Abstract
The Gram-negative bacterial outer membrane (GNB-OM) is asymmetric in its lipid composition with a phospholipid-rich inner leaflet and an outer leaflet predominantly composed of lipopolysaccharides (LPS). LPS are polyanionic molecules, with numerous phosphate groups present in the lipid A and core oligosaccharide regions. The repulsive forces due to accumulation of the negative charges are screened and bridged by the divalent cations (Mg2+ and Ca2+) that are known to be crucial for the integrity of the bacterial OM. Indeed, chelation of divalent cations is a well-established method to permeabilize Gram-negative bacteria such as Escherichia coli. Here, we use X-ray and neutron reflectivity (XRR and NR, respectively) techniques to examine the role of calcium ions in the stability of a model GNB-OM. Using XRR we show that Ca2+ binds to the core region of the rough mutant LPS (RaLPS) films, producing more ordered structures in comparison to divalent cation free monolayers. Using recently developed solid-supported models of the GNB-OM, we study the effect of calcium removal on the asymmetry of DPPC:RaLPS bilayers. We show that without the charge screening effect of divalent cations, the LPS is forced to overcome the thermodynamically unfavorable energy barrier and flip across the hydrophobic bilayer to minimize the repulsive electrostatic forces, resulting in about 20% mixing of LPS and DPPC between the inner and outer bilayer leaflets. These results reveal for the first time the molecular details behind the well-known mechanism of outer membrane stabilization by divalent cations. This confirms the relevance of the asymmetric models for future studies of outer membrane stability and antibiotic penetration.
Introduction
The outer membrane (OM) of Gram-negative bacteria (GNB) is a critical barrier to overcome in the search for new antibiotics, as molecules unable to cross the OM are rendered ineffective.1 Furthermore, some bacteria acquire antibiotic resistance by modifying the permeability of their OM.2 Developing a molecular level understanding of OM structure, dynamics, and interactions with other agents is thus of great importance for both basic and applied science. The GNB-OM is highly asymmetric with a phospholipid-rich inner leaflet and an outer leaflet that is comprised of lipopolysaccharides (LPS),3 complex macromolecules that can be divided into three structural components, lipid A, the core oligosaccharide, and the O-antigen. Lipid A is embedded in the hydrophobic core of the OM and consists of a phosphorylated diglucosamine group and four to seven acyl chains. Lipid A is covalently connected to the core oligosaccharide region, which is thus localized near the vicinity of the hydrophobic membrane. It is a chain of 8–12 sugars that can also be divided into the inner and outer core regions; the former is highly phosphorylated and carboxylated and therefore strongly anionic in nature. Connected to this and facing the extracellular environment is the largest part of the molecule, the O-antigen, a chain of variable sugars that act as the hydrophilic coating of the GNB outer surface.4−6 Bacterial mutant strains that do not have the O-antigen are termed “rough” due to the appearance of their colonies on Petri dishes, whereas O-antigen-expressing cell are “smooth”.
The outer membrane of GNB is an effective barrier for many harmful agents. Charged macromolecules are unable to penetrate the hydrophobic OM bilayer, while most hydrophobic molecules have a limited permeability thanks to the dense hydrophilic sugar region formed by the LPS core oligosaccharide and O-antigen in the outer leaflet.7 In addition, LPS molecules are linked electrostatically via divalent cations (in particular, Mg2+ and Ca2+), which bind to the anionic phosphate groups in the inner core,8 significantly contributing to resistance against hydrophobic antimicrobial agents.
