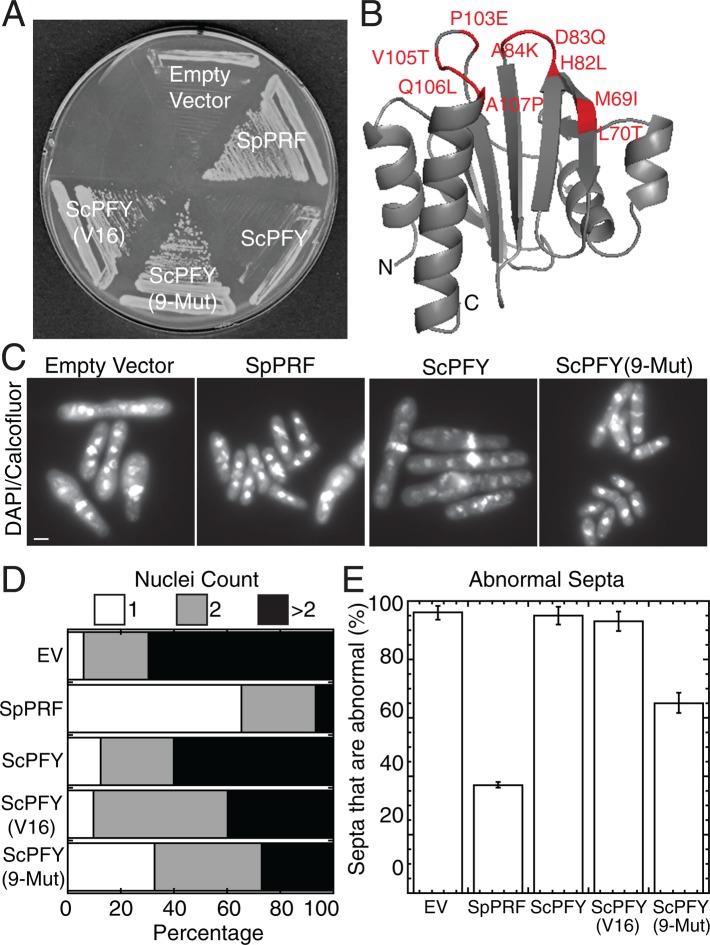
A budding yeast profilin ScPFY mutant library was engineered and used to select for mutants that complement the fission yeast profilin temperature-sensitive mutant cdc3-124. ScPFY(9-Mut) rescues the ability of profilin to facilitate actin assembly by the cytokinesis formin Cdc12 in vitro and allow contractile ring assembly in vivo.
Abstract
The evolutionarily conserved small actin-monomer binding protein profilin is believed to be a housekeeping factor that maintains a general pool of unassembled actin. However, despite similar primary sequences, structural folds, and affinities for G-actin and poly-l-proline, budding yeast profilin ScPFY fails to complement fission yeast profilin SpPRF temperature-sensitive mutant cdc3-124 cells. To identify profilin's essential properties, we built a combinatorial library of ScPFY variants containing either WT or SpPRF residues at multiple positions and carried out a genetic selection to isolate variants that support life in fission yeast. We subsequently engineered ScPFY(9-Mut), a variant containing nine substitutions in the actin-binding region, which complements cdc3-124 cells. ScPFY(9-Mut), but not WT ScPFY, suppresses severe cytokinesis defects in cdc3-124 cells. Furthermore, the major activity rescued by ScPFY(9-Mut) is the ability to enhance cytokinesis formin Cdc12-mediated actin assembly in vitro, which allows cells to assemble functional contractile rings. Therefore an essential role of profilin is to specifically facilitate formin-mediated actin assembly for cytokinesis in fission yeast.
INTRODUCTION
Cells are poised to rapidly assemble actin filaments because they maintain a large reserve of actin monomers bound to the small, ∼14-kDa protein profilin (Carlsson et al., 1977; Pollard and Cooper, 1984). Profilin prevents unwanted spontaneous actin assembly by strongly inhibiting nucleation, but profilin-actin readily adds to free barbed ends once new filaments are formed by an expanding list of nucleation and elongation factors (Chhabra and Higgs, 2007; Chesarone and Goode, 2009; Dominguez, 2009). These factors include proteins like formin and Ena/VASP, which contain continuous stretches of proline residues that profilin can bind simultaneously with actin (Schutt et al., 1993; Reinhard et al., 1995; Chang et al., 1997; Evangelista et al., 1997; Mahoney et al., 1997, 1999). In addition, most profilins increase the nucleotide exchange rate of actin monomers (Mockrin and Korn, 1980), which is essential in yeast (Wolven et al., 2000; Lu and Pollard, 2001). Profilin also binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2; Goldschmidt-Clermont et al., 1990), although the physiological significance is less clear.
Formins are specialized nucleation factors that drive rapid profilin-actin assembly for processes such as division, motility, and polarization (Higgs, 2005; Kovar, 2006; Breitsprecher and Goode, 2013). Formins are large proteins with two internal actin assembly formin homology (FH) domains FH1 and FH2, which are flanked by regulatory domains. The FH1 domain binds profilin-actin and transfers it to the FH2 domain–associated barbed end (Vavylonis et al., 2006). This direct transfer mechanism has been proposed to explain how the FH1 and FH2 domains cooperate to nucleate filaments that elongate profilin-actin up to 12-fold faster than control filaments (Romero et al., 2004; Kovar et al., 2006; Vavylonis et al., 2006; Paul and Pollard, 2008; Vidali et al., 2009). However, this model is oversimplified, as we discovered that the FH2 domain plays a critical role in allowing formins to drive the rapid assembly of profilin-actin (Neidt et al., 2009; Scott et al., 2011). We hypothesized that profilin may interact directly with the FH2 domain, which might help facilitate dissociation of profilin from the barbed end in preparation for the next round of actin monomer addition (Neidt et al., 2009; Scott et al., 2011).
Given their similar biochemical properties and general role in maintaining a common pool of ATP-loaded G-actin (Witke, 2004; Jockusch et al., 2007), it is not surprising that diverse profilins can often substitute for each other. For example, human profilin 1 complements a budding yeast profilin-null mutant (Ostrander et al., 1999), the plant profilins AtPRF1 and AtPRF3 partially rescue a fission yeast profilin temperature-sensitive (ts) mutant (Christensen et al., 1996), and bovine profilin 1 moderately rescues growth and cytokinesis defects in profilin-minus Dictyostelium cells (Schluter et al., 1998). However, multiple profilin isoforms within the same cell interact differently with particular actin filament nucleators to carry out different cellular roles (Pilo Boyl et al., 2007; Khadka et al., 2009; Mouneimne et al., 2012), and some profilins cannot replace each other. For example, budding yeast profilin ScPFY fails to complement the fission yeast profilin SpPRF ts mutant cdc3-124 (Balasubramanian et al., 1994). Therefore ScPFY may not be capable of performing a critical function in fission yeast, which could reveal profilin's essential cellular functions. Fission yeast cells have at least four distinct actin structures, which each carries out specific cellular roles (La Carbona et al., 2006; Kovar et al., 2011). Arp2/3 complex makes highly dynamic branched filaments for endocytic actin patches, whereas formins make less dynamic straight filaments for the cytokinetic contractile ring (Cdc12), polarized actin cables (For3), and mating (Fus1; Kovar et al., 2011).
To elucidate the essential function(s) of profilin in fission yeast, we created a combinatorial library of budding yeast profilin ScPFY variants and selected for mutations that complement the fission yeast profilin ts mutant cdc3-124 (Nurse et al., 1976). To determine the fundamental properties rescued by the isolated ScPFY mutants, we used a combination of in vitro biochemistry and live-cell time-lapse fluorescence microscopy.
