Abstract
Auxin signaling through the SCFTIR1-Aux/IAA-ARF pathway is one of the best-studied plant hormone response pathways. Components of this pathway, from receptors through to transcription factors, have been identified and analyzed in detail. Although we understand elementary aspects of how the auxin signal is perceived and leads to a transcriptional response, many questions remain about the in vivo function of the pathway. Two crucial issues are the tissue-specificity of the response, i.e. how distinct cell types can interpret the same auxin signal differently, and the response to a signaling gradient, i.e. how a graded distribution of auxin can elicit distinct expression patterns along its range. Here, we speculate on how signaling through the canonical SCFTIR1-Aux/IAA-ARF pathway may achieve divergent responses.
Introduction
Auxin plays a central role in nearly every aspect of plant development as well as in numerous adaptations to environmental cues. How this growth signal is differentially perceived and interpreted by individual cells to yield the plethora of varying molecular, physiological and growth responses is a central and historic question in plant biology (Darwin and Darwin 1888). Indole-3-acetic acid (henceforth also auxin) is the predominant form of auxin and is found across plant – and algal – species (Lau et al. 2009 for review). Other endogenous auxinic compounds have also been discovered, but these molecules are not as well-studied (i.e. indole-3-butyric acid, phenylacetic acid and 4-chloroindole-3-acetic acid) (Simon and Petrasek 2011 for review). Auxin distribution is highly regulated, especially within the embryo and growth centers of the apical and cambial meristems where auxin concentration gradients are formed that guide cellular differentiation (see Bhalerao and Bennett 2003, Bennett and Scheres 2010, Ha et al. 2010, Peris et al. 2010 for reviews). Furthermore, auxin distribution is dynamically modified in response to external stimuli such as light or gravity (Swarup et al. 2005, Ding et al. 2011). Such changes in auxin input elicit a distinct output in different tissues. For example, increased auxin levels lead to cell elongation in the hypocotyl whereas expansion is inhibited in the root. Thus, cells need to be able to discretely sense and interpret changes in auxin distribution that occur both through developmental time and as a result of reactive fluctuations.
Indole-3-acetic acid is a simple aromatic carboxylic acid that is found in the nanomolar to micromolar range in planta (Edlund et al. 1995, Uggla et al. 1996, Petersson et al. 2009). In the apoplast, auxin is principally in its protonated form, allowing diffusion across the plasma membrane, whereas the relatively alkaline cytosolic conditions lead to acidification and intracellular accumulation of the molecule. Auxin import is further facilitated by the influx carriers AUXIN RESISTANT 1/LIKE AUXIN RESISTANT (AUX1/LAX) (Bennett et al. 1996), as well as the P-GLYCOPROTEIN 4 (AtPGP4) (Santelia et al. 2005) and the NITRATE TRANSPORTER 1 (NRT1.1) (Krouk et al. 2010). Auxin efflux can only be achieved by active transport and is mediated by plasma membrane localized members of the PINFORMED family of efflux carriers (PIN) (Petrasek et al. 2006), as well as AtPGP1 and 19 (reviewed in Geisler and Murphy 2006). It is the cell type-specific polar localization of the exporters (as well as AtPGP4 and NRT1.1) that generates a flux of auxin within tissues and actively regulates auxin distribution. Environmental and developmental cues relayed by additional small signaling molecules, as well as auxin itself, lead to altered expression and localization of PIN efflux carriers and hence a redistribution of local auxin levels (see Vanneste and Friml 2009 for review). Auxin elicits both immediate (non-transcriptional) responses, such as cell-wall acidification and effects on endocytosis (Robert et al. 2010, Takahashi et al. 2012), as well as a vast array of transcriptomic responses consisting of thousands of auxin-responsive genes (Goda et al. 2008, Paponov et al. 2008, Chapman et al. 2012).
In this article, we review our current understanding of how auxin might elicit diverse outputs in different cell types and discuss how a graded distribution of auxin concentration can lead to defined responses along its range. We will focus in particular on regulation of the transcriptional response downstream of the nuclear SCFTIR1-Aux/IAA co-receptor complex (Calderon Villalobos et al. 2012).