The divalent cations within the LPS inner core region are thus essential for outer-leaflet integrity, and indeed, many agents that permeabilize the OM, such as cationic antibiotics or EDTA,7 disrupt these important electrostatic cross-links. In vivo studies on the interactions of chelating agents with GNB have revealed the profound effect that these have on OM structure: in the presence of EDTA, the OM loses its structural integrity and vast quantities of LPS are released into solution,9 amounting for up to 50% of the bacterial LPS in some cases.7 Furthermore, phospholipid patches are thought to form in the outer leaflet,10 causing ruptures on the membrane surface that render the bacteria more susceptible to bactericidal antibiotics.11 In the laboratory, a combination of EDTA and lysozyme is commonly used to disrupt GNB. EDTA first permeabilizes the OM, which then permits lysozyme to cross into the periplasmic space, where it degrades the peptidoglycan cell wall, destabilizing the bacterial cell.12 The cells may then burst due to osmotic pressure or, if prepared in isosmotic buffers, inner membrane-only spheroplasts can be prepared.
The structural complexity of LPS and the small size of bacteria make it difficult to obtain detailed molecular information on the interactions between divalent cations and the OM. However, insights into this interaction have been recently provided by biophysical studies of LPS monolayers.8,13−15 Air/liquid interfacial monolayers composed of the deep rough mutant Re-LPS were examined in the presence of both mono- and divalent cations,15 showing that in the presence of Ca2+ ions the rigidity of the monolayer was increased due to cross-linking of the phosphate and carboxyl groups in core sugar chains by the divalent cations. Using the same rough mutant LPS, Schneck et al.8 were able to show that divalent cations displace monovalent cations from the core oligosaccharide regions of LPS monolayers. The conformation of the O-antigen from smooth LPS was studied in the absence and presence of calcium in the solution subphase, showing that the O-antigen formed a shorter, denser layer in the presence of Ca2+.13
Simulations of the OM have suggested that divalent cations bind to both phosphate and carboxylate groups present on LPS. Wu et al.16 found the calcium predominantly in an octahedral complex when bound with the anionic groups present on the LPS sugar groups, with more than 50% of the coordination sites around the Ca2+ occupied by water. Lam et al.17 used a coarse grain model to show how divalent cations in the OM rigidified the model LPS layer. EDTA was found to disrupt this charge distribution by removing the charge screening effect of the divalent cations, which lead to electrostatic repulsion between adjacent LPS molecules. Pink et al.18 showed that Ca2+ was able to block the binding of protamine, a cationic antimicrobial peptide, to LPS and therefore reduce its antimicrobial activity.
Studying the molecular details of bacterial outer membranes under biologically relevant conditions is still difficult due to their small size, and recently, in an attempt to recreate the complex bacterial outer membrane, we have developed a new solid-supported GNB-OM model19 that is predominantly composed of phospholipids in the inner leaflet and LPS in the outer leaflet, mimicking the asymmetric lipid composition of bacterial OM’s. Here, we use this GNB-OM model to investigate the essential stabilizing role of divalent cations.
Materials and Methods
Materials
Ra mutant rough strain LPS (RaLPS) from EH100 Escherichia coli was obtained from Sigma-Aldrich (Dorset, UK). DPPC and tail-deuterated DPPC [d-DPPC, 1,2-dipalmitoyl(d62)-sn-glycero-3-phosphocholine] were obtained from Avanti Polar Lipids (Alabaster, AL). All phospholipid and LPS samples were used without further purification. All other chemicals were sourced from Sigma-Aldrich.
Solid-Supported Bilayer Deposition
Rough LPS containing GNB-OM models was deposited on the surface of single silicon crystals using a purpose-built Langmuir–Blodgett (LB) trough (KSV-Nima, Biolin Scientific, Finland).19−21 LB deposition was used to deposit the inner leaflet of the membrane on the silicon surface, and Langmuir–Schaeffer (LS) deposition was used for the outer leaflet.22 For the LB deposition of the inner bilayer leaflet, h-DPPC or d-DPPC was deposited from chloroform onto a clean, nonbuffered water subphase cooled to 10 °C containing 5 mM CaCl2. The phospholipid film was then compressed to a surface pressure of 35 mN m–1. A submerged silicon crystal was then lifted through the air/water interface at a speed of 4 mm/min while the surface pressure was kept constant. The LB trough was then cleaned and an air/liquid interfacial monolayer of RaLPS was deposited again on to the cleaned surface of a nonbuffered water subphase cooled to 10 °C containing 5 mM CaCl2. The RaLPS was deposited from an LPS suspension (2 mg/mL in 60% CH3Cl, 39% MeOH, and 1% H2O v/v) and compressed to 35 mN m–1. For the LS deposition of the bilayer outer leaflet the silicon crystal containing the LB-deposited DPPC monolayer on its surface was placed in a holder above the air/liquid interface with the angle of crystal adjusted using a purpose-built leveling device to make the crystal face parallel to the water surface. The silicon crystal (and LB film) was then dipped through the interface at a constant speed of 4 mm/min and lowered into a purpose-built sample cell in the well of the trough.