RESULTS
Isolation of an evolved budding yeast profilin (ScPFY) that supports life in fission yeast
Despite 72% similar amino acid sequences, similar structural folds, and similar affinities for monomeric actin and poly-l-proline (Table 1; Eads et al., 1998; Lu and Pollard, 2001), budding yeast profilin (ScPFY) fails to complement fission yeast profilin ts mutant cdc3-124 cells at the restrictive temperature of 36°C (Figure 1A; Balasubramanian et al., 1994). To identify the function that ScPFY is not capable of performing in fission yeast, we carried out directed evolution to convert ScPFY into fission yeast profilin (SpPRF) by single-residue editing and selected for ScPFY variants that make cdc3-124 cells viable at the restrictive temperature (see Materials and Methods).
TABLE 1:
Comparison of properties of SpPRF, WT ScPFY, and ScPFY(9-Mut).
| Profilin | Protein stabilitya (D1/2, M urea) | Poly-l-proline bindingb (Kd, μM) | Cdc12(FH1) bindingc (Kd, μM) | Muscle G-actin bindingd (Kd, μM) | Fission yeast G-actin bindinge (actin band intensity at 5 μM profilin, a.u.) | Nucleotide exchangef (time to one-half exchange at 0.5 μM profilin, s) | PtdIns(4,5)P2 bindingg (percentage unbound profilin in flowthrough, 100 μM PtdIns(4,5)P2) |
|---|---|---|---|---|---|---|---|
| SpPRF | 4.5 | 59 | 6.4 | 0.8 | 6.4 | 59.3 ± 6.7 | 34.1 ± 1.6 |
| ScPFY | 3.3 | 224 | 24.7 | 0.7 | 10.1 | 29.0 ± 3.9 | 57.4 ± 2.8 |
| ScPFY (9-Mut) | 2.6 | 195 | 22.7 | 0.4 | 9.4 | 16.3 ± 3.9 | 18.0 ± 2.9 |
aSupplemental Figure S3A.
bSupplemental Figure S4A.
cSupplemental Figure S4B.
dSupplemental Figure S3, B and C.
eSupplemental Figure S3, D and E.
fSupplemental Figure S3, F and G.
gSupplemental Figure S4C.
FIGURE 1:
Nine mutations in budding yeast profilin, ScPFY(9-Mut), partially complement fission yeast profilin-ts cells. (A) Complementation of the fission yeast profilin-ts strain cdc3-124 after 3 d at 36°C with either an empty vector control or vectors expressing the listed profilins under the control of the P81nmt1 promoter. The ScPFY(V16) mutant was identified during an initial selection (Supplemental Table S1). (B) Structure of ScPFY (Eads et al., 1998) highlighting nine mutations that lie within or border the actin-binding region and allow ScPFY to support life in fission yeast. (C–E) Fission yeast profilin cdc3-124 ts cells after 10 h at 36°C in liquid minimal medium, fixed with methanol, and stained with DAPI and calcofluor to visualize nuclei and septa. (C) Representative fluorescence images. Scale bar, 5 μm. (D) Number of nuclei in cells: 1 (white), 2 (gray), and >2 (black). (E) Percentage of septa that are abnormal (too thick, misplaced, misoriented, or multiple septa in a single cell). Error bars, SD, n = 3 sets of ≥50 cells each.
We initially targeted 16 of the 39 divergent residues between ScPFY and SpPRF (Supplemental Figure S1, A and B). Residues were selected because they 1) are invariant in profilins from the fission yeast genus Schizosaccharomyces, 2) are spread throughout the ScPFY structure, and 3) have single nucleotide degeneracy between the ScPFY and SpPRF residue (Supplemental Figure S1, A and B). Selection of ∼500,000 variants identified 17 unique ScPFY mutants containing 3–11 residue changes that are capable of allowing cdc3-124 cells to grow at the restrictive temperature (Supplemental Figure S1C and Supplemental Table S1). All but one of the 17 mutants included the M69I mutation (Supplemental Table S2). Although M69I is neither necessary nor sufficient to complement cdc3-124 cells, we found that the surrounding area that includes the actin-binding region is generally important for complementation (Supplemental Figure S2). There are nine amino acid differences between SpPRF and ScPFY in this region, of which only five were initially selected for mutation. We therefore made ScPFY(9-Mut), in which all nine residues were edited to their SpPRF counterpart (red residues in Figure 1B, red boxes in Supplemental Figure S1A).
ScPFY(9-Mut) functions better in fission yeast cells than either wild-type (WT) ScPFY or the originally isolated ScPFY variants but not as well as SpPRF (Figure 1, A and C–E). Cdc3-124 cells expressing ScPFY(9-Mut) from a plasmid under control of the weakest P81nmt1 promoter are significantly healthier than an empty vector control, WT ScPFY, or an original ScPFY(V16) mutant (Figure 1, C–E). For example, the percentage of cdc3-124 cells with greater than two nuclei dropped from 60% for WT ScPFY to 28% for ScPFY(9-Mut) (Figure 1D), and the percentage of abnormal septa decreased from 96% for WT ScPFY to 65% for ScPFY(9-Mut) (Figure 1E).
Comparison of ScPFY(9-Mut)'s general biochemical properties
To determine the rescued fundamental activity that allows complementation of cdc3-124 cells, we purified ScPFY(9-Mut) and investigated its general biochemical properties. We found that most of the properties of ScPFY(9-Mut) are not closer to SpPRF than to ScPFY and are therefore unlikely the reason for ScPFY(9-Mut)'s ability to rescue growth in cdc3-124 cells (Table 1; described in detail in the Supplemental Results). 1) The stability of ScPFY(9-Mut), as determined by urea denaturation, is lower than that of either ScPFY or SpPRF (Supplemental Figure S3A). 2) The affinity of ScPFY(9-Mut) for muscle G-actin and ability to prevent spontaneous actin assembly are similar to those of both ScPFY and SpPRF (Supplemental Figure S3, B and C). 3) The affinities of both ScPFY(9-Mut) and ScPFY for fission yeast G-actin are slightly higher than that of SpPRF (Supplemental Figure S3, D and E). 4) Both ScPFY(9-Mut) and ScPFY increase the rate of G-actin nucleotide exchange significantly more than does SpPRF (Supplemental Figure S3, F and G). 5) The affinity of ScPFY(9-Mut) and ScPFY for the proline-rich ligands poly-l-proline (PLP) and the formin Cdc12 FH1 domain are ∼3.5-fold lower than that of SpPRF (Supplemental Figure S4, A and B; Chang et al., 1997).
Conversely, ScPFY(9-Mut) binds PtdIns(4,5)P2 micelles twofold better than SpPRF and fivefold better than WT ScPFY (Table 1 and Supplemental Figure S4C). However, we do not believe that ScPFY(9-Mut)'s increased affinity for PtdIns(4,5)P2 explains its ability to complement cdc3-124 cells. First, human profilin HsPRO1 also binds PtdIns(4,5)P2 micelles very well (Kovar et al., 2001; Skare and Karlsson, 2002), yet it fails to complement cdc3-124 cells (see later discussion of Figure 4A; Ezezika et al., 2009). Second, plant profilin AtPRF1 weakly complements cdc3-124 cells (see Figure 4A; Christensen et al., 1996), yet it binds PtdIns(4,5)P2 poorly (Skare and Karlsson, 2002).
