CARM1-mediated H3R17 methylation in hESC-derived embryoid bodies regulates astrocyte fate commitment–associated genes and miRNAs directly by methylating the promoter or indirectly through miRNAs. This is the first report of the involvement of CARM1 in neural development, the absence of which is observed as sensory response defects in zebrafish embryos.
Abstract
Coactivator-associated arginine methyltransferase (CARM1/PRMT4)–mediated transcriptional coactivation and arginine methylation is known to regulate various tissue-specific differentiation events. Although CARM1 is expressed in the neural crest region in early development, coinciding with early neuronal progenitor specification, the role of CARM1 in any neuronal developmental pathways has been unexplored. Using a specific small-molecule inhibitor of CARM1-mediated H3R17 methylation in human embryonic stem cell line, we find that H3R17 methylation contributes to the maintenance of the astroglial cell population. A network of regulation was observed on the miR92a promoter by which H3R17-responsive Nanog bound to the miR92a promoter decreased upon inhibition, resulting in an abnormal gene expression program influencing the glial lineage. This was also true in zebrafish, in which, with the help of CARM1 inhibitor and CARM1 morpholinos, we show that inhibition of H3R17 methylation results in defective glial cell morphology and a sensory defect in a subpopulation. A gain-of-function strategy in which mCARM1 was introduced in the morpholino-treated embryos exhibited recovery of the sensory defect phenotype. This study thus establishes the functional cooperation between arginine methylation and microRNA expression in the neuronal developmental process, with potential implications in sensory development pathways.
INTRODUCTION
Chromatin modifications have emerged as key regulators of gene expression. These modifications act as signaling platforms and modulate the dynamic nature of chromatin, thereby influencing transcriptional responses. Several chromatin-modifying enzymes are also transcriptional coactivators, facilitating transcription by aiding in the recruitment process. Thus their role in the physiological scenario is multifaceted and includes repair, replication, and differentiation, among others. These enzymes also play critical roles in the process of development.
Transcriptional coactivators like coactivator-associated arginine methyltransferase (CARM1) and p300 are known to have a restricted pattern of expression around the embryonic day 10.5 (E10.5) stage of mouse development (Bhattacherjee et al., 2009). CARM1-associated methylation of arginine 17 and 26 residues of H3 have also been observed to occur in this specific stage (Torres-Padilla et al., 2007), and these marks have been shown to be essential for ICM specification. However, the E10.5 stage, which is associated with early somitogenesis and neural specification, has not yet been directly correlated with a functional association with CARM1 or H3R17/H3R26 methylation. The role of CARM1 in the process of differentiation has been shown to be exerted at multiple levels and in different tissues. Although it has been shown that CARM1-mediated H3R17 and H3R26 methylation is essential for the expression of Oct4, Nanog, and Sox2 in the maintenance of pluripotency (Wu et al., 2009), the transcriptional coactivation and histone methyltransferase functions of CARM1 have been linked to its role in promoting the differentiation of cells into multiple lineages (Chen et al., 2002; Kim et al., 2004; Yadav et al., 2008; Ito et al., 2009). The methyltransferase activity of CARM1 promotes thymocyte and chondrocyte differentiation, and the coactivation function promotes myogenesis and adipogenesis. Hence CARM1 seems to be involved in maintenance of pluripotency, as well as in multiple differentiation programs. However, this can be related to the spatiotemporal expression of CARM1 during early developmental stages and subsequent tissue-specific expression. CARM1 expression in early mouse embryo development is observed from the E9.5 stage, with maximal expression in the limb bud region, and remains consistent in the pharyngeal arch region, which corroborates earlier findings of CARM1 in muscle and skeletal differentiation events. The most striking is the expression in the brain region, where it peaks in E10.5 in all the brain regions (EMBRYS database, embrys.jp/embrys/html/MainMenu.html), which is coincident with somitogenesis and early brain development. The expression of CARM1 in this stage of development suggests a potential role in neuronal development.
The CARM1-knockout model has led to the identification of many developmental roles of CARM1, but there are no reports on defects in the neuronal lineage. We earlier identified a CARM1 inhibitor that inhibits H3R17 methylation specifically (Selvi et al., 2010). A microarray analysis (Supplemental Figure S1) of the perturbed gene expression upon inhibitor treatment followed by a Gene Ontology (GO) analysis led to enrichment of the neuronal lineage. This observation implies that the neuronal lineage may be regulated by H3R17 methylation, and to identify the role of CARM1 in this pathway, we adopted a knockdown approach in parallel with an inhibitor-based approach in zebrafish and human embryonic stem cells (hESCs).
In the present study, by using a H3R17 methylation–specific inhibitor of CARM1, TBBD, which inhibits CARM1-mediated methylation at H3R17 both in vitro and in vivo, we observed that glial cell maintenance is controlled by H3R17 methylation. A systematic approach investigating the effect of H3R17 methylation in hESC-derived embryoid bodies (EBs) led to the identification of a specific perturbation of the neuroectodermal lineage. Further analysis revealed an intricate network of transcriptional and posttranscriptional regulation of the astroglial fate commitment responsive to H3R17 methylation. Mechanistically, H3R17 methylation regulates Nanog levels, thereby influencing the miR17-92 network. Knocking down CARM1 expression in zebrafish resulted in glial cell–specific defects during development. Thus we conclude that there is a transcriptional network controlled by CARM1 that regulates genes such as NF1 and microRNAs (miRNAs) that target the genes associated with glial cell maintenance, thereby leading to maintenance of the astroglial cell population. The inhibition of CARM1-mediated H3R17 methylation disturbs this balance, which manifests itself as a loss in sensory response.