The SCFTIR1-Aux/IAA auxin-receptor complex regulates ARF activity
The TIR1 (TRANSPORT INHIBITOR RESISTANT 1) component of the SCFTIR1-Aux/IAA auxin receptor complex was first identified in an Arabidopsis forward-genetic screen for auxin-transport-inhibitor resistant mutants (Ruegger et al. 1997). In Arabidopsis, TIR1 is a member of a six-gene clade of F-box proteins that also includes AUXIN SIGNALING F-BOX PROTEIN 1 through 5 (AFB1–5) (Dharmasiri et al. 2005). tir1 mutants show retarded growth and diminished stature in addition to a resistance to auxin (Ruegger et al. 1998). Successive elimination of AFB1, AFB2 and AFB3 in tir1 mutants leads to exacerbation of the growth defects and increased auxin resistance (Dharmasiri et al. 2005). In vitro assays and mutant analysis have shown that AFB4 and AFB5 are also involved in auxin perception (Greenham et al. 2011).
TIR1 and the AFBs are F-box components of a nuclear SCF-type E3 ubiquitin ligase, which target the Aux/IAA (AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE) proteins for degradation by the 26S proteasome via polyubiquitination (Gray et al. 2001, Petroski and Deshaies 2005, dos Santos Maraschin et al. 2009). In addition to a conserved F-box domain, the TIR1/AFB proteins contain a leucine-rich-repeat (LRR) domain that binds the Aux/IAA transcriptional repressors (Gray et al. 1999, Tan et al. 2007). Crucially, the binding of Aux/IAAs to the TIR1/AFBs, is dependent on auxin (Gray et al. 2001, Dharmasiri et al. 2005, Kepinski and Leyser 2005, Tan et al. 2007). Auxin in effect works as molecular glue between the TIR1 LRR binding pocket and the recognition domain (DII) in the Aux/IAA proteins. In addition, biochemical studies indicate that both TIR1 and the Aux/IAA proteins are required for auxin binding and therefore function as co-receptors (Calderon Villalobos et al. 2012). Auxin promotes the formation of the co-receptor complex which in turn facilitates ubiquitination and degradation of the Aux/IAAs and consequent de-repression of the transcriptional auxin response (Figure 1).
Fig. 1.
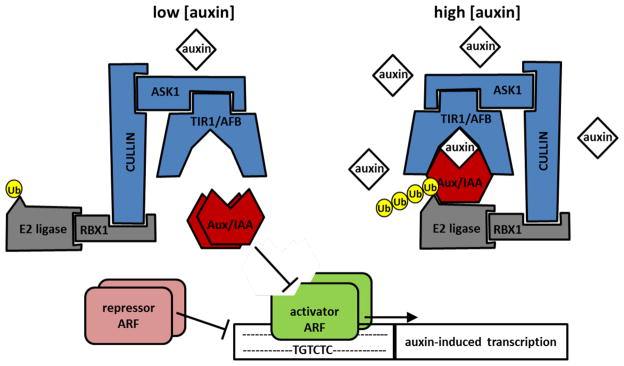
The SCFTIR1-Aux/IAA-ARF auxin-response pathway. A cartoon of the interactions in the canonical nuclear auxin receptor complex and its effects on auxin-induced transcription. The TIR1/AFB interaction with the Aux/IAA co-receptor/transcriptional repressors increases at higher auxin concentrations, promoting Aux/IAA ubiquitination and degradation. The transcriptional activity of activator ARFs is modulated by the levels of Aux/IAAs and repressor ARFs.