X-ray Reflectivity Measurements on LPS monolayers at the Air/Liquid Interface
X-ray reflectivity (XRR) and grazing incidence X-ray diffraction (GIXD) measurements were conducted at the 9-ID beamline at the Advanced Photon Source, Argonne National Laboratories (Argonne, IL) using a liquid surface scattering spectrometer operated with an X-ray wavelength of λ = 1.284 Å.
For XRR measurements, a custom-built Langmuir trough was placed in a hermetically sealed case which was backfilled with hydrated helium to reduce beam damage to the interfacial monolayer and background scattering. Preparation of LPS monolayers was conducted as described by us previously.23 Briefly, LPS monolayers were produced by depositing a RaLPS suspension in 60% CH3Cl, 39% MeOH, and 1% H2O (v/v) onto a cleaned air/liquid interface of 20 mM HEPES pH 7.2 buffer solution containing with 5 mM CaCl2 or 3 mM EDTA. The resulting film was then compressed to a series of surface pressures, at which XRR and GIXD analysis of the monolayer took place.
The GIXD generates a 2D map of the positions of the Bragg rods and peaks in the horizontal plane, Qxy [=(4π/λ) sin(2θxy/2)], and in the vertical plane, Qz [=(2π/λ) sin(αf)]. From the in-plane peak positions the lattice spacing, d, can be determined from
| 1 |
In the case of hexagonal packing, the unit cell dimensions are a = b and γ = 120° and relate to the lattice spacing by
| 2 |
where h and k are Miller indices.24 The unit cell area is then Acell = ab sin γ.
Neutron Reflectometry Measurements on Solid-Supported Bilayers
Specular neutron reflectometry (NR) measurements were carried out using the white beam INTER reflectometer25 at the Rutherford Appleton Laboratory (Oxfordshire, UK), using neutron wavelengths from 1 to 16 Å. The reflected intensity was measured at two glancing angles of 0.7° and 2.3° as a function of the momentum transfer, Qz [=(4π sin θ)/λ, where λ is wavelength and θ is the incident angle].
Purpose-built liquid flow cells for analysis of the silicon/liquid interface were placed on a variable angle sample stage in the NR instrument, and the inlet to the liquid cell was connected to a liquid chromatography pump (L7100 HPLC pump, Merck, Hitachi), which allowed for easy exchange of the solution isotopic contrast within the (3 mL volume) solid–liquid sample cell. For each isotopic contrast change a total of 22.5 mL of 20 mM pH/D 7.2 HEPES buffer solution was pumped through the cell (7.5 cell volumes) at a speed of 1.5 mL/min.
Reflectivity Data Analysis
Neutron and X-ray reflectivity data were analyzed using the in-house software, RasCal (version 1, A. Hughes, ISIS Spallation Neutron Source, Rutherford Appleton Laboratory), which employs an optical matrix formalism (described in detail by Born and Wolf26) to fit Abeles layer models to the interfacial structure. In this approach the interface is described as a series of slabs, each of which is characterized by its scattering length density (SLD), thickness, and roughness. Interfacial roughness was implemented in terms of an error function, according to the approach by Nevot and Croce.27 The reflectivity for the model starting point is then calculated and compared with the experimental data. A least-squares minimization is used to adjust the fit parameters to reduce the differences between the model reflectivity and the data. In all cases the simplest possible model (i.e. least number of layers), which adequately described the data, was selected.