RESULTS
Inhibition of H3R17 methylation affects ectodermal lineage
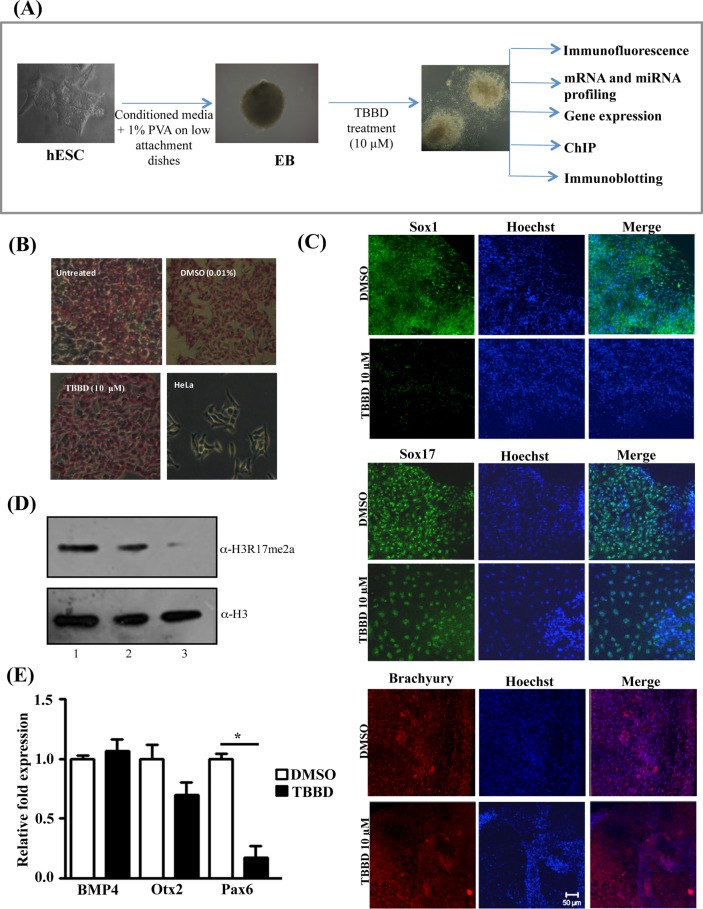
CARM1 has been associated with the maintenance of pluripotency. However, CARM1 has also been shown to influence differentiation of several mesoderm-derived tissues. To gain insight into the physiological and spatiotemporal role of CARM1 mediated histone methylation in lineage determination, we subjected hESCs and hESC-derived EBs to treatment with the H3R17 methylation–specific inhibitor TBBD (Figure 1A). The effect of TBBD treatment on pluripotency of BG01V cells was tested by examining the expression of alkaline phosphatase. There were no obvious differences observed upon inhibitor treatment, as shown in Figure 1B. This could be due to either a shorter treatment time or the specific inhibition of H3R17 methylation. It has been shown that methylation of both H3R17 and H3R26 is essential for the expression of pluripotency-associated genes in mouse ESCs. We hypothesized that a change in H3R17 methylation status might also have detectable effects on specific lineage markers. Hence we subjected hESC-derived EBs to TBBD treatment for 48 h from day 5 to day 7 and further processed them for immunofluorescence staining for the three germ-layer markers, Sox1 (ectoderm), Sox 17 (endoderm), and Brachyury/T (mesoderm). Remarkably, Sox1 expression was greatly reduced in the presence of the inhibitor, whereas there were only minimal effects on the expression of the other two marker genes (Figure 1C). The H3R17 methylation inhibition was evident at the tested concentration in the EBs (Figure 1D and Supplemental Figure S2). Significantly, the transcription factor Sox1 was previously shown to be important for early neuronal progenitor specification. To verify the specificity of the inhibitor toward the neuroectodermal lineage, we tested another classical neuroectodermal marker, Pax6, and observed significant down-regulation in this case whereas, levels of other ectodermal markers, like Otx2 and BMP4, were similar in the control and TBBD-treated samples (Figure 1E). These observations suggest that H3R17 methylation has a role in the neuroectodermal lineage.
FIGURE 1:
Inhibition of H3R17 methylation affects ectodermal lineage. (A) Experimental design using the embryonic stem cell system. (B) Alkaline phosphatase staining of untreated ES cells (I) and ES cells treated with DMSO (II) or TBBD (III). HeLa cells were used as negative control (IV). (C) Immunofluorescence analysis of germline marker expression in embryoid bodies (EBs) treated with DMSO or TBBD (10 μM) for 48 h. Expression of Sox1, Sox17, and Brachyury was analyzed. (D) Immunoblotting of human ES cell–derived EB lysates with antibodies against H3R17me2a and H3. The EBs were treated with DMSO (lane 1) or 5 μM (lane 2) or 10 μM (lane 3) TBBD for 48 h. A dose-dependent decrease of H3R17 methylation is observed with increasing concentration of TBBD. (E) qRT-PCR analysis of TBBD-treated EBs for ectodermal markers Pax6, BMP4, and Otx2 compared with DMSO-treated EBs. Significant down-regulation of Pax6 was observed with very minimal effects on BMP4 and Otx2 (n = 3; t-test p values: Pax6, 0.0159; BMP4, 0.5649; Otx2, 0.1988).
CARM1 influences neural differentiation pathways by regulating miRNA expression
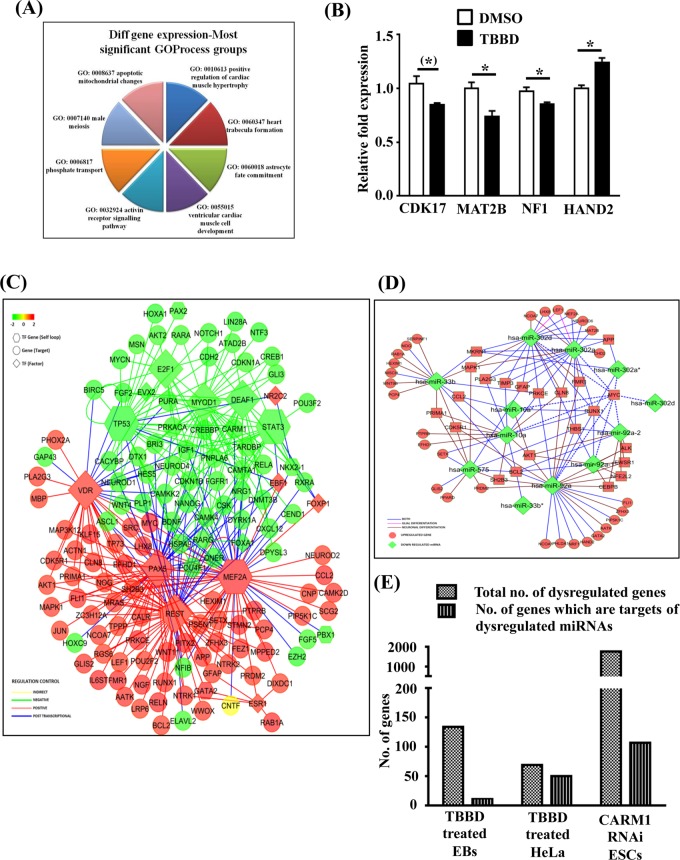
To gain insight into the global gene expression changes associated with H3R17 methylation in hESC-derived EBs, we performed gene expression and miRNA expression profiling of the EBs treated with TBBD. The number of genes that were down-regulated was higher than those with up-regulated expression. Of interest, the pathway alteration analysis led to the identification of several astroglial fate commitment genes that were altered upon TBBD treatment, along with the already reported role of CARM1 in muscle differentiation (Figure 2A). The down-regulation of one important astroglial lineage–specific gene, NF1 (neurofibromatosis 1), was validated by real-time expression analysis (Figure 2B, lane 3). Other genes whose expressions were validated include those that encode a cyclin-dependent kinase (CDK17) and MAT2B alpha, an enzyme involved in the SAM biosynthesis pathway (Figure 2B, lanes 1 and 2). Of the differentially up-regulated gene category, a transcription factor that is also a developmental regulator, HAND2, was validated (Figure 2B, lane 4). In the miRNA expression analysis, we found that there were almost no miRNAs that were up-regulated; instead, a few miRNAs known to be involved in carcinogenesis, pluripotency, and differentiation pathways exhibited a down-regulation of expression (Supplemental Figure S3). Because the H3R17 methylation mark has been associated with transcriptional activation and the inhibition of this mark led to differential regulation of both gene expression and miRNA expression, we performed a network analysis with these data sets in order to visualize the broader network of the factors perturbed upon TBBD treatment (Figure 2, C and D). Because EBs display similarities to the early developmental stages, we expected a highly interconnected transcriptional program with few important transcription factor nodes. Although there were only a few neural-glial differentiation/function–related transcription factors, such as RE-1 silencing transcription factor (REST) and p53 in this network, their potential targets were numerous, including several significant ones, including NeuroD, CAMK4, Nanog, and so on (Figure 2C). Of interest, this network analysis also led to the visualization of the self-regulatory transcription