Auxin-induced transcriptional changes are mediated by AUXIN RESPONSE FACTOR (ARF) B3-type transcription factors, which bind to the auxin response elements (AuxRE) in the promoters of auxin-responsive genes (Ulmasov et al. 1997, see Guilfoyle and Hagen 2007 for review). Based on protein sequence homology, the family of ARFs can be broadly subdivided into “Q-rich” activator-ARFs and “non-Q-rich” repressor-ARFs, which promote and inhibit transcription at AuxRE, respectively (Guilfoyle and Hagen 2007). At low auxin levels, the (activating) ARFs are bound by Aux/IAAs (Tiwari et al. 2001), which recruit the TOPLESS co-repressor and associated chromatin-modification machinery, thereby inhibiting transcription (Szemenyei et al. 2008). The transcriptional repression of (activator-)ARF activity is released upon auxin-induced Aux/IAA degradation, leading to an induction of gene expression (Guilfoyle and Hagen 2007). ARF activation of transcription is further modulated independently of auxin levels by repressor-ARFs, presumably also via chromatin modification (Ulmasov et al. 1999, see Tiwari 2003 for review, Causier et al. 2012). It is worth noting that the ARF family members have been divided into so-called activators and repressors based on their behavior in transient and heterologous assays with a limited number of promoters, both groups of proteins may display more complex behaviors in planta.
Several direct ARF target genes have been identified using different methods, including transient expression in protoplasts, chromatin immunoprecipitation, transcriptomic analysis of auxin signaling mutants, activation of gene expression in the presence of translational inhibitors, or simply because they are up-regulated by auxin treatment and contain an AuxRE in their promoter (Ulmasov et al. 1997, Hardtke et al. 2004, Okushima et al. 2005, Okushima et al. 2007, Cole et al. 2009, Schlereth et al. 2010). Direct ARF-mediated, auxin-responsive genes include genes involved in auxin signaling itself (e.g. the Aux/IAA co-receptors and GRETCHEN HAGEN 3 (GH3) auxin amido-conjugases) (Hagen and Guilfoyle 1985, Nemhauser et al. 2006) as well as transcription factors implicated in the high-order regulation of development (e.g. LATERAL ORGAN BOUNDARY DOMAIN CONTAINING PROTEINs and TARGET OF MONOPTEROS 7) (Okushima et al. 2007, Schlereth et al. 2010). Intriguingly, some auxin-responsive genes show diffuse, graded expression patterns, e.g. select Aux/IAAs, whereas others display defined expression domains, e.g. certain LBDs (Okushima et al. 2007, Vernoux et al. 2011). Moreover, some are induced ubiquitously by auxin treatment while the induction of others is restricted to specific tissues (Paponov et al. 2008).
How could the SCFTIR1-Aux/IAA-ARF pathway mediate divergent responses in different cell types?
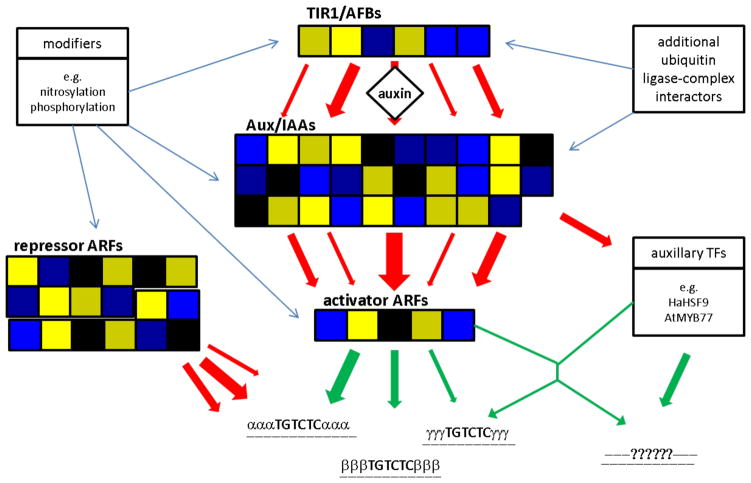
Auxin elicits differing responses depending on the cellular context in which it is perceived (reviewed in Kieffer et al. 2010); either promoting or inhibiting particular cellular processes, such as cell division and cell expansion, and inducing or repressing the transcription of specific genes contingent on cell-identity and developmental age. It is posited that the SCFTIR1-Aux/IAA-ARF pathway is modular and that mixing-and-matching of the different members within the three families of proteins generates an auxin response tailored to a specific cell type (reviewed in Weijers and Jurgens 2004, De Smet et al. 2010, Calderon Villalobos et al. 2012, Rademacher et al. 2012). Indeed, individual family members differ in their spatial expression patterns. Transcriptional reporters indicate that the TIR1/AFBs are widely expressed throughout the plant, but translational fusions suggest that post-transcriptional regulation of TIR1/AFB restricts spatial protein expression levels (Parry et al. 2009). This may be partly due to the action of the small RNA miR393 (Vidal et al. 2010, Chen