Abstract
A fundamental question in the field of medical mycology is the origin of virulence in those fungal pathogens acquired directly from the environment. In recent years, it was proposed that the virulence of certain environmental animal-pathogenic microbes, such as Cryptococcus neoformans , originated from selection pressures caused by species-specific predation. In this study, we analyzed the interaction of C. neoformans with three Paramecium spp., all of which are ciliated mobile protists. In contrast to the interaction with amoebae, some Paramecium spp. rapidly ingested C. neoformans and killed the fungus. This study establishes yet another type of protist-fungal interaction supporting the notion that animal-pathogenic fungi in the environment are under constant selection by predation.
Keywords: Paramecium, virulence factors, Cryptoccocus neoformans, environmental predators, evolution
Introduction
Cryptococcus neoformans is a free-living pathogenic fungus that primarily inhabits soil contaminated with decaying organic matter and bird excreta [1,2]. C. neoformans is unusual among the pathogenic fungi in that it has several well characterized virulence factors such as a polysaccha-ride capsule, degradative enzymes, and melanin pigment production [3]. The existence of well-defined mammalian virulence factors in C. neoformans is of particular interest since the organism normally inhabits soil and it is uncertain how those traits were selected through evolution. C. neoformans is a non-specific pathogen that has been reported to infect and/or cause disease in a myriad of taxonomically diverse organisms from mammals to birds to such invertebrates as insects and worms [4–8].
One of the enduring questions in the field of medical mycology is the origin and maintenance of virulence by microbes, such as C. neoformans , which have no requirement to inhabit vertebrate hosts for replication or survival. One theory to explain this phenomenon posits that selective pressures in the environment, possibly from protists, selected for the emergence of traits that could also function for mammalian virulence [9,10]. Supporting this view, the intracellular pathogenic strategy of C. neoformans for Acanthomoeba castellanii and Dictyostelium discoideum is strikingly similar to that observed with mammalian macrophages [11,12].
Thus far, the study of C. neoformans interactions with protista has been limited to amoeba species and social amoebae of slime molds [2,11,13,14]. In an effort to ascertain the nature of the interaction of C. neoformans with other unicellular protists, we explored the outcome of C. neoformans interactions with three species of paramecia. Our goal was to determine whether such interactions favored the paramecium or the fungus and to investigate the suitability of these hosts for the study of cryptococcal pathogenesis. Unlike A. castellanii and D. discoideum , which obtain nutritients by phagocytosis, Paramecium spp. are rapidly swimming grazers with a prodigious capacity for ingesting bacteria and fungi [15]. Here we show that in contrast to the interaction of C. neoformans with the amoeboid predators, A. castellanii and D. discoideum , where the fungus is rarely killed by the amoeba, certain species of paramecia can reduce fungal cell numbers.
Materials and methods
Organisms and culture conditions
Stocks of Paramecium multimicronucleatum and Paramecium aurelia (Carolina Biological Supply, Burlington, NC) were grown as per company instructions. Paramecium tetraurelia (stock 51) cultures were obtained from the American Type Culture Collection (ATTC, Manassas, VA) and grown in cereal grass media containing Enterobacter aero-genes (ATTC # 13048). Stocks of Paramecium tetraurelia were subcultured every two weeks in media containing Enterobacter aerogenes . A volume of 50 μl of a bacterial culture was supplemented weekly to sustain paramecial growth. C. neoformans strains H99 (serotype A), 24067 (serotype D), and NIH 112 (serotype B) were grown for 24 – 48 h in Difco Sabouraud Dextrose Broth (BD) at 30ºC and 150 rpm.
Ingestion assays
Light microscopy. C. neoformans cells were labeled with 1% Uvitex 2B solution in PBS (Polyscience, Inc Warrington, PA) for 10–30 min at room temperature (RT). Cells were then washed twice with spring water (Carolina Biological Supply, Burlington, NC) and approximately 30–35 cells of P. multimicronucleatum were added to 96-well tissue culture plates in cereal grass culture media. Paramecia were allowed to acclimate for 30 minat RT. Afterwards, 1 × 105 Uvitex 2B C. neoformans -labeled cells were added to the protist suspension. Plates were then incubated for 2 hat RT. Following incubation, the paramecia were fixed with 2.5% formaldehyde and visualized with a DAPI filter-equipped Zeiss Axiophot microscope.