factor modules (represented as hexagons in the figure), as well as those that do not possess this feedback ability (represented as rhombi). One of the most striking observations was the predominance of self-regulated transcription factor nodes such as p53 and STAT3 in the negatively correlated cluster. Tumor suppressor p53 is a well-established modulator of the ES cell differentiation pathway, and several studies have shown that p53 resists the self-renewal property and promotes differentiation (Ungewitter and Scrable, 2010; Jain et al., 2012; Li et al., 2012). Similarly, the LIF-STAT pathway has been shown to be essential for regulation of ES cell self-renewal status (Raz et al., 1999). Thus the presence of both p53 and STAT3-associated network suggests perturbation of the self-renewal versus differentiation pathway. However, the overall influence of H3R17 methylation on both these pathways exhibits significant down-regulation, and hence they cluster together in the negative correlation cluster. H3R17 methylation is indeed important for p53-responsive gene expression (Selvi et al., 2010), and protein arginine methylation has also been linked to the STAT signaling pathway (Kleinschmidt et al., 2008; Bedford and Clarke, 2009). On the other hand, the presence of differentiation-associated transcription factors such as REST, Pax, and Myc in the positive correlation cluster signifies the changes present in EB formation that may or may not be influenced by H3R17 methylation directly. However, there were a few interconnected nodes among these differentially expressed genes where the transcription factor and the target showed inverse expression patterns (blue lines in Figure 2C). This inverse correlation implied regulation at the posttranscriptional level. An indirect mode of regulation with respect to ciliary neurotrophic factor (CNTF) was also evident from the network. Cell culture models suggest a role for CNTF in promoting glial maturation (Murphy et al., 1997).
FIGURE 2:
CARM1 influences neural differentiation pathways by regulating miRNA expression. (A) Pie chart representation of differentially regulated genes upon TBBD treatment (10 μM) of BG01V-derived EBs for 48 h. (B) Validation of differentially regulated genes upon TBBD treatment of the human ES cell line BG01V-derived EBs compared with DMSO-treated EBs. CDK17, MAT2B, and NF1 show down-regulation of gene expression with similar folds as observed in the microarray, whereas HAND2 exhibits up-regulation of expression (n = 3; t-test p values: CDK17, 0.0517; MAT2B, 0.0261; NF1, 0.0412; and HAND2, 0.0103). (C) Integrated network analysis of gene expression. Down-regulated transcription factors are represented in green, and transcription factors with up-regulated expression are represented in red. Indirect regulation is depicted in yellow, and posttranscriptional regulatory nodes are represented in blue. Hexagons represent transcription factors, and circles represent gene targets. Rhomboids depict self-regulatory transcription factors. (D) Posttranscriptional regulation of gene expression. Green indicates down-regulation; red represents up-regulation. Pink lines denote the miRNA–mRNA network of glial differentiation, and red lines represent the neuronal differentiation network. Blue lines depict the involvement of the miRNA and mRNA network associated with both glial and neuronal cells. (E) Graphical representation of genes regulated by H3R17 methylation through modulation of miRNA expression, with the gene list obtained from microarray expression analysis of hESC-derived EBs treated with TBBD, the gene list from the microarray data set obtained upon treating HeLa cells with TBBD, and the gene list obtained from the microarray data set of RNA interference of CARM1 in mouse ES cells (Wu et al., 2009).
On mapping the posttranscriptional regulatory network (Figure 2D), we found the targets of all the down-regulated miRNAs to be up-regulated, including very significant candidates, such as GFAP and MAPK1. Considering that one of the significant enriched terms in the GO analysis was astrocyte fate commitment and the network map provided evidence for glial cell maturation with a predominance of posttranscriptional modulation, we decided to perform a meta-analysis on perturbed miRNAs that are specific to the astroglial and neuronal lineage (Figure 2D). A majority of the down-regulated miRNAs and the corresponding mRNAs were common for both the astroglial and neuronal lineages, which is expected since the EB model does not proceed to any specific neural precursor. However, there was a significant up-regulation of neuronal lineage– associated genes as compared with the glial lineage, which suggests that the H3R17 methylation inhibition possibly leads to skewing of the pathway toward the neuronal lineage by suppressing the astroglial lineage.
The overall effect of H3R17 methylation on gene expression at both the transcriptional and posttranscriptional levels was analyzed by determining the overlap between genes that are differentially regulated upon inhibitor treatment in hESC-derived EBs (Figure 2, A and E, lane 1), inhibitor-treated HeLa cells (Supplemental Figure S1 and Figure 2E, lane 2), and the data set from CARM1 RNA interference in mouse ESCs (Wu et al., 2009; Figure 2E, lane 3). When these changes were correlated with the miRNA changes associated with R17 methylation inhibition (Supplemental Figure S3), a significant proportion was found to be related to miRNA-associated regulation, implying an important contribution of posttranscriptional processing events.