et al. 2011, Si-Ammour et al. 2011). The family of Aux/IAAs (consisting of 29 members in Arabidopsis) (Overvoorde et al. 2005) also appears to have divergent tissue-specific gene expression. However, a systematic analysis of spatial reporter-gene expression for this family has not been conducted and visualization of native, basal Aux/IAA protein expression levels is hampered by the high turnover rate of translational reporter fusions (Lokerse and Weijers 2009 for review). A more comprehensive analysis of spatial ARF gene-expression in the seedling root and embryo demonstrates that this family (consisting of 23 members in Arabidopsis) is differentially expressed in various cell types and developmental zones (Rademacher et al. 2011). ARF protein expression levels may also be expected to be different from the relative promoter activities as several ARFs have been shown to be subject to post-transcriptional or post-translational regulation (Fahlgren et al. 2006, Salmon et al. 2008, Rosado et al. 2012). Thus, there are definite differences in the cellular composition of the SCFTIR1-Aux/IAA-ARF pathway and these may form the basis of the specificity of the transcriptional response.
If differences in the cellular composition of the SCFTIR1-Aux/IAA-ARF pathway help determine the specificity of the response, there must be family member-dependent differences in the component interactions. TIR1/AFB-Aux/IAA interactions have been tested by in vitro co-immunoprecipitation as well as auxin-dependent yeast 2-hybrid assays (Gray et al. 2001, Greenham et al. 2011, Calderon Villalobos et al. 2012, Yu et al. 2013). Results indicate that the affinity of TIR1 and the AFBs for the Aux/IAA co-receptors differs for the various Aux/IAA family members (Calderon Villalobos et al. 2012). A number of approaches, particularly yeast 2-hybrid tests, have documented homo- and heterodimeric interactions between members of the Aux/IAA and ARF families (reviewed in Guilfoyle and Hagen 2012). One large-scale study suggested that, in general, the Aux/IAAs bind more strongly to the activating ARFs than the repressing ARFs (Vernoux et al. 2011). However, this study is subject to the usual caveats concerning yeast 2-hybrid data. In fact, a recent study indicates that ARF9, a member of the “non-Q-rich” repressor class, interacts with IAA10 in plant cells, and that this interaction has an important function during embryogenesis (Rademacher et al. 2012). Ultimately, the specificity of the SCFTIR1-Aux/IAA-ARF-mediated transcriptional response must lie in the DNA-binding site preference of the different ARFs; this implies that there is more to ARF-DNA binding than recognition of the AuxRE (TGTCTC) alone. Furthermore, homotypic and heterotypic interactions of the ARFs indicate that multiple recognition sequences in close proximity may be involved in the binding of particular ARF dimers to the DNA (Ulmasov et al. 1999). Thus, there are three interaction levels in the SCFTIR1-Aux/IAA-ARF pathway where the expression and interaction properties of divergent family members within the separate modules could theoretically direct the cellular transcriptional response elicited by changes in auxin levels (Figure 2).
Fig. 2.
Fine-tuning the transcriptional auxin response through the SCFTIR1-Aux/IAA-ARF pathway. The modular nature of the members of the SCFTIR1-Aux/IAA-ARF pathway may allow for specific outputs of pathway activation, depending on the cellular expression of family members with distinct interaction affinities. In Arabidopsis there are 6 auxin F-box receptors (TIR1 and AFB1–5), 29 Aux/IAA co-receptors, 5 activator ARFs (ARF5–8 and ARF19) and 18 repressor ARFs (of which ARF2, 12, 14, 15, 19, 20, 21 and 22 contain recognized transcriptional repression domains and the remaining ten may have as yet undescribed transcription factor activities). Red and green arrows indicate theoretical negative and positive regulation, respectively.