For NR data, the systems under study were asymmetrically deposited bilayers composed of DPPC (inner leaflet):RaLPS (outer leaflet), where we were able to take advantage of contrast variation via the exchange of hydrogen for deuterium in the DPPC tails and in the buffer solutions. Each isotopically labeled bilayer was examined under three solution contrasts [D2O, silicon-matched water (SMW, 38% D2O:62% H2O), and H2O] to yield six reflectivity profiles for each structure examined. The six reflectivity profiles were constrained to fit to a single profile of layer thickness and roughness for the silicon deposited bilayer, but the data fits from each isotopic contrast were allowed to vary in the SLD of each individual layer in order to account for hydration/volume fraction. The parameter fit values and the scattering length density profiles that these describe were then used to determine the bilayer structure across and surface coverage (i.e., volume fraction of bilayer defects across the surface22) and interfacial roughness. The lipid asymmetry was determined from the SLD of the tail regions of the d-DPPC-labeled bilayer using previously described linear equations.19
The volume fractions of the RaLPS and DPPC in the headgroup layers of the bilayer structures were not able to be determined due to the minimal isotopic contrast between the DPPC headgroups and the LPS core oligosaccharide region. Therefore, the percentages of DPPC, LPS, and water quoted in this paper are describing the lipid tail regions of the each leaflet within the bilayer.19
Error analysis of the fitted parameters was carried out using Rascal’s “bootstrap” error algorithm. The parameter value distributions were obtained and propagated through the rest of the derived parameters according to standard error treatment methods.
Results and Discussion
Structural Studies on RaLPS monolayers
XRR measurements were obtained for RaLPS monolayers deposited at the air/liquid interface on a HEPES buffered solution subphase containing either 5 mM CaCl2 or 3 mM EDTA. The purpose of these measurements was to examine the accumulation of divalent cations (in this case Ca2+) from solution by the LPS located at the air/liquid interface by comparing the density profile of the monolayers with and without calcium cations present.
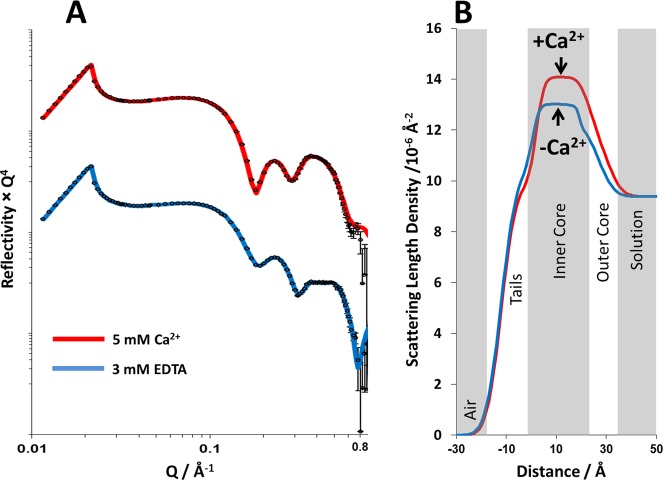
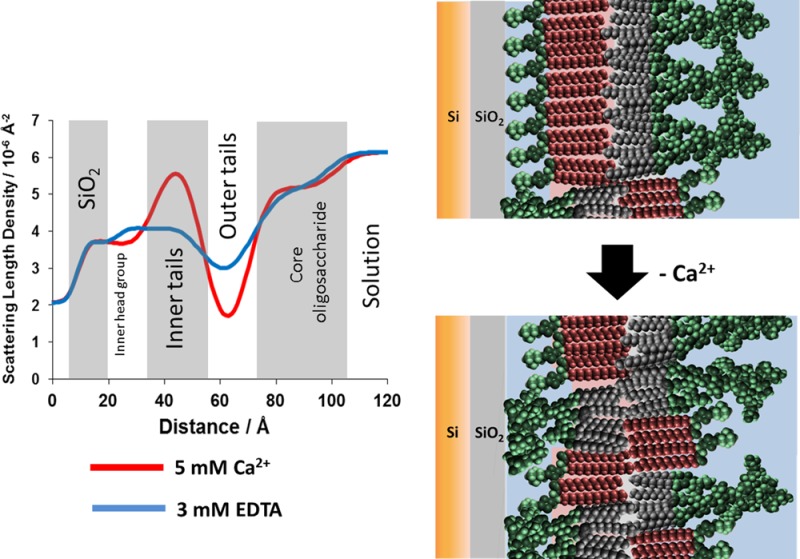