C. neoformans killing assays . Approximately 1 × 105 C. neoformans cells were added to 100 cells of P. tetraurelia in a 96-well tissue culture plate for a 1:1000 effector to target ratio. C. neoformans cells in cereal grass media alone served as a control. Killing of C. neoformans H99 cells by P. tetraurelia cells was determined by counting colony-forming units (CFU) of C. neoformans and comparing colony counts at 0 and 24 hof co-incubation. At each time interval, the number of colonies associated with and without paramecia was determined. The protozoa were lysed by forcibly pulling the culture through a 30.5 gauge needle 10 times. This procedure lysed 86% of the paramecia cells and allowed intracellular C. neoformans to be counted. The procedure of pulling C. neoformans through a 30.5 gauge needle had no effect on C. neoformans viability (data not shown). The suspension containing the C. neoformans was then plated on Sabouraud Dextrose Agar (BD) plates containing 10 μg/ml of penicillin-streptomycin to inhibit bacterial growth. All plates were incubated at 30°C for 48 h All CFU counts were done in triplicate.
Transmission Electron Microscopy . C. neoformans was added to P. tetraurelia at a ratio of 1:1000 effector to target cells and was incubated at RT for 2 h The samples were then fixed with 2.5% glutaraldehyde, 2.0% paraformalde-hyde in 0.1 M sodium cacodylate buffer, post-fixed with 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries, Burlington VT). Ultrathin sections were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kv.
Statistical analysis
Statistical analysis was performed using two-way ANOVA (factors were time and presence/absence of Paramecium spp.). Comparison between the 24 hcontrol group (C. neoformans alone) and 24 hexperimental group (C. neoformans Paramecium spp) was done by the Student's t -test. All statistical analyses were performed using Microsoft Excel 2003. A P-value of 0.05 was considered statistically significant.
Results
Paramecium spp. ingest C. neoformans
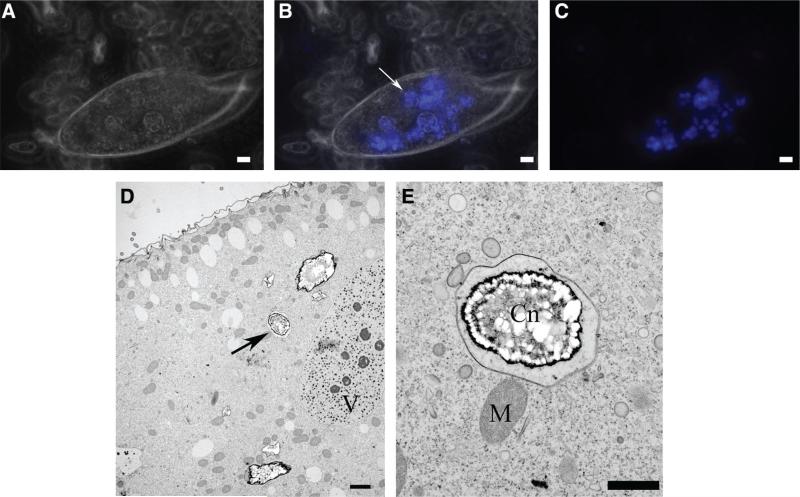
To ascertain whether fungal cells were ingested by paramecia, ingestion assays were performed using P. multimicronucleatum and C. neoformans 24067. This assay involved labeling C. neoformans with the cell wall stain Uvitex 2B and observing the paramecium using fluorescent microscopy. The results showed that this Paramecium species engulfed C. neoformans cells during a 2 hincubation (Fig. 1A,C). Transmission electron microscopy experiments using C. neoformans H99 and P. tetraurelia confirmed this result by providing visual evidence of digested C. neoformans within P. tetraurelia food vacuoles (Fig. 1D,E). When viewed under the microscope in real time, ingestion of all C. neoformans by paramecia cells caused a transient decrease in the rate of protozoal swimming common to all Paramecium spp. during grazing (data not shown). When viewed at 24 h paramecia cells did not contain C. neoformans cells, further implying digestion of previously ingested cells (data not shown).
Fig. 1.
Incubation of Paramecium spp. with Cryptococcus neoformans strains 24067 and H99 results in the ingestion of fungal cells. (A) Light microscopy, 400× magnification. (C) Corresponding fluorescence microscopy showing 1% Uvitex 2B labeled 24067. (B) Merged light and fluorescent microscopy images showing 24067 present within P. mulitmicronucleatum (arrow). (D) Transmission electron micrograph of H99 infected Paramecium tetraurelia (arrow). 2000× magnification, V=vacuole. (E) Magnified view of image (D). After a 2 h incubation, P. tetraurelia begins to digest C. neoformans in a membrane-bound food vacuole. 5000× magnification, Cn=C. neoformans, M=mitochondria. (A–C) Bar=10 μm. (D) Bar=2 μm. (E) Bar=1 μm.