Aside from the divergent expression of TIR1/AFB, Aux/IAA, and ARF family members, cell type-specific auxin responses may also be influenced by additional regulators of the SCFTIR1-Aux/IAA-ARF pathway. For instance, TIR1 activity was recently shown to be modulated by nitric oxide-mediated S-nitrosylation, leading to increased TIR1-Aux/IAA interaction (Terrile et al. 2012). There are reports that the Aux/IAAs and ARFs are phosphorylated (Colon-Carmona et al. 2000, Vert et al. 2008). Furthermore, both Aux/IAAs and ARFs may interact with additional transcription factors to modify the expression of their targets, e.g. MYB77 has been shown to interact with ARF7 (Shin et al. 2007), KANADI proteins interact with ARF3 (Kelley et al. 2012) and HEATSHOCK FACTOR 9 interacts with IAA27 in sunflower (Carranco et al. 2010).
Finally, many tissue-specific auxin-responsive genes may be indirect auxin targets, regulated by auxin-induced or -repressed transcription factors. Direct tissue-specific regulation of just a few secondary transcription factors could lead to highly divergent outcomes in overall transcriptional responses. Alternatively, the SCFTIR1-Aux/IAA-ARF pathway may directly and broadly regulate secondary transcription factors that then relay specificity by interacting with tissue-specifically expressed co-factors.
How could an auxin gradient elicit distinct expression domains through the SCFTIR1-Aux/IAA-ARF pathway?
Auxin gradients give positional information and guide cellular maturation in growth centers such as the vascular cambium and the root apical meristem. These concentration gradients are thought to function to drive distinct expression patterns in the cells along their range to stage development from cell division through to cell expansion and differentiation (see Bhalerao and Bennett 2003 for review). According to this thesis the relationship between local auxin concentration levels and auxin-responsive gene expression is not a simple linear one; higher auxin levels may not necessarily elicit stronger induction or repression respectively for all auxin-sensitive genes. This suggests that separate branches of the SCFTIR1-Aux/IAA-ARF pathway have different sensitivity to auxin and may be discretely activated along a concentration gradient.
Notably, assessment of the interaction of TIR1 and AFBs with Aux/IAAs using auxin-dependent yeast-2-hybrid assays has shown that distinct F-box-Aux/IAA pairs indeed have different auxin dose-response dependencies in their interactions (Calderon Villalobos et al. 2012). In vitro analysis of the dissociation constants for auxin binding among different TIR1/AFB-Aux/IAA co-receptor pairs has demonstrated a broad spectrum (encompassing orders of magnitude) of auxin-sensing capability that is mostly governed by the Aux/IAA component of the complex (Calderon Villalobos et al. 2012). In addition, heterologous reconstitution of the TIR1/AFB-mediated degradation of fluorescently tagged Aux/IAAs in yeast has recently demonstrated that there are major differences in the degradation dynamics of Aux/IAAs in vivo (Havens et al. 2012). These findings suggest that different local auxin concentrations result in distinct relative levels of the Aux/IAAs present in a particular cell. Assuming that there are differences among the interaction specificities of the Aux/IAAs and ARFs expressed in cells along a concentration range, different sets of ARF target genes may be activated at different local auxin concentrations. Further investigation of TIR1/AFB-Aux/IAA interactions has identified putative determinants in this differential binding. Residues outside of the DII of Aux/IAAs are likely involved, as the interaction and degradation-rates of full-length Aux/IAA proteins is markedly different from that of truncations or the isolated DII (Havens et al. 2012). A screen for tir1 mutants with altered binding properties performed using the auxin-dependent yeast 2-hybrid interaction assay has recently pinpointed two specific mutations on the outside of the LRR binding pocket that lead to increased auxin-mediated TIR1/AFB-Aux/IAA interaction and caused an auxin-hypersensitive phenotype when expressed in planta (Yu et al. 2013). These studies indicate that differences in the auxin-dependent, TIR1/AFB-assisted degradation dynamics of Aux/IAAs could generate distinct thresholds of expression along an auxin gradient and that features outside of the known interaction interfaces of the TIR1/AFB-Aux/IAA co-receptor complex may modulate the sensitivity to auxin in an as yet unknown manner.