H3R17 methylation regulates miR17-92 activity through regulation of Nanog expression
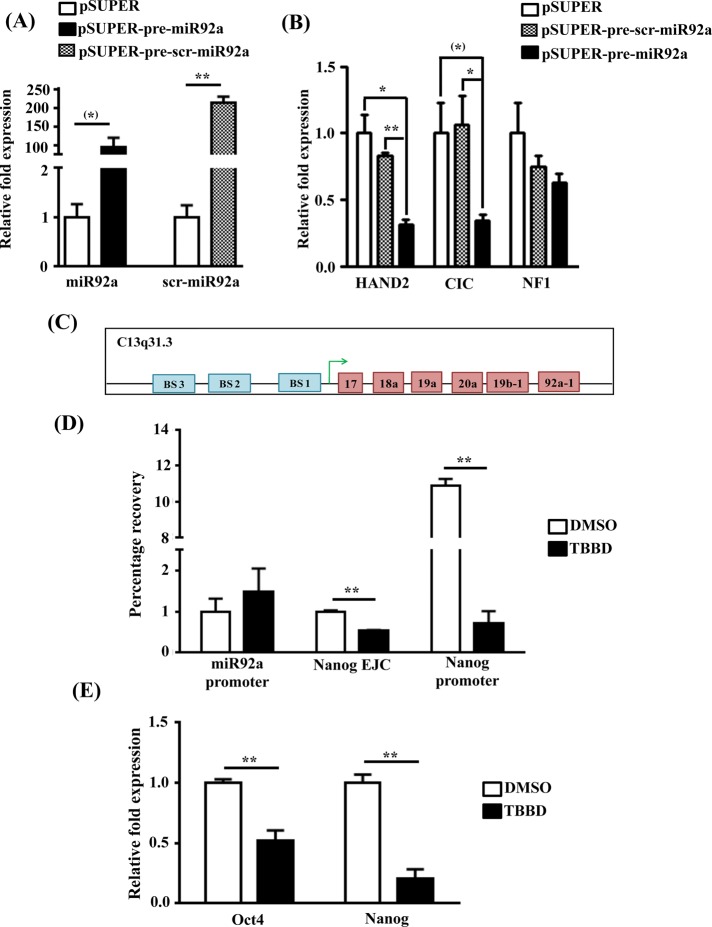
From the network, apart from miR10a and miR575, which are known to be up-regulated in glial cells, another important candidate, miR92a, which regulates various genes involved in neural differentiation, was also observed. Although the role of this miRNA is established in carcinogenesis, its relation to differentiation is relatively less explored. Integrating the miRNA expression analysis and the gene expression data sets, we found that putative miR92a-regulated genes such as CIC and HAND2 were up-regulated upon TBBD treatment as observed in the gene expression analysis; the up-regulation of HAND2 was validated (Figure 2B). These genes are transcriptional regulators involved in neural development (Lee et al., 2002; Vincentz et al., 2012). To validate the correlation that miR92a regulates these targets, we cloned the pre-miR92a construct, which, when transfected in SH-SY5Y cells, showed severalfold up-regulation of the intended miRNA (Figure 3A and Supplemental Figure S4). The up-regulation of miR92a led to 60% down-regulation of expression of each of its putative targets (Figure 3B) in comparison to the empty vector and scrambled RNA controls, which were unchanged. As internal control, we also checked for the expression of NF1, which, incidentally, does not have miR92a-binding sites, and, as expected, no change was observed in the NF1 levels in the miR92a-overexpressing cells. Taken together, these observations indicate that CARM1 seems to be an important regulator of the neural–glial cell differentiation switch by exerting posttranscriptional modulation on genes such as CIC and HAND2 through miR92a.
FIGURE 3:
H3R17 methylation regulates miR17-92 activity through regulation of Nanog expression. (A) qRT-PCR analysis showing the overexpression of mature miR92a and mature scrambled miRNA after transfection of pSUPER-pre-miR92a and pSUPER-pre-scr-miR92a in SH-SY5Y cells compared with vector-transfected control cells (n = 3; t-test p values: miR92a, 0.0644; scr-miR92a, 0.0063. (B) Expression analysis of HAND2, CIC, and NF1 genes upon overexpression of pre-miR92a compared with vector control–transfected cells and scrambled pre-miR92a–transfected cells (n = 3; *p < 0.05, **p < 0.01). (C) Schematic representation of the miR17-92 cluster promoter with three Nanog binding sites (BS). (D) Chromatin immunoprecipitation and qRT-PCR from EBs treated with DMSO or TBBD. Lane 1, H3R17 enrichment in Nanog binding region in the miR17-92 promoter cluster. Lane 2, enrichment across the exon–intron junction in Nanog (p = 0.0030). Lane 3, H3R17 enrichment in the promoter region of Nanog (p = 0.0022). (E) Real-time PCR quantification of Oct4 (Pou5f1) and Nanog expression in BG01V cells treated with TBBD (10 μM) for 24 h, taking DMSO-treated cells as control (n = 3, t-test p values: Oct4, 0.0063; Nanog, 0.0014).
The down-regulation of miR92a expression upon TBBD treatment could either be a cause of H3R17 methylation associated transcriptional regulation of the miR92a promoter or an indirect effect through other transcriptional regulators influenced by H3R17 inhibition. To delineate the direct versus indirect effects of H3R17 methylation on miR92a promoter, we performed a chromatin immunoprecipitation analysis using antibodies against methylated H3R17, followed by quantitative real-time PCR (qRT-PCR) analysis. Surprisingly, no enrichment of H3R17 methylation was observed on the miR92a promoter (Figure 3D lane 1), and hence we reanalyzed the promoter region of the miR17-92 cluster to which miR92a belongs. Of interest, we identified that the promoter region had Nanog binding sites (Figure 3C and Supporting Sequence Information), which is regulated by CARM1 during differentiation (Wu et al., 2009). During the preparation of our manuscript, Garg et al. (2013) reconfirmed the involvement of Nanog in miR92a expression and showed the presence of three Nanog binding sites in the miR17-92 cluster promoter region. It was shown earlier that CARM1 regulates Nanog expression by binding and methylating its promoter and also indirectly through the binding of Oct4 and Sox2 to its enhancer, the levels of Oct4 and Sox2 also being directly regulated by CARM1 binding to their promoters (Wu et al., 2009). Because Nanog levels have been shown to be responsive to H3R17 methylation levels (Wu et al., 2009), we analyzed the Nanog promoter and found a significant decrease in the enrichment of H3R17 methylation in response to TBBD treatment (Figure 3D, lane 3). This enrichment was very specific to the promoter since H3R17 methylation levels at the first exon–intron junction away from the promoter were found to be minimal but still affected by TBBD treatment (Figure 3D, lane2). This decrease in the promoter H3R17 levels was also reflected in Nanog expression. TBBD-mediated inhibition of H3R17 methylation at a concentration of 10 μM resulted in down-regulation of the expression of the pluripotency-associated genes Oct4/Pou5f1 and Nanog (Figure 3E). This correlates well with earlier reports in which CARM1 was shown to regulate the expression of pluripotency-related genes, with their expression reduced drastically upon CARM1 knockdown (Wu et al., 2009). Thus CARM1-mediated H3R17 methylation regulates astroglial fate commitment through a complex regulatory network involving transcriptional regulation of Nanog, which influences the posttranscriptional profile by modulating miR92a levels.