FIGURE 4:
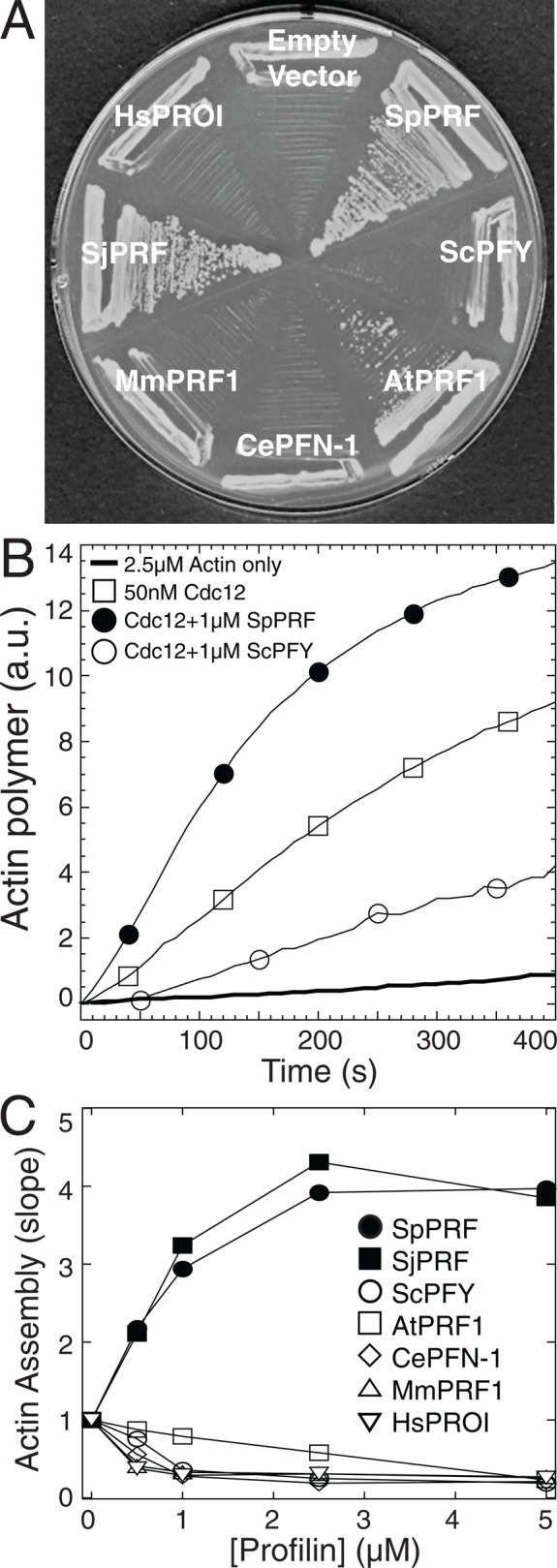
Enhancement of actin assembly with formin Cdc12 correlates with profilin's ability to complement profilin mutant cdc3-124 cells. (A) The fission yeast profilin mutant cdc3-124 strain after 3 d at 36°C with either a control empty vector or a vector expressing various profilins under control of the P81nmt1 promoter. Profilin abbreviations: SpPRF, S. pombe; ScPFY, Saccharomyces cerevisiae; AtPRF1, Arabidopsis thaliana; CePFN-1, Caenorhabditis elegans; MmPRF1, Mus musculus; SjPRF, Schizosaccharomyces japonicus; and HsPROI, Homo sapiens. (B, C) Utilization of purified profilins by formin Cdc12 in vitro. Spontaneous assembly of 2.5 μM Mg-ATP actin monomers (20% pyrene-labeled), 50 nM Cdc12(FH1FH2), and a range of concentrations of the indicated profilins. (B) Time course of the polymerization of actin monomers in the absence (thick curve) or presence of Cdc12(FH1FH2) (□) and 1 μM SpPRF (•) or ScPFY (◯). (C) Dependence of the actin assembly rate (slope) in the presence of 50 nM Cdc12(FH1FH2) on the concentration of SpPRF (•), SjPRF (▪), ScPFY (◯), AtPRF1 (□), CePFN-1 (⋄), MmPRF1 (▵), or HsPROI (▿).
ScPFY(9-Mut) suppresses cytokinesis defects in profilin-mutant cdc3-124 cells
Because WT ScPFY and ScPFY(9-Mut) bind similarly to their general ligands G-actin and proline-rich peptides, the basis for ScPFY(9-Mut) complementing cdc3-124 cells is likely more complicated. Vegetatively growing fission yeast cells primarily have three actin-dependent structures, each of which is dependent on a specific actin assembly factor: endocytic actin patches (Arp2/3 complex), cytokinetic contractile rings (formin Cdc12), and polarizing actin cables (formin For3; Kovar et al., 2011). However, actin cables are not required for viability (Feierbach and Chang, 2001). Fission yeast profilin mutant cells are primarily defective in cytokinesis (Balasubramanian et al., 1994, 1996; Chang et al., 1996; Lu and Pollard, 2001), although endocytic actin patch defects have also been reported (Pelham and Chang, 2001, 2002). Therefore we used fluorescence imaging to determine whether defects in either Arp2/3 complex–mediated endocytosis or formin Cdc12-mediated cytokinesis are the primary reasons that cdc3-124 cells are not viable. Visualizing the internalization of the integral membrane marker green fluorescent protein GFP-Syb1 (Basu and Chang, 2011) revealed that compared with WT cells treated with the Arp2/3 complex inhibitor CK-666 (Nolen et al., 2009), cdc3-124 cells do not have major endocytosis defects at permissive (25°C), semipermissive (32°C), or restrictive (36°C) temperature (Figure 2). It is therefore unlikely that cdc3-124 cells are inviable due to endocytosis defects. Conversely, depletion of profilin in fission yeast leads to dysfunctional formin Cdc12-mediated contractile rings that fail to constrict during cytokinesis (Figure 3). Contractile rings were labeled with the myosin-II regulatory light chain Rlc1-GFP to follow their assembly and constriction (Figure 3A; Le Goff et al., 2000). Time-lapse fluorescence microscopy revealed that 100% of dividing WT cells form a functional contractile ring that begins constricting by 35 min, whereas after 2 h at the semirestrictive temperature of 32°C, no dividing cdc3-124 cells form constricting rings by 35 min (Figure 3B and Supplemental Video S1). Therefore profilin mutant cdc3-124 cells have significant functional defects in cytokinesis but not endocytosis.
FIGURE 2:
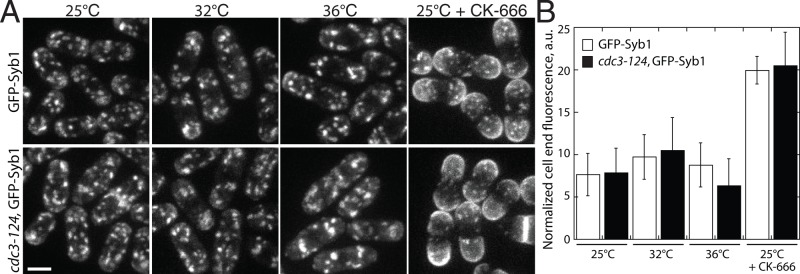
Profilin mutant cdc3-124 cells do not have significant endocytosis defects. Endocytosis in WT and cdc3-124 fission yeast cells. (A, B) Endocytosis in cells expressing integral membrane marker GFP-Syb1. When GFP-Syb1 is localized to the plasma membrane, endocytosis is impaired. (A) Fluorescence micrographs of WT or cdc3-124 cells at permissive (25°C), semipermissive (32°C), or restrictive (36°C) temperature for 2 h before imaging. Cells treated with 100 μM CK-666 for 30 min are used as a positive control for endocytic defects. Scale bar, 5 μm. (B) Mean cell-end GFP-Syb1 fluorescence for WT or cdc3-124 cells normalized to background fluorescence. Error bars, SD; n = 10 cells.