P. tetraurelia and P. aurelia kill C. neoformans
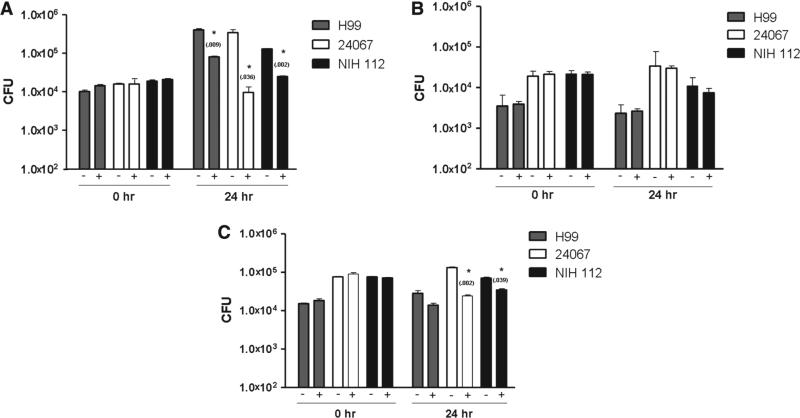
To ascertain the outcome of the interaction between different serotypes of C. neoformans and Paramecium spp., combinations of these species were co-incubated for 24 hand fungal colony counts were determined at t=0 hand 24 h Two-way ANOVA revealed that CFU counts were affected by both time and the presence of both P. tetraurelia and P. aurelia (P < 0.001) but not by the presence of P. multimicronucleatum . After 24 h in the presence of P. tetraurelia, H99, 24067, and NIH 112 CFU counts decreased by 80%, 97%, and 81%, respectively, when compared to the 24 hcryptococcal CFU in the absence of paramecia (Fig. 2A). The CFU counts decreased by 82% for 24067 and by 51% for NIH 112 after 24 hincubation with P. aurelia when compared to the 24 hcontrol, as well (Fig. 2C). In the comparison amongst C. neoformans strains, both P. tetraurelia and P. aurelia killed strain 24067 cells while P. multimicronucleatum had no impact on the growth or survival of these cells (Fig. 2A–C). NIH 112 was killed by P. aurelia, however, the presence of P. tetraurelia merely reduced its growth relative to conditions without paramecia (Fig. 2A,C). The presence of P. multimicronucleatum did not affect the growth of NIH 112 when compared to conditions without the paramecia (Fig. 2B). The growth and survival of H99 was not affected by the presence of either P. multimicronucleatum or P. aurelia , but its growth was reduced in the presence of P. tetraurelia relative to conditions without paramecia (Fig. 2A–C).
Fig. 2.
Twenty-four hour incubation of Cryptococcus neoformans strains H99, 24067 and NIH 112 with Paramecium tetraurelia, P. mulitmicronucleatum, and P. aurelia. The histograms show average CFU+95% CI at time 0 and 24 h (A) Incubation of C. neoformans strains with P. tetraurelia. (B) Incubation of C. neoformans strains with P. multimicronucleatum. (C) Incubation of C. neoformans strains with P. aurelia. A plus (+) sign indicates the presence of a Paramecium species, whereas a minus (−) sign indicates the absence of a Paramecium sp.s. *indicates statistical significance (P < 0.05). Numbers in parenthesis denote the P-value when comparing the 24 h control group and 24 h experimental group. P-values were obtained using the Student's t-test.
C. neoformans presence is not lethal to Paramecium spp
All three Paramecium spp. survived their interaction with C. neoformans with no evidence of protist killing after a 24-h incubation in the presence of C. neoformans .
Discussion
Incubation of C. neoformans with Paramecium spp. led to rapid ingestion of fungal cells by the protozoan. After the initial ingestion, there was a noticeable transient slowing of swimming motion by Paramecium spp. cells [15], but no visible damage to the protozoal cells occurred. Cells belonging to C. eoformans serotypes A, B, and D (with the exception of H99 incubated with P. aurelia) that were co-incubated with either P. tetraurelia or P. aurelia, either exhibited a reduction in growth, or were killed relative to conditions that contained no protozoal cells, evidenced by a reduction in colony counts. We interpret this result as indicating that some Paramecium spp. can prey on fungal cells and reduce fungal growth by killing cryptococcal cells. However, P . multimicronucleatum does not reduce CFU numbers for any of the C . neoformans strains studied (Fig. 2B). One possible explanation for this result is the fact that this species is the largest amongst all Paramecium spp. (~200–350 μm) [15]. The size of its cell body relative to C. neoformans may cause the paramecia to filter the fungus out of its feeding apparatus, as described [15]. Our data suggests that Paramecium spp. interact with C. neoformans differently than the interaction reported by Bunting et al . and Steenbergen et al. regarding Acanthamoeba polyphaga and A. castellani, respectively [11,16]. Unlike the interactions with A. castellanii and macrophages, interactions of C. neoformans with Paramecium spp. do not result in death of the protozoan after a 24-h incubation period [17]. In this regard, the interaction between cryptococci and paramecia also differs from the interactions of cryptococcal cells with C. elegans [18].