CARM1-mediated H3R17 methylation regulates neural developmental pathway in zebrafish
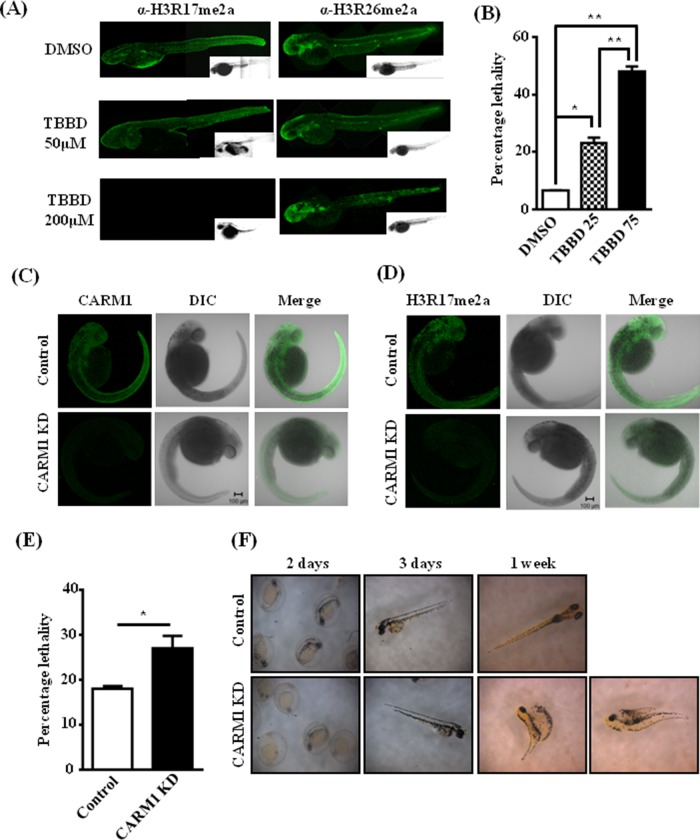
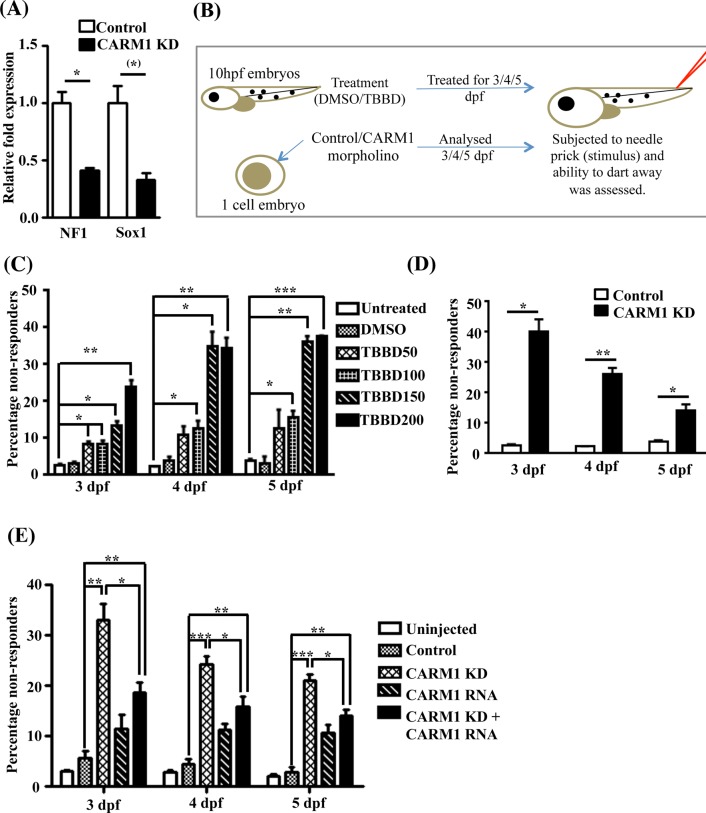
H3R17 methylation seemingly regulates astroglial commitment during neural development. Because EBs are an in vitro developmental model system with limited potential, we decided to further validate our observations with zebrafish (Zf) embryos. Treatment with the inhibitor led to no obvious morphological changes, but immunostaining followed by whole-mount imaging showed specific and drastic inhibition of H3R17 methylation, with H3R26 methylation being unaffected (Figure 4A). Although there were no evident morphological changes, the treatment showed a dose-dependent increase in lethality in treated embryos (Figure 4B). The treated embryos also showed decreased Sox1 expression, similar to the effect in embryoid bodies (Supplemental Figure S5). To ensure that the effects observed upon TBBD treatment in zebrafish are due to inhibition of CARM1-mediated histone methylation and to rule out any off-target effects of the compound, we resorted to a knockdown-based approach in zebrafish embryos. We performed a knockdown of CARM1 expression in zebrafish embryos using morpholinos (Figure 4C), which was confirmed by dramatic down-regulation of H3R17 methylation (Figure 4D), and assayed the effect on viability (Figure 4E). As expected, a marginal effect of lethality was observed upon knockdown. Surprisingly, there were no gross developmental changes/arrest in growth at any specific stage in comparison to the control morpholino–injected embryos, as opposed to the neonatal lethality observed in mice (Yadav et al., 2003). However, these results corroborated well with the absence of lethality observed in earlier CARM1-knockdown studies in zebrafish (Batut et al., 2011). Remarkably, few of the embryos showed gross morphological defects typical of embryos with severe neuromuscular developmental defects and died after 1 wk of development (Figure 4F).
FIGURE 4:
CARM1-mediated H3R17 methylation regulates neural developmental pathway in zebrafish. (A) Immunofluorescence analysis of zebrafish embryos treated with different concentrations of DMSO or TBBD at 10 h postfertilization and stained using H3R17me2a and H3R26me2a antibodies. (B) Percentage lethality upon TBBD treatment. A dose-dependent increase in the lethality was observed. The numbers indicate TBBD concentration (micromolar) (n = 100; *p < 0.05, **p < 0.01). (C) Morpholino-mediated CARM1 knockdown in zebrafish. Control/CARM1 morpholino–injected Zf were immunostained with CARM1 antibody; a whole- mount confocal image represents the extent of knockdown. (D) Control/CARM1 morpholino– injected Zf were immunostained with α-H3R17me2a; a whole-mount confocal image indicates the decrease in H3R17 levels. (E) Percentage lethality observed in CARM1 morpholino–injected embryos compared with control morpholino–injected embryos (n = 180; p = 0.039). (F) The phenotype of the morphants was observed at regular intervals, and the defects seen in a few of the embryos after 1 wk are represented.