FIGURE 3:
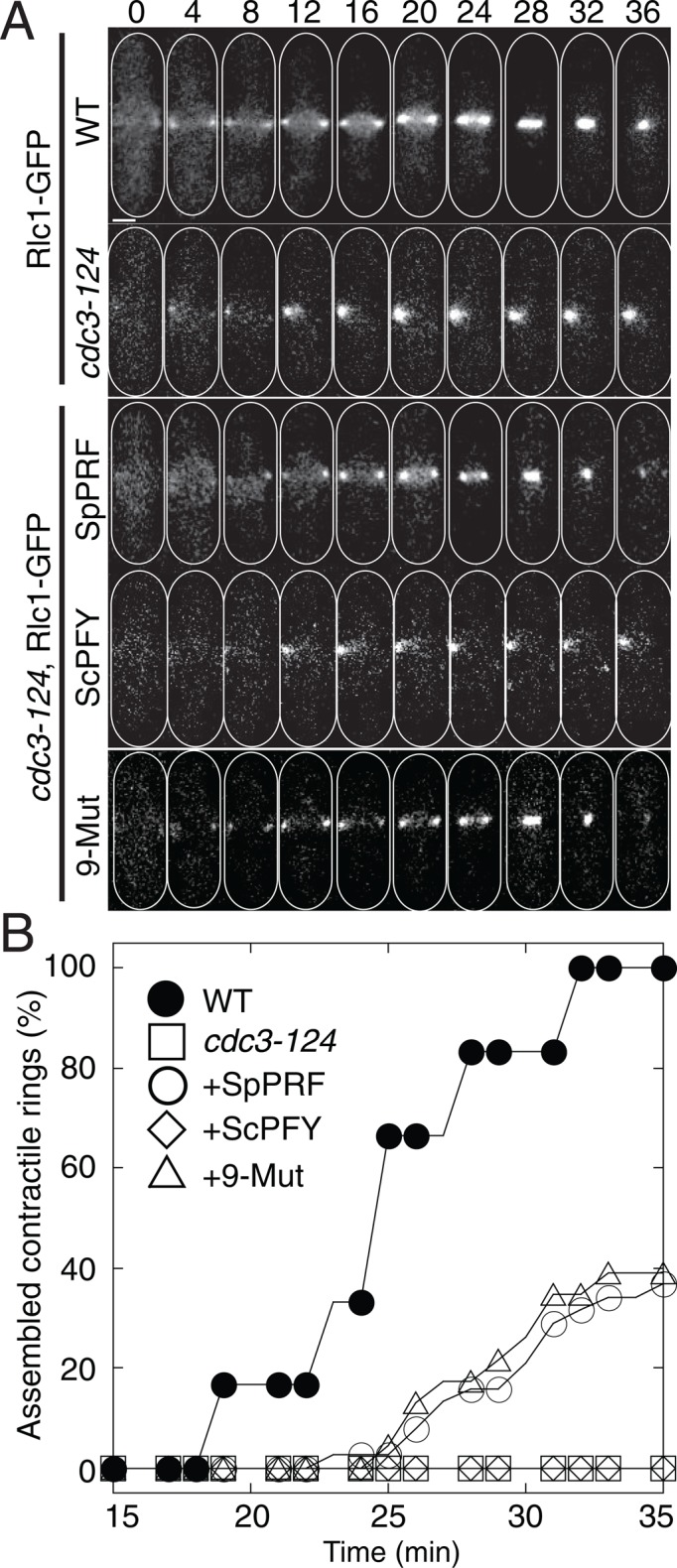
ScPFY(9-Mut) rescues cytokinesis defects of cdc3-124 cells. (A, B) Cytokinesis in WT, cdc3-124, or cdc3-124 cells expressing SpPRF, WT ScPFY, or ScPFY(9-Mut) from the ura4 locus under control of the P81nmt1 promoter. (A) Time-lapse micrographs of representative cells forming a contractile ring labeled with Rlc1-GFP (Supplemental Videos S1 and S2). Scale bar, 2 μm. (B) Plot of the cumulative frequency of constricting contractile rings over time for WT (•), cdc3-124 (□), or cdc3-124 cells expressing SpPRF (◯), WT ScPFY (⋄), or ScPFY(9-Mut) (⋄).
We next determined whether contractile ring assembly defects in cdc3-124 cells are suppressed by expression of ScPFY(9-Mut). We constructed cdc3-124 strains with SpPRF, WT ScPFY, or ScPFY(9-Mut) integrated at the ura4 locus (one copy per cell) under control of the P81nmt1 thiamine-repressible promoter. After 2 h at 32°C, no functional contractile rings are assembled after 35 min in cdc3-124 cells expressing WT ScPFY (Figure 3, A and B, and Supplemental Video S2). However, expression of either SpPRF or ScPFY(9-Mut) increases the percentage of functional rings to ∼40% by 35 min. The decreased expression level of SpPRF from the weakest P81nmt1 promoter likely accounts for <100% functional contractile rings (Supplemental Figure S5). Therefore profilin's essential role in formin Cdc12-mediated cytokinetic contractile ring assembly is properly performed only by profilins that support life in fission yeast (SpPRF and ScPFY(9-Mut)).
Profilins that do not support life in fission yeast cells do not stimulate formin Cdc12-mediated actin assembly in vitro
Profilin's capacity to facilitate contractile ring assembly appears to be critical for viability. Formin Cdc12 is essential for cytokinesis (Chang et al., 1997), and its ability to rapidly assemble actin filaments for contractile ring assembly requires profilin (Chang et al., 1997; Kovar et al., 2003; Yonetani et al., 2008). Human profilin HsPRO1 does not complement cdc3-124 cells and is utilized poorly by Cdc12 in vitro (Ezezika et al., 2009). Therefore we determined whether there is a correlation between the ability of diverse profilins to complement cdc3-124 cells and to enhance actin assembly in vitro mediated by a constitutively active Cdc12 construct containing the formin homology FH1 and FH2 domains (Figure 4; Kovar et al., 2003, 2006; Kovar and Pollard, 2004; Neidt et al., 2008; Scott et al., 2011). We found that only profilins from the fission yeast genus Schizosaccharomyces (SpPRF, SjPRF) complement cdc3-124 cells well (Figure 4A) and increase the in vitro rate of Cdc12-mediated actin assembly over the rate of formin alone (Figure 4, B and C). Budding yeast (ScPFY), mammalian (HsPRO1, MmPRF1), and worm (CePFN-1) profilins do not complement cdc3-124 cells (Figure 4A) and decrease the rate of Cdc12-mediated actin assembly over a range of profilin concentrations (Figure 4, B and C; Ezezika et al., 2009; Neidt et al., 2009). Plant profilin AtPRF1 somewhat complements cdc3-124 cells (Figure 4A; Christensen et al., 1996) and, compared with the noncomplementing profilins, decreases the rate of Cdc12-mediated actin assembly more modestly (Figure 4C). Therefore there is a general correlation between the ability of diverse profilins to support life in fission yeast and to facilitate formin Cdc12-mediated actin assembly.