Based on our observations, the paramecium feeding apparatus and digestive system are not disabled by the virulence factors of C. neoformans serotypes A, B, and D. Consequently, the C. neoformans-Paramecium spp. interaction provides a fundamentally different system for studying fungal-host interactions than the amoeba, slime mold, and worm systems described previously. Additionally, our results describe another type of interaction that can be expected to constitute a major selective pressure for C. neoformans and other related yeasts in the environment.
Acknowledgements
We thank Andre Nicola for helping with the fluorescent microscopy work and for providing Uvitex 2B. We also thank Susana Frases-Carvajal and Nareen Abboud for their insight and advice regarding each of the experimental procedures.
This work was supported by National Institutes of Health grants AI33774, AI33142, and HL59842-01 to A.C. CJC was supported by Training Program in Cellular and Molecular Biology and Genetics, T32 GM007491. SZF is supported by the Dr Marshall Horwitz Fellowship of Yeshiva University.
Footnotes
Declaration of interest: None.
References
- 1.Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; Washington, DC: 1998. [Google Scholar]
- 2.Steenbergen JN, Casadevall A. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 2003;5:667–675. doi: 10.1016/s1286-4579(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 3.Ma H, May RC. Virulence in Cryptococcus species. Adv Appl Microbiol. 2009;67:131–190. doi: 10.1016/S0065-2164(08)01005-8. [DOI] [PubMed] [Google Scholar]
- 4.Goodchild LM, Dart AJ, Collins MB, et al. Cryptococcal meningitis in an alpaca. Aust Vet J. 1996;74:428–430. doi: 10.1111/j.1751-0813.1996.tb07559.x. [DOI] [PubMed] [Google Scholar]
- 5.McNamara TS, Cook RA, Behler JL, Ajello L, Padhye AA. Cryptococcosis in a common Anaconda (Eunectes murinus). J Zoo Wildlife Med. 1994;25:128–132. [Google Scholar]
- 6.Pfeiffer TJ, Ellis DH. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J Med Vet Mycol. 1992;30:407–408. [PubMed] [Google Scholar]
- 7.Wolf AM. Fungal diseases of the nasal cavity of the dog and cat. Vet Clin North Am Small Anim Pract. 1992;22:1119–1132. doi: 10.1016/s0195-5616(92)50304-7. [DOI] [PubMed] [Google Scholar]
- 8.Hough I. Cryptococcosis in an eastern water skink. Aust Vet J. 1998;76:471–472. doi: 10.1111/j.1751-0813.1998.tb10183.x. [DOI] [PubMed] [Google Scholar]
- 9.Casadevall A, Steenbergen JN, Nosanchuk JD. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi – the Cryptococcus neoformans paradigm. Curr Opin Microbiol. 2003;6:332–337. doi: 10.1016/s1369-5274(03)00082-1. [DOI] [PubMed] [Google Scholar]
- 10.Levitz SM. Does amoeboid reasoning explain the evolution and maintenance of virulence factors in Cryptococcus neoformans? Proc Natl Acad Sci USA. 2001;98:14760–14762. doi: 10.1073/pnas.261612398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen KP, Schleicher M. Dictyostelium discoideum : a genetic model system for the study of professional phagocytes. Profilin, phosphoinositides and the lmp gene family in Dictyostelium. Biochim Biophys Acta. 2001;1525:228–233. doi: 10.1016/s0304-4165(01)00108-8. [DOI] [PubMed] [Google Scholar]
- 13.Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun. 2003;71:4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neilson JB, Ivey MH, Bulmer GS. Cryptococcus neoformans : pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun. 1978;20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wichterman R. The Biology of Paramecium. Blakiston; New York: 1953. [Google Scholar]
- 16.Bunting LA, Neilson JB, Bulmer GS. Cryptococcus neoformans : gastronomic delight of a soil ameba. Sabouraudia. 1979;17:225–232. doi: 10.1080/00362177985380341. [DOI] [PubMed] [Google Scholar]
- 17.Feldmesser M, Tucker S, Casadevall A. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 2001;9:273–278. doi: 10.1016/s0966-842x(01)02035-2. [DOI] [PubMed] [Google Scholar]
- 18.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA. 2002;99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]