CARM1 is involved in sensory response
The neuromuscular defects observed in the knockdown embryos are characteristic of NF1 mutations. Of interest, NF1 has been shown to disorient the astroglial lineage balance (Dasgupta and Gutmann, 2005) and has been associated with defects in spinal curvature and in learning and other specialized processes (Crawford et al., 2007). As observed in TBBD-treated EBs, down-regulation of Sox1 and NF1 expression was also seen in the CARM1 morphants (Figure 5A), confirming the role of CARM1 in Sox1 and NF1 expression in another model system, implicating differential levels of CARM1 regulation leading to defects in the astroglial lineage. The extent of down-regulation was greater than that observed upon TBBD treatment, indicating the possible involvement of the other functions of CARM1 in the regulation of expression of these genes. Examination of CARM1 expression in mouse suggests that its expression is strong in diencephalon, which is involved in specialized learning processes and sensory effects, but disappears in the middle region. Other factors that were dysregulated, like the miR92a target, HAND2, are also known to be involved in the development of the sympathetic neural system (Vincentz et al., 2012). Hence we decided to investigate the effect of TBBD treatment on sensory response as represented in Figure 5B using the embryos that survived. About 40% of the TBBD-treated embryos showed a decreased response to needle prick, which is a significant phenotype (Figure 5C). Remarkably, this was not reduced with time, probably due to the stability of the inhibitor and constant availability of the inhibitor in the treatment solution. Approximately 50% of the morphants also exhibited a defect in their sensory response (Figure 5D). This phenotype was more pronounced at early stages of the knockdown, with an observable reduction over time, indicating a possible turnover mechanism. The high turnover rate of glial cells could be an added reason for the reversal of the phenotype at later stages. To provide conclusive evidence for the role of CARM1 in the sensory response, we performed a rescue experiment in which mCARM1 was injected into the CARM1 morphants (Supplemental Figure S6). This led to a significant recovery of the sensory response (Figure 5E). Thus it is evident that the arginine methyltransferase CARM1 regulates the sensory response, possibly through H3R17 methylation.
FIGURE 5:
CARM1 is involved in sensory response. (A) Gene expression analysis of NF1 and Sox1 in CARM1 morphants in comparison with control (n = 3; t-test p values: NF1, 0.0293; Sox1, 0.0536. (B) Assay to measure response to stimulus. About 200 embryos injected with control/CARM1 morpholino at single-cell stage and 200 embryos each treated with DMSO/TBBD at 10 h postfertilization were observed for response to stimulus (100 embryos each were analyzed on 3, 4, and 5 d postfertilization [dpf]). (C) Comparison of the response to stimulus between DMSO-treated embryos and embryos treated with increasing concentrations of TBBD on 3, 4, and 5 dpf (n = 100; *p < 0.05, **p < 0.01, ***p < 0.005). The percentage of nonresponders showed a drastic increase at higher concentrations of inhibitor, especially for the 4- and 5-dpf treatments. (D) Comparison of the response to stimulus between control and CARM1 morphants on 3, 4, and 5 dpf (n = 100; p values: day 3, 0.0113; day 4, 0.007; day 5, 0.0378). The effect on the sensory response is more pronounced in the 3-dpf samples and is diluted out over the developmental stage. (E) Rescue of sensory response upon injection of in vitro–transcribed CARM1 RNA. The loss in sensory response observed in CARM1 morphants was rescued to a greater extent on 3 dpf. The effect of the morpholino and the RNA decreased on 4 and 5 dpf owing to dilution with time.
Histone H3R17 methylation inhibition leads to abnormal astroglial cells
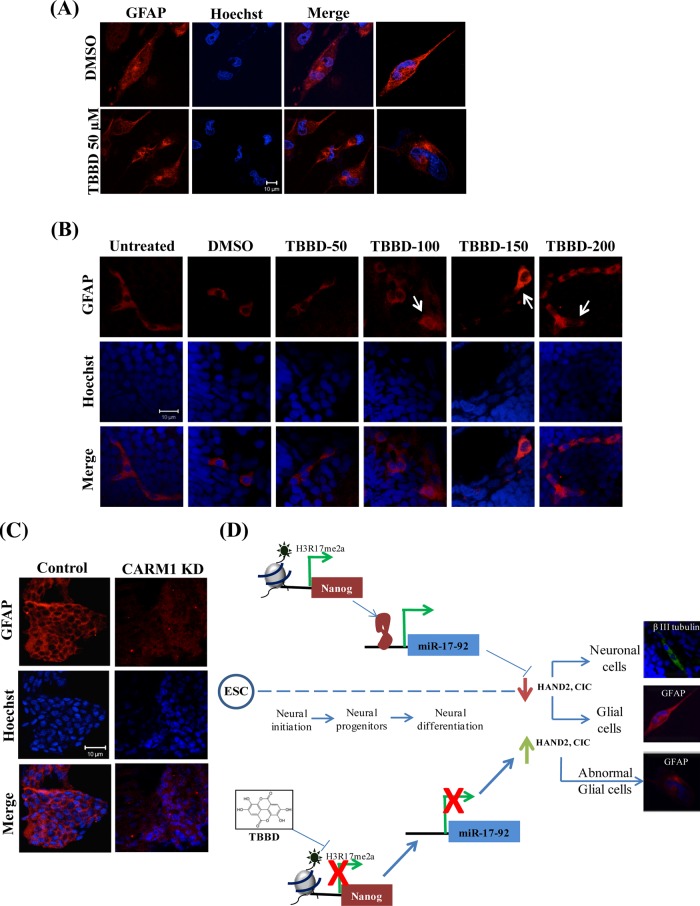
The loss in sensory response associated with inhibitor treatment or knockdown had a diluting effect, indicating a possible turnover mechanism. Astroglial cells are characterized by a relatively rapid turnover rate. The integration of the gene expression and the miRNA expression pattern suggested abnormal up-regulation of a bona fide glial cell marker, the structural protein glial fibrillary acidic protein (GFAP), which can potentially trigger this turnover. Hence we treated glial cells with TBBD and looked for GFAP expression. Remarkably, drastic mislocalization of GFAP expression was observed in TBBD-treated cells as compared with the control (Figure 6A and Supplemental Figure S7). Immunostaining of the cryosectioned embryos that survived the treatment revealed mislocalization of GFAP in TBBD-treated embryos in comparison to untreated and dimethyl sulfoxide (DMSO; solvent control)–treated embryos (Figure 6B), reflecting altered expression and nuclear mislocalization, whereas the neuronal marker β-III-tubulin expression and localization were unaffected even on treatment with 200 μM TBBD (Supplemental Figure S8). Immunostaining of cryosectioned CARM1-morphant embryos revealed an abnormal pattern of GFAP expression (Figure 6C), and this effect was much more drastic than that observed upon TBBD treatment, possibly due to the severity of a knockdown as compared with the specific inhibitor–based approach. However, the neural specific marker β-III-tubulin exhibited normal expression, indicating a glial cell–specific effect (Supplemental Figure S9). Activation of certain kinase pathways, such as the Jun N-terminal kinase (JNK) and mitogen-activated protein kinase (MAPK)families, can also lead to GFAP accumulation, thereby forming abnormal astrocytes (Tang et al., 2006). Hence TBBD-treated cells were tested for any involvement of signaling pathways, which revealed a marginal perturbation of phospho-ERK upon TBBD treatment (Supplemental Figure S10).