Figure 1 shows the XRR profiles, model fits, and scattering length density profiles obtained for RaLPS monolayers at 35 mN m–1 on solution subphases containing either 5 mM CaCl2 (data shown in blue) or 3 mM EDTA (data in red). The density profiles were determined by fitting the monolayers to a simple three-layer model of the LPS structure. Moving from air to solution (see Figure 1B), these layers were the lipid tails followed by the inner and outer core oligosaccharide regions. This structure was found to be consistent with the model used previously on rough mutant LPS monolayers by Le Brun et al.23
Figure 1.
A comparison of the X-ray reflectometry profiles and model data fits (A) and the scattering length density profiles these fits describe (B) for air/liquid interface containing an RaLPS monolayer held at 35 mM m–1 in the presence of 20 mM HEPES pH 7.2 H2O buffer with either 5 mM CaCl2 (red) or 3 mM EDTA (blue). The air/liquid interface was set to be between the tails and inner-core region of the LPS monolayer.
Model fitting revealed differences in both monolayer thickness and SLD profile (see Figure 1) in the presence and complete absence of Ca2+. The prominent difference was in the SLD of the inner core oligosaccharide region, where the monolayer on the calcium cation containing subphase had a significantly higher scattering length density than the same monolayer on the divalent cation free solution (see Table 1). The difference of 1 × 10–6 Å–2 in SLD can be attributed to the binding of Ca2+ to the anionic phosphate and carboxylate groups within the core oligosaccharide region.7 From a consideration of the change in SLD and ionic radius of Ca2+, it is possible to calculate the number of calcium ions that would be required per RcLPS headgroup to produce the observed change. After converting the SLD change to an electron density and using an area of 128 Å2 per RaLPS as determined by GIXD, there are 5.3 Ca2+ per RaLPS headgroup, in good agreement with the stoichiometry determined by plasma emission spectroscopy on LPS from E. coli rough mutants which found four to five divalent metal ions per LPS.28 See the Supporting Information for details of the calculation.
Table 1. Parameters Obtained from Fits of XRR Data from RaLPS Monolayers Deposited at the Air/Liquid Interface of a 20 mM HEPES Buffered Solution Subphase Containing either 5 mM CaCl2 or 3 mM EDTA.