ScPFY(9-Mut) is utilized better than wild-type ScPFY by formin Cdc12 in vitro
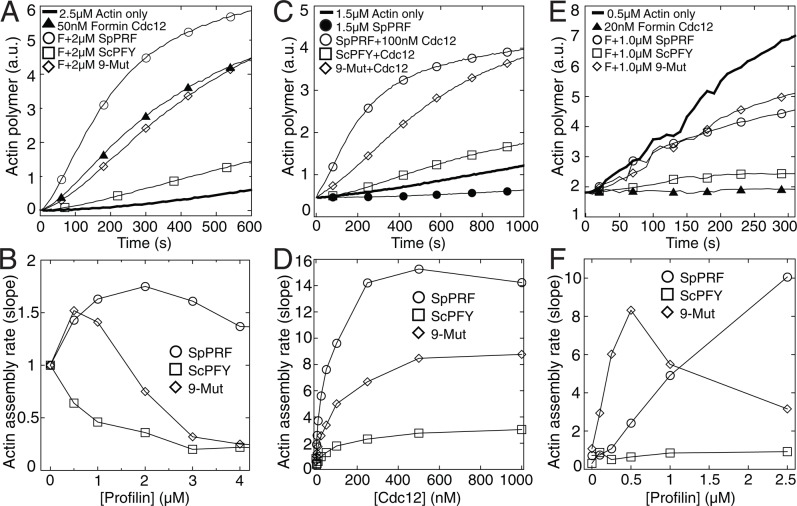
We predicted that the property rescued by ScPFY(9-Mut) is the ability to enhance formin Cdc12-mediated actin assembly. Therefore we used three different pyrene actin assays to determine profilin's general effect on Cdc12(FH1FH2)-mediated actin filament assembly (Figure 5). First, we followed the spontaneous assembly of 2.5 μM actin monomers with 50 nM Cdc12 and a range of profilin concentrations (Figure 5, A and B). Over a large range of profilin concentrations, SpPRF increases Cdc12-mediated actin assembly, whereas WT ScPFY decreases it. ScPFY(9-Mut) has an intermediate ability to stimulate Cdc12-mediated actin assembly, increasing the rate at lower concentrations and gradually decreasing the rate at higher concentrations. Second, we examined the ability of a range of Cdc12 concentrations to stimulate the assembly of equimolar concentrations of actin and profilin (1.5 μM; Figure 5, C and D). Cdc12 assembles actin associated with ScPFY(9-Mut) at a level intermediate between SpPRF and WT ScPFY. Third, we followed the effect of a range of profilin concentrations on the elongation of preassembled actin filament barbed ends associated with Cdc12 (Figure 5, E and F). Both SpPRF and ScPFY(9-Mut) increase the seeded assembly rate, whereas WT ScPFY does not.
FIGURE 5:
ScPFY(9-Mut) is utilized better than WT ScPFY by the formin Cdc12. (A, B) Spontaneous assembly of 2.5 μM Mg-ATP muscle actin monomers (20% pyrene-labeled) over a range of profilin concentrations. (A) Time course of the polymerization of actin monomers alone (thick curve) or with 50 nM formin Cdc12(FH1FH2) (▴) or Cdc12(FH1FH2) and 2 μM SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄). (B) Dependence of spontaneous actin assembly rate (slope) on the concentration of SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄). (C, D) Spontaneous assembly of 1.5 μM Mg-ATP muscle actin monomers (20% pyrene-labeled) and 1.5 μM profilin over a range of Cdc12(FH1FH2) concentrations. (C) Time course of the polymerization of actin monomers alone (thick curve) or with 1.5 μM SpPRF (•) or 100 nM Cdc12(FH1FH2) and 1.5 μM of SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄). (D) Dependence of spontaneous actin assembly rate (slope) on the concentration of Cdc12(FH1FH2) in the presence of 1.5 μM SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄). (E, F) Seeded elongation assay; barbed-end addition of 0.5 μM Mg-ATP actin monomers (10% pyrene-labeled) to 0.5 μM preassembled actin filament seeds. (E) Time course of actin alone (thick curve) or with 20 nM Cdc12(FH1FH2) (▴) or Cdc12(FH1FH2) and 1 μM SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄). (F) Dependence of seeded actin assembly rate (slope) on the concentration of SpPRF (◯), WT ScPFY (□), or ScPFY(9-Mut) (⋄).
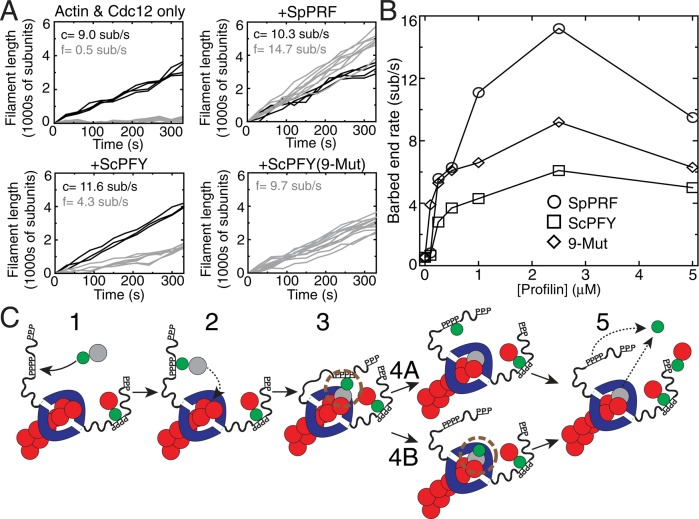
We also used total internal reflection fluorescence (TIRF) microscopy to visualize and quantify the specific effect of profilin on the barbed-end elongation rate of individual actin filaments (Figures 6, A and B, Supplemental Figure S6, and Supplemental Videos S3–S6). In the absence of profilin, Cdc12-associated filaments elongate at ∼0.5 subunit/s, whereas internal control filaments elongate at ∼9.0 subunits/s (Figure 6A; Kovar et al., 2003; Neidt et al., 2008; Skau et al., 2009; Scott et al., 2011). All three profilins increase the elongation rate of Cdc12-associated filaments but by significantly different amounts (Figure 6, A and B). SpPRF increases Cdc12-mediated barbed-end elongation by ∼30-fold, WT ScPRF by only ∼10-fold, and ScPFY(9-Mut) by an intermediate ∼20-fold. Therefore the fission yeast cytokinesis formin Cdc12 assembles actin associated with ScPFY(9-Mut) significantly better than actin associated with WT ScPFY, likely explaining why ScPFY(9-Mut) allows Cdc12-mediated contractile ring assembly in cells.
FIGURE 6:
TIRF microscopy of formin-mediated assembly of profilin-actin. (A, B) TIRF microscopy visualization of the spontaneous assembly of unlabeled 1.0 μM Mg-ATP actin with 0.5 μM Mg-ATP actin labeled with Oregon Green (Supplemental Videos S3–S6). (A) Plots of individual filament lengths over time with 2.5 nM Cdc12(FH1FH2) alone or Cdc12(FH1FH2) and 2 μM SpPRF, WT ScPFY, or ScPFY(9-Mut). Control (c) and formin-associated (f) filaments are denoted by black and gray lines, respectively, with average elongation rates indicated. (B) Dependence of the Cdc12(FH1FH2)-mediated barbed end elongation rate on the concentration of SpPRF (◯), WT ScPFY (□), and ScPFY(9-Mut) (⋄). (C) Model for formin-mediated actin filament elongation of profilin-actin. 1) Profilin-actin binds to proline-rich stretches in the formin FH1 domain. 2) The FH1-profilin-actin complex interacts with the FH2 domain–associated barbed end of the elongating actin filament. 3) Profilin-actin incorporates as the new ultimate actin subunit of the actin filament, creating a “ring” complex between formin-profilin-actin. Profilin and the FH2 domain may interact (brown circle). 4) Interaction between the FH2 domain and profilin may drive profilin dissociation from the barbed end before (4A and 5) or after profilin dissociates from the FH1 domain (4B and 5).