FIGURE 6:
Histone H3R17 methylation inhibition leads to abnormal astroglial cells. (A) Analysis of GFAP expression in U373 MG cells treated with TBBD for 24 h. DMSO-treated cells were used as control. The quantification of abnormal GFAP localization is included in the Supplemental Information. (B) Immunofluorescence analysis of GFAP expression in zebrafish embryos treated with different concentrations of TBBD from 10 h postfertilization for 48 h depicts an abnormal GFAP staining at increased concentrations. (C) Immunostaining of cryosectioned control and morphant embryos using antibody against GFAP shows abnormal GFAP localization in the morphants. (D) Schematic representation of the role of H3R17 methylation in specifying the astroglial lineage.
Thus the astroglial fate commitment pathway is regulated by CARM1-mediated H3R17 methylation by a network of transcriptional and posttranscriptional regulation involving Nanog and miR92a. An additional level of regulation in this fate commitment is brought about by the kinase signaling pathways (Figure 6D).
DISCUSSION
Although involvement of CARM1 in differentiation has been well established for muscle tissues and adipocytes, a role for CARM1 in neural function has not been previously demonstrated. A microarray analysis was carried out with HeLa cells treated with an H3R17 methylation–specific inhibitor, TBBD (Supplemental Figure S1). The analysis indicated up-regulation of several genes in different signaling pathways involved in the process of cancer manifestation. An interesting subset included several genes implicated in neurodegenerative diseases, which were also up-regulated as a consequence of TBBD treatment.
The effect of TBBD treatment could be a result of regulation at multiple levels, including posttranscriptional regulation of gene expression. It was shown that CARM1-mediated H3R17 methylation regulates a few myogenic miRNA promoters, thereby influencing the process of muscle differentiation (Mallappa et al., 2011). This observation corroborates previous studies on the necessity of CARM1 in muscle differentiation (Chen et al., 2002). Hence, to determine whether there are any potential miRNAs that are regulated by H3R17 methylation, we performed microarray profiling of the miRNAs upon TBBD treatment of the EBs. However, the inhibition of H3R17 methylation and the silencing of CARM1 expression led to a global alteration in the gene profile, with nearly equal numbers of genes being up-regulated or down-regulated. Of these, there were various genes involved in neural differentiation that were dysregulated, including Sox1 and NF1. Our present data regarding miRNA profile alteration upon inhibition of H3R17 methylation emphasizes the role of this modification in regulating miRNA expression, which could inversely influence gene expression. Of interest, we observed down-regulation of miR10a and miR575, which are specifically up-regulated in glial cells and hence have been hypothesized to play a role in the maintenance of the astroglial lineage (Cho et al., 2011). Similarly, members of the miR302 family, which are known to be involved in maintenance of pluripotency, were also down-regulated. Of interest, miR92a, which is involved in neural development (Bian et al., 2013), as well as in carcinogenesis (Olive et al., 2010), was down-regulated. This is in agreement with the gene expression analysis done with HeLa cells treated with TBBD, in which dysregulation of various genes involved in both tumor progression and tumor suppression was observed (Supplemental Figure S11). Further, these results also corroborated well with the contradictory roles of CARM1 in maintenance of pluripotency and in various differentiation pathways, exemplified by down-regulation of miRNAs controlling both pathways.
Collectively these data suggest that, under the normal conditions of early neuronal development, CARM1-mediated H3R17 methylation regulates the transcription of Sox1 and NF1, which are genes involved in early neuronal progenitor specification and astroglial lineage balance maintenance, respectively. A further level of regulation is exerted by H3R17 methylation on miRNA promoters specific for the glial lineage, such as miR10a, miR575, and miR92a.
On integrating the gene expression and miRNA expression data, it was observed that most of the miRNAs in the network target Myc. Although the role of Myc in neuronal differentiation is not well elucidated, this suggests a potential role for this factor in neuronal differentiation. On TBBD treatment and subsequent H3R17 methylation inhibition, expression of the tumor suppressor NF1 is reduced, leading to an imbalance in the astroglial lineage. It was previously shown that absence of NF1 leads to increased glial population, which would exhibit abnormal accumulation of GFAP. Remarkably, upon TBBD treatment, abnormal accumulation of GFAP is observed in different cell types as well as in zebrafish. Thus this complex regulatory network provides a glimpse into the molecular factors responsible for the glial cell–specific defects observed upon TBBD treatment as depicted in Figure 2, C and D. The glial population is further increased by activation of certain kinase pathways, such as the JNK and MAPK families, contributing to abnormal GFAP accumulation and leading to formation of abnormal astrocytes.
Thus, with the small-molecule inhibitor TBBD, which specifically inhibits H3R17 methylation, we have been able to elucidate a role for H3R17 methylation in the astroglial lineage. This was also manifested as a defect in the sensory response, which could be overcome by performing a rescue experiment. CARM1 RNA alone exhibited a phenotype that could be an effect of overexpression of a developmental regulator or due to toxicity effects. However, when expressed in the morphants, it exhibits a recovery from the observed nonresponder phenotype, indicating involvement of CARM1 in this response, as well as an absolute requirement for an optimal dosage of CARM1 expression and the associated methylation for this process. Because glial cells are abundant and are involved in structural maintenance, it is possible that these defects may not result in drastic phenotype. However, it is possible that there could be minor defects with very specific changes. In light of the available information that the H3R17 methylation mark is not a highly abundant modification, the observation that the inhibition of this modification leads to a perturbation in the astroglial lineage by regulating crucial transcriptional regulators is indeed significant. The extent of these abnormalities may be different in a scenario in which the R17 methylation mark is constantly inhibited throughout the process of development and needs further investigation.
MATERIALS AND METHODS
BG01V cells (GSC-1103; GlobalStem) were maintained in a feeder-free culture system. Routine maintenance, embryoid body formation, and alkaline phosphatase staining were done as per the detailed methodology in the Supplemental Methods.
Zebrafish routine maintenance was performed according to standard protocols (Westerfield, 1995), and details of morpholinos were adapted from Batut et al. (2011); protocols are described in the Supplemental Methods.
For cryosectioning of zebrafish whole embryos, the embryos were fixed in paraformaldehyde and cryoprotected using an increasing concentration of sucrose (5, 7.5, 12.5, 20%), then a 2:1 ratio of 20% sucrose and OCT, and finally 100% OCT. Cryoblocks of these embryos were made by freezing them in OCT, and they were sectioned at −30°C using a cryostat (Barthel and Raymond, 1990).