Additionally, feedback regulation of the SCFTIR1-Aux/IAA-ARF pathway could lead to non-linear dose-response curves for auxin-sensitive transcription. The expression of several Aux/IAA genes is strongly induced by auxin and thus forms a regulatory negative-feedback loop (Tiwari et al. 2001). Differences in Aux/IAA protein synthesis and degradation rates at varying auxin levels may therefore affect complex input to output relations. Some ARFs are also induced by auxin, potentially refining the response (Lau et al. 2011). Together with additional auxin-responsive regulators of the SCFTIR1-Aux/IAA-ARF pathway (e.g. MYB77, whose gene expression is also induced by auxin) (Shin et al. 2007), the picture that emerges is one of a complex self-regulating network that could produce diverse transcriptional interpretations of gradual changes in local auxin levels.
Perspective
Although the SCFTIR1-Aux/IAA-ARF pathway is a relatively well-studied plant hormone signaling pathway, there are still many questions to be addressed in order to better understand its in vivo function. For example, the current model does not account for direct auxin-mediated repression of transcription. Generally, auxin down-regulated genes are affected at later time-points, compared to auxin-induced genes, suggesting that repression may be mostly indirect and the result of the primary induction of repressors (Paponov et al. 2008). Further investigation is required to ascertain if (and how) the pathway acts to directly repress transcription. An additional key issue concerns the specificity of the different ARF family members. The phenotypes of individual arf mutants vary considerably, indicating redundant and non-overlapping function (Hardtke et al. 2004). Further studies will assess whether these differences are, at least in part, due to differences in ARF target genes. Another intriguing observation that merits more attention is the finding that separate cell-lineages in the (root) meristem may maintain individual and highly divergent auxin concentration gradients, as visualized by the recently developed DII-Venus biosensor (Brunoud et al. 2012). This may indicate that the regulation of cellular maturation by auxin is not generic across cell types but may involve cell lineage-specific interpretation of isolated signaling gradients.
Recently, cell type-specific analysis of genome-wide auxin responses in four different tissue-specific GFP-marker lines of the seedling root was accomplished by the use of fluorescence activated cell sorting (Bargmann et al. 2013). This study demonstrated that different cell types have both divergent and parallel transcriptomic responses to auxin treatment. Interestingly, the AuxRE was found to be over-represented only in the promoters of genes with relatively equal induction in all four cell types analyzed, suggesting that the more cell type-specific auxin responses may be mediated by alternate promoter elements. Cross reference of the auxin-response data with spatial expression data in the root showed that a gene’s response to auxin treatment predicts its endogenous spatial expression along the longitudinal axis of the root, with auxin-induced genes generally displaying high meristematic expression and vice versa for auxin-repressed genes. This correlation could be indicative of meristematic auxin distribution in the root apex being a determinant in the spatial expression of thousands of genes. Further analysis of large-scale auxin- and spatial-expression datasets as well as transcription factor-target sets may be able to dissect cell type-specific and graded auxin responses to reveal separate modes of gene regulation.
Interestingly, recent studies indicate that AUXIN BINDING PROTEIN 1 (ABP1) is a negative regulator of the SCFTIR1-Aux/IAA-ARF pathway (Tromas et al. 2013). ABP1 is a well-characterized cell surface receptor that has been implicated in a number of rapid non-transcriptional auxin responses (see Napier et al. 2002, Tromas et al. 2010, Sauer and Kleine-Vehn 2011 for reviews). Conditional inactivation of ABP1 results in a number of growth defects in seedlings and also affects the expression of auxin-regulated genes (Braun et al. 2008, Effendi et al. 2011). Strikingly this group shows that loss of ABP1 acts to suppress the phenotype caused by loss of the TIR1/AFB F-box proteins. ABP1 knockdown also increased Aux/IAA degradation. These results suggest that auxin perception at the cell surface may influence nuclear auxin-receptor sensitivity (Tromas 2013). These examples show there may be many additional inputs to the SCFTIR1-Aux/IAA-ARF pathway that could impart contextual specificity of the response.