DISCUSSION
An essential role of profilin is to facilitate formin-mediated actin assembly for cytokinesis
We report here that profilin's ability to enhance formin Cdc12-mediated actin filament elongation for assembly of the cytokinesis contractile ring is essential in fission yeast. Inefficient actin filament elongation presumably leads to a deficient number of appropriately long actin filaments. Actin filaments assembled by Cdc12 at the division site de novo (Wu et al., 2006; Vavylonis et al., 2008; Coffman et al., 2009; Laporte et al., 2011) and cortical filaments assembled by Cdc12 and transported to the division site (Huang et al., 2012) may both contribute to contractile ring assembly. Search, capture, pull, and release (SCPR) is a stochastic ring assembly model by which Cdc12 assembles ∼1-μm-long filaments de novo from precursor nodes that are captured by myosin-II motors on neighboring nodes (Vavylonis et al., 2008; Coffman et al., 2009; Laporte et al., 2011). Inadequate profilins may not allow Cdc12 to produce filaments quickly enough to facilitate sufficient connections between nodes (Vavylonis et al., 2008). In the absence of precursor nodes, Cdc12 assembles actin filaments for contractile rings de novo from one or a few spots via a pathway that depends on the septation initiation network (Hachet and Simanis, 2008; Huang et al., 2008; Roberts-Galbraith and Gould, 2008; Saha and Pollard, 2012). The actin assembly parameters of Cdc12 (number of filaments and their elongation rate) have not been well defined for node-independent pathways. However, it is reasonable to assume that these mechanisms also require a sufficient number of rapidly elongating actin filaments for proper ring assembly (Hachet and Simanis, 2008; Huang et al., 2008, 2012; Saha and Pollard, 2012).
What role in formin-mediated actin assembly is rescued by ScPFY(9-Mut)?
Profilin is believed to increase the elongation rate of formin-associated actin filaments by increasing the local actin concentration and orienting actin monomers to the barbed end of the growing actin filament (Figure 6C; Romero et al., 2004; Kovar et al., 2006; Vavylonis et al., 2006). FH1 and FH2 domains have been proposed to play distinct but complementary roles in the assembly of profilin-actin (Figure 6C; Kovar et al., 2006; Vavylonis et al., 2006; Paul and Pollard, 2008; Courtemanche and Pollard, 2012). The FH2 domain remains processively associated with the elongating F-actin barbed end, whereas the FH1 domain binds profilin-actin and transfers it to the FH2 domain–associated barbed end (Chang et al., 1997; Kovar et al., 2003; Yonetani et al., 2008). Profilin then dissociates from both the FH1 domain and the barbed end to allow additional rounds of actin monomer addition.
However, this model does not explain several experimental results that implicate a direct involvement of the FH2 domain in formin-mediated assembly of profilin-actin. First, both the FH1 and FH2 domains contribute to the preference of specific formin isoforms for actin bound to particular profilin isoforms (Neidt et al., 2009). Second, the FH2 domain influences the degree to which formins select against labeled actin in the presence of profilin (Scott et al., 2011). Third, it has been documented multiple times that the FH2 domain influences the elongation rate of profilin-actin (Paul and Pollard, 2008; Neidt et al., 2009; Vidali et al., 2009; Scott et al., 2011). Therefore the FH2 domain appears to have a direct influence on profilin-actin elongation, which we hypothesize occurs through a novel profilin–FH2 domain interaction (Figure 6C). Considering that the nine mutations in ScPFY(9-Mut) do not alter the affinity for either the proline-rich FH1 domain or G-actin (Table 1), we suspect that the mutations rescue an as-yet-undefined interaction with the FH2 domain.
We predict that profilin binds the FH2 domain quite weakly, which prevents free profilin or profilin-actin from associating with the FH2 domain but allows interaction when profilin-actin is tethered to the FH1 domain in the “ring complex” during processive elongation (Figure 6C). This is consistent with our failure to detect interactions between profilin and the FH2 by simple pull-down assays (unpublished data). It is possible that association of profilin or profilin-actin with the FH2 domain contributes to the addition of new actin monomers to the barbed end (Figure 6C, step 3) and/or contributes to the dissociation of profilin from the formin-associated barbed end (Figure 6C, steps 4 and 5). Aligning crystal structures of actin in complex with either profilin or the budding yeast formin Bni1 FH2 domain suggests that the FH2 domain and profilin may both contact the most recently added actin subunit at similar positions (Otomo et al., 2005; Vavylonis et al., 2006). If profilin remains bound to the ultimate actin subunit while the FH2 domain translocates to the barbed end, the backbones of the FH2 domain and profilin might be in close contact; such a clash could explain why mathematical models of formin-mediated elongation of profilin-actin require the use of a dissociation constant that is substantially higher than that measured for the isolated profilin-actin interaction (Vavylonis et al., 2006). Strikingly, this putative profilin–FH2 domain interaction is predicted to overlap with the site of the nine mutations in ScPFY(9-Mut). Thus this interface might be finely tuned to specific formin and profilin isoform pairs that are tailored for rapid actin assembly. The profilin-FH2 domain interaction is predicted to occur in the “stair-stepping” model of formin-mediated actin filament elongation (Xu et al., 2004; Otomo et al., 2005; Mizuno et al., 2011) but not the “stepping second” model, in which profilin dissociates before the FH2 domain translocates to the ultimate actin subunit (Paul and Pollard, 2009a, b).
Profilin also has a role in Arp2/3 complex–mediated endocytic actin patches
Although endocytosis is not significantly perturbed in profilin mutant cdc3-124 cells (Figure 2), the kinetics of Arp2/3 complex-mediated actin patches is disrupted. Time-lapse fluorescence microscopy of actin patches revealed that compared with wild-type cells, cdc3-124 cells at the semipermissive temperature of 33.5°C contain approximately threefold more F-actin and internalize one-third as well (Suarez et al., 2015). The accumulation of additional F-actin can explain slower actin patch migration, since cofilin-mediated filament severing is critical for actin patch internalization (Okreglak and Drubin, 2007; Berro et al., 2010). Given that actin patches in cdc3-124 cells contain more F-actin, we hypothesized that an additional role of profilin is to limit Arp2/3 complex–mediated actin assembly. Furthermore, an antagonistic relationship between profilin and Arp2/3 complex has been revealed genetically, by which mutations in the Arp2/3 complex components sop2, arp2, and arp3 suppress the profilin cdc3-124 mutation (Balasubramanian et al., 1996; McCollum et al., 1996; Morrell et al., 1999). We discovered that profilin directly inhibits actin branch formation by Arp2/3 complex, which is necessary for formin Cdc12-mediated contractile ring assembly (Suarez et al., 2015). Therefore, in addition to profilin's essential role in facilitating contractile ring assembly by Cdc12 for cytokinesis, profilin is also required for yeast cells to maintain the proper density and size of diverse homeostatic F-actin networks (Burke et al., 2014; Suarez et al., 2015).
Even though fission yeast cells have relatively few F-actin networks (Kovar et al., 2011), the single profilin isoform has at least two distinct roles. Higher eukaryotes express multiple profilin isoforms and contain a far higher number of different filament networks, which depend on a diverse array of nucleation factors (Chhabra and Higgs, 2007; Chesarone and Goode, 2009; Dominguez, 2009). Previous work in our lab revealed that the worm formin CYK-1 differently utilizes the three worm profilins in vitro (Neidt et al., 2009). There is also growing evidence that formins (Khadka et al., 2009) or Ena/VASP proteins (Mouneimne et al., 2012) utilize profilin isoforms differently in vivo. Thus profilin isoforms may be sophisticated regulators of actin nucleation factors for diverse cellular processes.