Immunofluorescence analysis was performed with the detailed methodology provided in the Supplemental Methods.
RNA isolation and gene expression analysis were performed as described earlier (Selvi et al., 2010). The primer sequences are provided in Supplemental Tables S1 and S2.
Immunoblotting analysis was performed as described earlier (Selvi et al., 2010); the details of the antibodies are included in the Supplemental Methods.
For miRNA expression, pre-miR92a and pre-scr-miR92a (where the seed sequence was scrambled) were cloned individually into pSUPER.puro vector as per manufacturer's instructions. The constructs were used to transfect SH-SY5Y cells, and the cells were harvested for RNA isolation after 36 h of transfection.
For quantification of miRNA expression, Invitrogen Ncode Vilo miRNA cDNA synthesis kit was used to make cDNA from total RNA isolated by the TRIzol method, following the manufacturer's instructions. The cDNA thus made was used for downstream qRT-PCR–based profiling.
Chromatin immunoprecipitation was performed as described in Karantzali et al. (2008) and Selvi et al. (2010).
Network analysis was performed with the detailed methodology provided in the Supplemental Methods.
Supplementary Material
Acknowledgments
Work in the authors’ laboratory is supported by funding from the Jawaharlal Nehru Centre for Advanced Scientific Research and the Department of Biotechnology, Government of India (Program Support: Chromatin and Disease). We thank MRS Rao, President, Jawaharlal Nehru Centre for Advanced Scientific Research, for support. Zebrafish work in R.K.M.'s lab is supported by a Centre of Excellence Epigenetics grant from the Department of Biotechnology. We thank B. S. Suma, Confocal Facility, Jawaharlal Nehru Centre for Advanced Scientific Research, for technical help. We acknowledge Lorraine Young, Centre for Biomolecular Sciences, Nottingham, United Kingdom, for initial training of R.S. on stem cell work. We thank K. Ravinder and Navneet Kaur Matharu for help and discussions on zebrafish experiments. A.S. is a Council of Scientific and Industrial Research Senior Research Fellow, and T.K.K. is a J.C.Bose National Fellow..
Abbreviations used:
- CARM1
coactivator-associated arginine methyltransferase 1
- EB
embryoid body
- GFAP
glial fibrillary acidic protein
- GO
Gene Ontology
- hESC
human embryonic stem cell
- miRNA
microRNA
- SAM
S-adenosyl methionine
- TBBD
ellagic acid
- Zf
zebrafish.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0019) on November 12, 2014.
REFERENCES
- Barthel LK, Raymond PA. Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. doi: 10.1177/38.9.2201738. [DOI] [PubMed] [Google Scholar]
- Batut J, Duboe C, Vandel L. The methyltransferases PRMT4/CARM1 and PRMT5 control differentially myogenesis in zebrafish. PLoS One. 2011;6:e25427. doi: 10.1371/journal.pone.0025427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee V, Horn KH, Singh S, Webb CL, Pisano MM, Greene RM. CBP/p300 and associated transcriptional co-activators exhibit distinct expression patterns during murine craniofacial and neural tube development. Int J Dev Biol. 2009;53:1097–1104. doi: 10.1387/ijdb.072489vb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- Cho JA, Park H, Lim EH, Lee KW. MicroRNA expression profiling in neurogenesis of adipose tissue-derived stem cells. J Genet. 2011;90:81–93. doi: 10.1007/s12041-011-0041-6. [DOI] [PubMed] [Google Scholar]
- Crawford AH, Parikh S, Schorry EK, Von Stein D. The immature spine in type-1 neurofibromatosis. J Bone Joint Surg Am. 2007;89((Suppl 1)):123–142. doi: 10.2106/JBJS.F.00836. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Gutmann DH. Neurofibromin regulates neural stem cell proliferation, survival, and astroglial differentiation in vitro and in vivo. J Neurosci. 2005;25:5584–5594. doi: 10.1523/JNEUROSCI.4693-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Po A, Miele E, Campese AF, Begalli F, Silvano M, Infante P, Capalbo C, De Smaele E, Canettieri G, et al. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. 2013;32:2819–2832. doi: 10.1038/emboj.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, Taniguchi N, Hashimoto M, Tsuchiya M, Ozaki T, et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9:47. doi: 10.1186/1471-213X-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E, Schulz H, Hummel O, Hubner N, Hatzopoulos A, Kretsovali A. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, Bedford MT. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279:25339–25344. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Chan WI, Cheung M, Cheng YC, Appleby VJ, Orme AT, Scotting PJ. CIC, a member of a novel subfamily of the HMG-box superfamily, is transiently expressed in developing granule neurons. Brain Res Mol Brain Res. 2002;106:151–156. doi: 10.1016/s0169-328x(02)00439-4. [DOI] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C, Hu YJ, Shamulailatpam P, Tae S, Sif S, Imbalzano AN. The expression of myogenic microRNAs indirectly requires protein arginine methyltransferase (Prmt)5 but directly requires Prmt4. Nucleic Acids Res. 2011;39:1243–1255. doi: 10.1093/nar/gkq896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Dutton R, Koblar S, Cheema S, Bartlett P. Cytokines which signal through the LIF receptor and their actions in the nervous system. Prog Neurobiol. 1997;52:355–378. doi: 10.1016/s0301-0082(97)00020-8. [DOI] [PubMed] [Google Scholar]
- Olive V, Jiang I, He L. mir-17–92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R, Lee CK, Cannizzaro LA, d’Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi BR, Batta K, Kishore AH, Mantelingu K, Varier RA, Balasubramanyam K, Pradhan SK, Dasgupta D, Sriram S, Agrawal S, Kundu TK. Identification of a novel inhibitor of coactivator-associated arginine methyltransferase 1 (CARM1)-mediated methylation of histone H3 Arg-17. J Biol Chem. 2010;285:7143–7152. doi: 10.1074/jbc.M109.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Xu Z, Goldman JE. Synergistic effects of the SAPK/JNK and the proteasome pathway on glial fibrillary acidic protein (GFAP) accumulation in Alexander disease. J Biol Chem. 2006;281:38634–38643. doi: 10.1074/jbc.M604942200. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010;24:2408–2419. doi: 10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, VanDusen NJ, Fleming AB, Rubart M, Firulli BA, Howard MJ, Firulli AB. A Phox2- and Hand2-dependent Hand1 cis-regulatory element reveals a unique gene dosage requirement for Hand2 during sympathetic neurogenesis. J Neurosci. 2012;32:2110–2120. doi: 10.1523/JNEUROSCI.3584-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 1995. The Zebrafish Book. [Google Scholar]
- Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, Langford CF, Glover DM, Zernicka-Goetz M. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.