There are many interesting avenues for further research on the transcriptional response to changes in auxin concentration levels through the SCFTIR1-Aux/IAA-ARF pathway. Availability of gain-of-function mutants of the different Aux/IAAs (Li et al. 2009) and ARFs (Krogan et al. 2012) makes it possible to study the (in)activation of particular branches of the SCFTIR1-Aux/IAA-ARF pathway and may help in the identification of distinctive transcriptional targets downstream of specific Aux/IAA and ARF family members. Large-scale systematic studies (e.g. chromatin immunoprecipitation analysis of the Aux/IAA and ARF families) will give insight on the range of variability in the specificity and overlap of this crucial auxin signal transduction pathway. However, the process of auxin perception is probably much more complex than we currently appreciate and also involves signal transduction through pathways not (yet) associated with the canonical pathway. It is likely that future study of this pathway and its interactions will yield fruitful outcomes for years to come.
Acknowledgments
The authors would like to thank Dr. Dior Kelly for critical reading of the manuscript and the reviewers for helpful comments and suggestions. Research in the authors’ lab is supported by grants from the NIH (GM46444), NSF (IOS 0744800), DOE (FG02-11ER16007), the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (GBMF 3038) to ME.
References
- Bargmann BO, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, Birnbaum KD. A map of cell type-specific auxin responses. Mol Syst Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B. Root development-two meristems for the price of one? Curr Top Dev Biol. 2010;91:67–102. doi: 10.1016/S0070-2153(10)91003-X. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Bennett MJ. The case for morphogens in plants. Nat Cell Biol. 2003;5:939–943. doi: 10.1038/ncb1103-939. [DOI] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell. 2008;20:2746–2762. doi: 10.1105/tpc.108.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J. Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc Natl Acad Sci USA. 2010;107:21908–21913. doi: 10.1073/pnas.1014856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One. 2012;7:e36210. doi: 10.1371/journal.pone.0036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Bao ML, Sun YZ, Yang YJ, Xu XH, Wang JH, Han N, Bian HW, Zhu MY. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol Biol. 2011;77:619–629. doi: 10.1007/s11103-011-9838-1. [DOI] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136:1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C, Darwin FE. The power of movement in plants. 1888. [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, Naudts M, Levesque MP, Ehrismann JS, Inze D, Luschnig C, Benfey PN, Weijers D, Van Montagu MC, Bennett MJ, Jurgens G, Beeckman T. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, Friml J. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol. 2011;13:447–452. doi: 10.1038/ncb2208. [DOI] [PubMed] [Google Scholar]
- dos Santos Maraschin F, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59:100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklof S, Sundberg B, Moritz T, Sandberg G. A Microscale Technique for Gas Chromatography-Mass Spectrometry Measurements of Picogram Amounts of Indole-3-Acetic Acid in Plant Tissues. Plant Physiol. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi Y, Rietz S, Fischer U, Scherer GF. The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. Plant J. 2011;65:282–294. doi: 10.1111/j.1365-313X.2010.04420.x. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 2006;580:1094–1102. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, Kiba T, Takatsuto S, Fujioka S, Asami T, Nakano T, Kato H, Mizuno T, Sakakibara H, Yamaguchi S, Nambara E, Kamiya Y, Takahashi H, Hirai MY, Sakurai T, Shinozaki K, Saito K, Yoshida S, Shimada Y. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, Estelle M. The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr Biol. 2011;21:520–525. doi: 10.1016/j.cub.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Fletcher JC. Shoot Apical Meristem Form and Function. 2010;91:103–140. doi: 10.1016/S0070-2153(10)91004-1. [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ. Rapid induction of selective transcription by auxins. Mol Cell Biol. 1985;5:1197–1203. doi: 10.1128/mcb.5.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development. 2012;139:1105–1109. doi: 10.1242/dev.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S. Defining auxin response contexts in plant development. Curr Opin Plant Biol. 2010;13:12–20. doi: 10.1016/j.pbi.