MATERIALS AND METHODS
Schizosaccharomyces pombe strains and cell microscopy
Supplemental Table S3 lists the Schizosaccharomyces pombe strains used in this study. Differential interference contrast (DIC) and epifluorescence images were collected on an Orca-ER camera (Hamamatsu, Bridgewater, NJ) on an IX-81 microscope (Olympus, Tokyo, Japan), with a 60×/1.4 numerical aperture (NA) Plan Apo objective. Confocal images were collected on a Cascade 512BT camera (Photometrics, Tucson, AZ) on a Zeiss Axioimager M1–equipped 63×/1.4 NA oil-immersion lens and a Yokogawa CSU-X1 spinning-disk unit (Solamere, Salt Lake City, UT). Nuclei and septa were visualized with 4′,6-diamidino-2-phenylindole (DAPI) (nuclei) and calcofluor (septa; Kovar et al., 2003) and quantified for at least 200 cells. Cells were grown for 48 h in EMM5S before visualization of contractile rings.
Integrated profilins under control of the P81nmt1 promoter were expressed by growing cells for 24 h in EMM5S after the removal of thiamine (5 μg/ml). Cells were grown for an additional 2 h at 32 or 36°C before imaging. For contractile ring movies, cells were imaged on EMM5S-agar (2.5%) pads containing 0.1 mM n-propyl gallate. Contractile rings from a single Z-plane were visualized with Rlc1-GFP every minute. Endocytosis defects were determined by acquiring confocal Z-series (0.50-μm intervals spanning the entire cell) of GFP-Syb1 after incubating cells for 2 h at the indicated temperatures. Stock solutions of 10 mM CK-666 (Sigma-Aldrich, St. Louis, MO) were prepared in dimethyl sulfoxide. A 500-μl amount of exponentially growing cells (OD of 0.2–0.6) was incubated in a 1.5-ml tube for 30 min with 100 μM CK-666. For experiments at restrictive temperature, cells were preincubated at the indicated temperature for 2 h, followed by an additional 30 min with 100 μM CK-666 (Burke et al., 2014). Fluorescence intensity at the ends of the cells was normalized to background fluorescence and quantified using ImageJ (National Institutes of Health, Bethesda, MD).
ScPFY mutant library construction
A combinatorial budding yeast profilin ScPFY mutant library was constructed by PCR mutagenesis (Yuan et al., 2005). ScPFY was divided into ten 60-mer primers (IDT, Coralville, IA) that overlapped by 20 base pairs at both ends. Primers contained a 50/50 mix of the ScPFY and SpPRF base for each of 16 variable positions (theoretical library size, 216 = 65,536). After an initial PCR with Taq polymerase (New England BioLabs, Ipswich, MA) and all 10 primers, the products were used as a template for a second clean-up PCR using an 8:2 ratio of Taq polymerase to Pfu Turbo (Agilent Technologies-Stratagene, Santa Clara, CA) with only the outermost primers. The final products were cloned into the S. pombe pREP81 expression vector by restriction digest and ligation.
The pREP81-ScPFY library was electroporated into S. pombe cdc3-124 cells, plated on EMM5S-leu media for 2 d at 25°C, and then transferred to 36°C for 3 d. Of 192 colonies recovered from ∼500,000 mutant profilins, 17 were selected for further study after retesting their ability to complement the cdc3-124 strain at 36°C. Recovered plasmids were sequenced (Table 1). ScPFY(9-Mut) was constructed by the same two-step PCR method. Bacterial expression constructs (pMW) for profilin were prepared by standard cloning procedures.
Immunoblotting
To determine level of detection of SpPRF, ScPFY, or ScPFY(9-Mut) profilins by α-SpPRF antibody (Lu and Pollard, 2001), 1.0 μg of each purified profilin was run on an SDS–PAGE gel, transferred onto a nitrocellulose membrane, and probed with α-SpPRF and IRDye 800 CW goat anti-mouse immunoglobulin G secondary antibody (LI-COR Biosciences, Lincoln, NE). Optical density normalized to Coomassie-stained gel was quantified using ImageJ. To determine profilin expression levels in fission yeast cells, wild-type cells, cdc3-124 cells, or cdc3-124 cells expressing integrated SpPRF, ScPFY, or ScPFY(9-Mutant) under the low-strength P81nmt1 promoter were grown overnight at 25°C, then rinsed three times with EMM5S without thiamine and grown in EMM5S without thiamine at 25°C for 24 h. Cells were then grown at 32°C for 2 h before being broken open by glass bead lysis in a FastPrep-24 (MP Biomedicals, Solon, OH) in profilin buffer (20 mM Tris-HCl pH 7.5, 150 mM KCl, 0.2 mM dithiothreitol [DTT]) and Complete protease inhibitors (Roche, Fishers, IN). A Bradford assay was used to determine total protein concentration with equal amounts loaded and separated on an SDS–PAGE gel, transferred onto a polyvinylidene difluoride (PVDF) membrane, and probed with α-SpPRF. Profilin amounts in different cell types were quantified relative to the wild-type using ImageJ (Burke et al., 2014).
Protein purification
Cdc12(FH1FH2), Cdc12(FH1), and profilins were purified from bacteria as described (Lu and Pollard, 2001; Kovar and Pollard, 2004; Neidt et al., 2009). Ca-ATP actin was purified from chicken skeletal muscle as described (Neidt et al., 2008). Gel-filtered actin was labeled on Cys-374 with pyrenyl iodoacetamide or Oregon Green iodoacetamide (Life Technologies-Invitrogen, Grand Island, NY; Kovar et al., 2003). Immediately before each experiment, Ca-ATP actin was converted to Mg-ATP actin by addition of 0.1 volume of 2 mM ethylene glycol tetraacetic acid (EGTA) and 0.5 mM MgCl2 for 2 min at 25°C. Extinction coefficients for Cdc12(FH1FH2) (51,255 M−1; Kovar et al., 2003), SpPRF (20,065 M−1; Lu and Pollard, 2001), SjPRF (19,940 M−1), ScPFY (19,940 M−1; Haarer et al., 1990), AtPRF1 (17,085 M−1; Christensen et al., 1996), CePFN-1 (18,450 M−1; Polet et al., 2006), MmPRF1 (21,600 M−1), and HsPRO1 (18,500 M−1; Metzler et al., 1994) were estimated from the amino acid composition.
Fluorescence spectroscopy
Spontaneous and seeded actin assembly was measured from the fluorescence of pyrene-actin (excitation at 364 nm and emission at 407 nm) with a Safire2 (Tecan, Durham, NC) fluorescence plate reader as described previously (Neidt et al., 2008). Final protein concentrations are indicated in the figure legends. Conditions were as follows: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.2 mM ATP, and 90 μM CaCl2. Profilin Kd values were determined by fitting the spontaneous actin assembly data to an exponential decay equation.
Profilin's affinity for PLP (Sigma-Aldrich) or Cdc12(FH1) was determined by measuring profilin's intrinsic tryptophan fluorescence with excitation at 295 nm and emission at 323 nm (Neidt et al., 2009). Profilin (1.0 μM) was incubated with a range of PLP or Cdc12(FH1) concentrations for 30 min at room temperature and the fluorescence read in a Safire2 plate reader. Plots of the dependence of the concentration of bound profilin on the concentration of PLP or Cdc12 (FH1) were fitted with a quadratic equation to determine Kd values. Profilin's stability was measured by denaturation with urea (Lu and Pollard, 2001). Profilin (1.0 μM) was incubated with a range of urea concentrations for 30 min at room temperature, and the fluorescence (excitation, 295 nm; emission, 370 nm) was read in the Safire2 plate reader. The reported values are an average of three independent experiments.
Profilin's effect on the exchange the nucleotide of the actin monomer was measured by the loss of εATP fluorescence with excitation at 360 nm and emission at 410 nm (Lu and Pollard, 2001). Muscle actin was incubated with AG1-x4 resin (Bio-Rad, Hercules, CA) at 4ºC for 30 min to remove ATP. εATP Mg-actin was made by adding 400 μM εATP, 200 μM EGTA, and 50 μM MgCl2 to the resin-treated actin for 50 min at 4ºC. Excess εATP was removed by incubation with AG1-x4 resin for 30 min at 4ºC. εATP fluorescence after the addition of 500 μM unlabeled ATP was plotted versus time. The time to half-maximal fluorescence was plotted versus profilin concentration.