| RaLPS
monolayer on Ca2+ containing subphase |
RaLPS
monolayer on EDTA containing subphase |
|||||
|---|---|---|---|---|---|---|
| layer | thickness/Å | roughness/Å | ρ/10-6 Å-2 | thickness/Å | roughness/Å | ρ/10-6 Å-2 |
| tails | 13.8 ± 0.1 | 4.4 ± 0.1 | 9.83 ± 0.1 | 12.7 ± 0.2 | 4.9 ± 0.1 | 10.7 ± 0.1 |
| inner core oligosaccharide | 23.5 ± 0.5 | 2.5 ± 0.1 | 14.0 ± 0.1 | 21.2 ± 0.5 | 1.8 ± 0.2 | 13 ± 0.1 |
| outer core oligosaccharide | 8.6 ± 0.5 | 4.7 ± 0.4 | 10.25 ± 0.1 | 8.5 ± 0.2 | 3.7 ± 0.5 | 10.6 ± 0.2 |
In addition, the monolayer was found to be slightly thicker in the presence of calcium than when this was removed, although these differences were found to be relatively small, accounting for a difference of only 1.1 Å in the thickness of the acyl chain region and 2.3 Å for the inner core oligosaccharide layer (the outer core thickness was the same within error). These differences in thickness are probably due to a difference in monolayer tilt under different conditions examined, with the LPS molecules within the monolayer having a higher tilt relative to the surface normal with divalent cations removed from the system than when they are present, yielding a thinner monolayer. Electrostatic repulsions between neighboring LPS molecules would force the headgroups apart, leading to increased tilt.
GIXD of monolayers at an air/liquid interface provides information along the plane of the interface for molecules packed with 2D crystallinity. In the case of LPS, as with phospholipids, only the hydrocarbon tails will have sufficient crystallinity to generate Bragg rods. For the case of RaLPS in the presence of Ca2+, a single Bragg rod at Qxy = 1.46 Å–1, consistent with ordered hexagonal packing of the acyl chains,29 was observable (see Figure 2) at 35 mN m–1. This was not observed on EDTA-containing subphases, suggesting a less ordered monolayer in the latter case. The unit cell dimensions in the Ca2+ case were calculated to be a = b = 4.970 Å and γ = 120°. This results in an area per unit cell of 21.4 Å2, which is consistent with monolayers of phospholipids at similar surface pressures.30 The unit cell only contains one hydrocarbon chain from the RaLPS molecule, and since the peak of the rod at Qz is 0 Å–1, there is no tilt, so the area per RaLPS is simply 128 Å2. GIXD observations for RaLPS differ from those observed for ReLPS and RcLPS (which have shorter core regions) and lipid A, which has no core. In these monolayers, distorted hexagonal packing (resulting in three Bragg rods) was observed at 30 mM m–1.15,23,31 However, the area per RaLPS is in agreement with the published work, where the area per LPS ranges from 108 to 127 Å2, depending on the LPS used. The lack of Bragg rods observed in the presence of EDTA is reminiscent of lipid A monolayers in the presence of the non-natural antimicrobial peptides acryl-lysyl octamer and arylamide foldamer (AA-1). The introduction of these antimicrobial peptides to monolayers of lipid A induces a disordered phase, and the Bragg rods completely disappear.32 This would suggest that the antimicrobial peptides interact with the lipid A tails, but in this case EDTA will only effect structural properties in the core region. X-ray reflectometry showed that AA-1 only resided in the polar headgroups of lipid A, showing that influences on headgroup structure can have effects in the structure across the whole molecule.
Figure 2.
