in prokaryotic repressors of the Arc family. Based on these data, we suggest that plant tissues express short-lived nuclear proteins as a primary response to IAA. We propose that these proteins act as activators or repressors of genes responsible for mediating the various auxin responses.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainley W. M., Walker J. C., Nagao R. T., Key J. L. Sequence and characterization of two auxin-regulated genes from soybean. J Biol Chem. 1988 Aug 5;263(22):10658–10666. [PubMed] [Google Scholar]
- Ashburner M. Puffs, genes, and hormones revisited. Cell. 1990 Apr 6;61(1):1–3. doi: 10.1016/0092-8674(90)90205-s. [DOI] [PubMed] [Google Scholar]
- Ballas N., Wong L. M., Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J Mol Biol. 1993 Oct 20;233(4):580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D. S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990 Mar 23;60(6):999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Carrington J. C., Freed D. D., Leinicke A. J. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell. 1991 Sep;3(9):953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Conner T. W., Goekjian V. H., LaFayette P. R., Key J. L. Structure and expression of two auxin-inducible genes from Arabidopsis. Plant Mol Biol. 1990 Oct;15(4):623–632. doi: 10.1007/BF00017836. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Odell G. M., Schubiger G. Cytoarchitecture and the patterning of fushi tarazu expression in the Drosophila blastoderm. Genes Dev. 1987 Dec;1(10):1226–1237. doi: 10.1101/gad.1.10.1226. [DOI] [PubMed] [Google Scholar]
- Evans M. L., Ray P. M. Timing of the auxin response in coleoptiles and its implications regarding auxin action. J Gen Physiol. 1969 Jan;53(1):1–20. doi: 10.1085/jgp.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Giudice L. C., Waxdal M. J., Weintraub B. D. Comparison of bovine and mouse pituitary glycoprotein hormone pre-alpha subunits synthesized in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4798–4802. doi: 10.1073/pnas.76.10.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. A., Zupan J. R., Citovsky V., Zambryski P. C. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell. 1992 Jan 10;68(1):109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- Jones M. D. Reverse transcription of mRNA by Thermus aquaticus DNA polymerase followed by polymerase chain reaction amplification. Methods Enzymol. 1993;218:413–419. doi: 10.1016/0076-6879(93)18033-9. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Bonifer C., Sippel A. E. Identification and characterization of the protein encoded by the human c-myb proto-oncogene. EMBO J. 1986 Aug;5(8):1903–1911. doi: 10.1002/j.1460-2075.1986.tb04443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leyser H. M., Lincoln C. A., Timpte C., Lammer D., Turner J., Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993 Jul 8;364(6433):161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988 Jun;8(6):2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. A., Hagen G., Brown C. S., Gee M. A., Guilfoyle T. J. Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell. 1989 Feb;1(2):229–239. doi: 10.1105/tpc.1.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R. L., Zokas L., Schreiber R. D., Verma I. M. Rapid induction of the expression of proto-oncogene fos during human monocytic differentiation. Cell. 1985 Jan;40(1):209–217. doi: 10.1016/0092-8674(85)90324-1. [DOI] [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller P. W., Keller J. A., Parks J. E., Silbert J. E., Theologis A. Structural characterization of the early indoleacetic acid-inducible genes, PS-IAA4/5 and PS-IAA6, of pea (Pisum sativum L.). J Mol Biol. 1993 Oct 20;233(4):789–798. doi: 10.1006/jmbi.1993.1555. [DOI] [PubMed] [Google Scholar]
- Osborne M. A., Silver P. A. Nucleocytoplasmic transport in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 1993;62:219–254. doi: 10.1146/annurev.bi.62.070193.001251. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992 Dec;100(4):1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo M. A., Freed D. D., Carrington J. C. Nuclear transport of plant potyviral proteins. Plant Cell. 1990 Oct;2(10):987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Kusaba M., Hiraoka Y., Nagata T. Characterization of the auxin-regulated par gene from tobacco mesophyll protoplasts. Plant J. 1991 Nov;1(3):327–332. doi: 10.1046/j.1365-313x.1991.t01-2-00999.x. [DOI] [PubMed] [Google Scholar]
- Theologis A., Huynh T. V., Davis R. W. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol. 1985 May 5;183(1):53–68. doi: 10.1016/0022-2836(85)90280-3. [DOI] [PubMed] [Google Scholar]
- Varagona M. J., Schmidt R. J., Raikhel N. V. Monocot regulatory protein Opaque-2 is localized in the nucleus of maize endosperm and transformed tobacco plants. Plant Cell. 1991 Feb;3(2):105–113. doi: 10.1105/tpc.3.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., Quail P. H. Native phytochrome: Inhibition of proteolysis yields a homogeneous monomer of 124 kilodaltons from Avena. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5272–5276. doi: 10.1073/pnas.79.17.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]