Fission yeast actin pull-down assay
Fission yeast cells were grown to OD595 of ∼1.7 at 25ºC. The cells were harvested, washed with water and phosphate-buffered saline (PBS), resuspended in PBS, and broken open by a FastPrep-24 benchtop homogenizer (MP Biomedicals, Santa Ana, CA) after adding ∼0.3 g of acid-washed glass beads (Sigma-Aldrich). The homogenate was clarified at 13,000 rpm for 15 min at 4ºC and at 50,000 rpm for 30 min at 4ºC. The supernatant was incubated with 25 μl of bed volume PLP-Sepharose beads and a range of concentrations of purified recombinant profilin for 2 h at 4ºC. Beads were collected, washed three times, boiled in protein sample buffer, and run on a 15% SDS–PAGE gel. Western Blot confirmed the actin band with an anti–β-actin antibody (Santa Cruz Biotechnology, Dallas, TX).
PtdIns(4,5)P2 binding
Profilin's ability to bind PtdIns(4,5)P2 micelles was assayed by microfiltration (Haarer et al., 1993; Lambrechts et al., 1997). PtdIns(4,5)P2 (Sigma-Aldrich) micelles were prepared at a concentration of 1 mg/ml in water by water bath sonication for 5 min at room temperature. A range of PtdIns(4,5)P2 micelle concentrations was incubated with 2.5 μM profilin for 2 h at 4ºC. The samples were then transferred to Amicon Ultracel membranes (Millipore, Billerica, MA) with a molecular weight cutoff of 30,000 Da. The samples were centrifuged through the membranes for 1 min at 2000 rpm. The flowthrough was collected, run on a 15% SDS–PAGE gel, and stained with Coomassie Blue. The intensity of the profilin bands was measured by an Odyssey infrared imager (LI-COR).
TIRF microscopy
TIRF images of Oregon Green–labeled Mg-ATP actin excited by evanescent wave fluorescence were acquired every 10 s on an IX-71 microscope (Olympus) fitted with through-the-objective TIRF illumination and an iXon electron-multiplying charge-coupled device camera (Andor Technology, South Windsor, CT) as described previously (Neidt et al., 2008). A mixture of 1.0 μM unlabeled Mg-ATP actin and 0.5 μM (33%) Mg-ATP Oregon Green actin was mixed with 2× TIRF buffer (1×: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% [500 cp] methylcellulose) and formin alone or with profilin and transferred to a flow cell for imaging.
Supplementary Material
Acknowledgments
We thank Michael Glotzer (University of Chicago, Chicago, IL) for helpful suggestions, members of the Kovar lab for technical assistance, and Ben Glick for the use of SnapGene for plasmid construction. This work was supported by National Institutes of Health Molecular and Cellular Biology Training Grant T32 GM007183 (to A.J.B., J.R.C., and T.A.B.), National Science Foundation Graduate Student Fellowship DGE-1144082 (to J.R.C.), American Heart Association Grant 11SDG5470024 (to V.S.), and National Institutes of Health Grant RO1 GM079265 (to D.R.K.).
Abbreviations used:
- FH
formin homology
- TIRF
total internal reflection fluorescence
- ts
temperature sensitive
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0281) on November 12, 2014.
*These authors contributed equally.
REFERENCES
- Balasubramanian MK, Feoktistova A, McCollum D, Gould KL. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Chang F. Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr Biol. 2011;21:905–916. doi: 10.1016/j.cub.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro J, Sirotkin V, Pollard TD. Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol Biol Cell. 2010;21:2905–2915. doi: 10.1091/mbc.E10-06-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Christensen HE, Ramachandran S, Tan CT, Surana U, Dong CH, Chua NH. Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 1996;10:269–279. doi: 10.1046/j.1365-313x.1996.10020269.x. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem. 2012;287:7812–7820. doi: 10.1074/jbc.M111.322958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors–structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284:2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Petzold AS, Brown SS. Mutational analysis of yeast profilin. Mol Cell Biol. 1993;13:7864–7873. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Huang J, Huang Y, Yu H, Subramanian D, Padmanabhan A, Thadani R, Tao Y, Tang X, Wedlich-Soldner R, Balasubramanian MK. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J Cell Biol. 2012;199:831–847. doi: 10.1083/jcb.201209044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- Khadka DK, Liu W, Habas R. Non-redundant roles for Profilin2 and Profilin1 during vertebrate gastrulation. Dev Biol. 2009;332:396–406. doi: 10.1016/j.ydbio.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Sirotkin V, Lord M. Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21:177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Yang P, Sale WS, Drobak BK, Staiger CJ. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. J Cell Sci. 2001;114:4293–4305. doi: 10.1242/jcs.114.23.4293. [DOI] [PubMed] [Google Scholar]
- La Carbona S, Le Goff C, Le Goff X. Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex. Biol Cell. 2006;98:619–631. doi: 10.1042/BC20060048. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X, Motegi F, Salimova E, Mabuchi I, Simanis V. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J Cell Sci. 2000;113:4157–4163. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, Almo SC. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol. 1999;6:666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Metzler WJ, Bell AJ, Ernst E, Lavoie TB, Mueller L. Identification of the poly-L-proline-binding site on human profilin. J Biol Chem. 1994;269:4620–4625. [PubMed] [Google Scholar]
- Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- Mockrin SC, Korn ED. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5’-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- Morrell JL, Morphew M, Gould KL. A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast. Mol Biol Cell. 1999;10:4201–4215. doi: 10.1091/mbc.10.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–630. doi: 10.1016/j.ccr.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1035. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, Ernst EG, Lavoie TB, Gorman JA. Polyproline binding is an essential function of human profilin in yeast. Eur J Biochem. 1999;262:26–35. doi: 10.1046/j.1432-1327.1999.00354.x. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2 formins. J Biol Chem. 2009a;284:12533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009b;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat Cell Biol. 2001;3:235–244. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Pilo Boyl P, Di Nardo A, Mulle C, Sassoe-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V, Perlas E, Massimi M, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polet D, Lambrechts A, Ono K, Mah A, Peelman F, Vandekerckhove J, Baillie DL, Ampe C, Ono S. Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil Cytoskeleton. 2006;63:14–28. doi: 10.1002/cm.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Saha S, Pollard TD. Characterization of structural and functional domains of the anillin-related protein Mid1p that contribute to cytokinesis in fission yeast. Mol Biol Cell. 2012;23:3993–4007. doi: 10.1091/mbc.E12-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter K, Schleicher M, Jockusch BM. Effects of single amino acid substitutions in the actin-binding site on the biological activity of bovine profilin I. J Cell Sci. 1998;111:3261–3273. doi: 10.1242/jcs.111.22.3261. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Scott BJ, Neidt EM, Kovar DR. The functionally distinct fission yeast formins have specific actin-assembly properties. Mol Biol Cell. 2011;22:3826–3839. doi: 10.1091/mbc.E11-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare P, Karlsson R. Evidence for two interaction regions for phosphatidylinositol(4,5)-bisphosphate on mammalian profilin I. FEBS Lett. 2002;522:119–124. doi: 10.1016/s0014-5793(02)02913-7. [DOI] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015 doi: 10.1016/j.devcel.2014.10.027. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PAC, Guerin C, Franco P, Li M, Burkart G, Augustine RC, Blanchoin L, Bezanilla M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA. 2009;106:13341–13346. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG. In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol. 2000;150:895–904. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Kurek I, English J, Keenan R. Laboratory-directed protein evolution. Microbiol Mol Biol Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.