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012;194:391–401. doi: 10.1111/j.1469-8137.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, Benkova E, Nacry P, Gojon A. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jurgens G. Auxin triggers a genetic switch. Nat Cell Biol. 2011;13:611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- Lau S, Shao N, Bock R, Jurgens G, De Smet I. Auxin signaling in algal lineages: fact or myth? Trends Plant Sci. 2009;14:182–188. doi: 10.1016/j.tplants.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Li H, Cheng Y, Murphy A, Hagen G, Guilfoyle TJ. Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol. 2009;149:1277–1288. doi: 10.1104/pp.108.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokerse AS, Weijers D. Auxin enters the matrix – assembly of response machineries for specific outputs. Curr Opin Plant Biol. 2009;12:520–526. doi: 10.1016/j.pbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Napier RM, David KM, Perrot-Rechenmann C. A short history of auxin-binding proteins. Plant Mol Biol. 2002;49:339–348. [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant. 2008;1:321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris CIL, Rademacher EH, Weijers D. Green Beginnings – Pattern Formation in the Early Plant Embryo. 2010;91:1–27. doi: 10.1016/S0070-2153(10)91001-6. [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 2009;21:1659–1668. doi: 10.1105/tpc.109.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, Skupa P, Benkova E, Perry L, Krecek P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zazimalova E, Friml J. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Rademacher E, Moller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68:597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G. Different Auxin Response Machineries Control Distinct Cell Fates in the Early Plant Embryo. Dev Cell. 2012;22:211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Covanova M, Hayashi K, Dhonukshe P, Yang Z, Bednarek SY, Jones AM, Luschnig C, Aniento F, Zazimalova E, Friml J. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143:111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA. 2012;109:19537–19544. doi: 10.1073/pnas.1214774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Ramos J, Callis J. Degradation of the auxin response factor ARF1. Plant J. 2008;54:118–128. doi: 10.1111/j.1365-313X.2007.03396.x. [DOI] [PubMed] [Google Scholar]
- Santelia D, Vincenzetti V, Azzarello E, Bovet L, Fukao Y, Duchtig P, Mancuso S, Martinoia E, Geisler M. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005;579:5399–5406. doi: 10.1016/j.febslet.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Sauer M, Kleine-Vehn J. AUXIN BINDING PROTEIN1: The Outsider. Plant Cell. 2011;23:2033–2043. doi: 10.1105/tpc.111.087064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Jr, Vazquez F. miR393 and Secondary siRNAs Regulate Expression of the TIR1/AFB2 Auxin Receptor Clade and Auxin-Related Development of Arabidopsis Leaves. Plant Physiol. 2011;157:683–691. doi: 10.1104/pp.111.180083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Petrasek J. Why plants need more than one type of auxin. Plant Sci. 2011;180:454–460. doi: 10.1016/j.plantsci.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Terrile MC, Paris R, Calderon-Villalobos LI, Iglesias MJ, Lamattina L, Estelle M, Casalongue CA. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012;70:492–500. doi: 10.1111/j.1365-313X.2011.04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB. The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paponov I, Perrot-Rechenmann C. AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci. 2010;15:436–446. doi: 10.1016/j.tplants.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Tromas A, Paque S, Stierle V, Quettier AL, Muller P, Lechner E, Genschik P, Perrot-Rechenmann C. Auxin-Binding Protein 1 is a negative regulator of the SCFTIR1/AFB pathway. Nat Commun. 2013;4:2496. doi: 10.1038/ncomms3496. [DOI] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA. 1996;93:9282–9286. doi: 10.1073/pnas.93.17.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc’h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C, Kepinski S, Bennett M, De Veylder L, Traas J. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutierrez RA. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Jurgens G. Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol. 2004;7:687–693. doi: 10.1016/j.pbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]