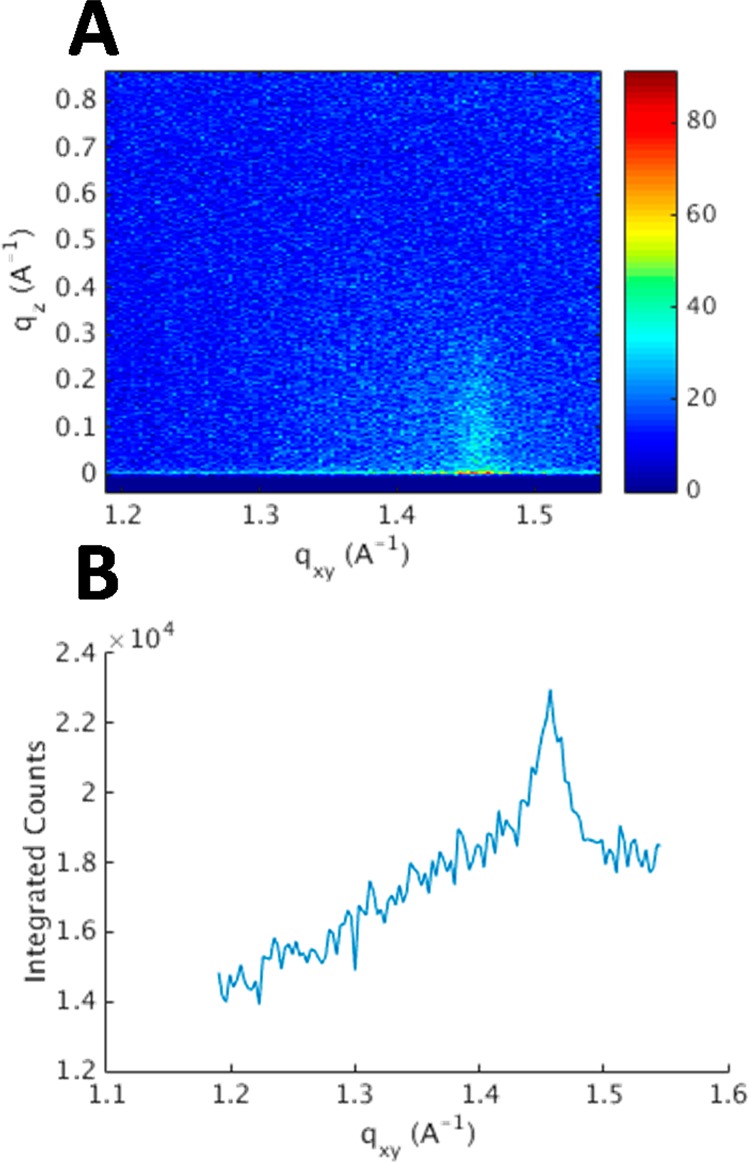
A GIXD contour plot obtained from an air/liquid interface containing an RaLPS monolayer held at 35 mM m–1 in the presence of 20 mM HEPES pH 7.2 H2O buffer with 5 mM CaCl2 (A). A plot of this data integrated over Qz is shown (B).
Structural Studies on DPPC: RaLPS Bilayers
To examine the effect divalent cations have on stabilizing the GNB-OM, the effect of Ca2+ removal upon bilayer OM models was examined. For this study, asymmetric DPPC (inner leaflet):RaLPS (outer leaflet) bilayers deposited at the silicon/water interface were initially examined in the presence of Ca2+ by NR.
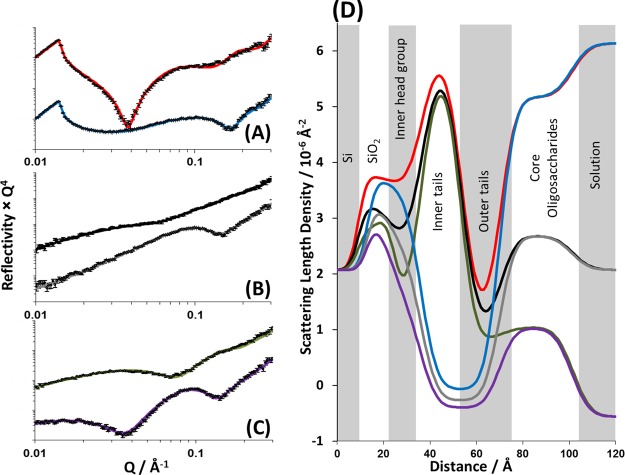
Figure 3 shows the neutron reflectivity profiles and model data fits obtained for the DPPC/RaLPS bilayer deposited on a silicon crystal surface in the presence of 5 mM CaCl2. Reflectivity profiles obtained from this bilayer were collected under six differing isotopic conditions, comprising two differing phospholipid isotopic labels of the bilayer (h- and d-DPPC), each examined under three differing solution isotopic conditions (100% D2O, SMW, and 100% H2O). A minimum of five layers is required to describe the structure of the interface; these layers are, moving sequentially from silicon to solution, a hydrated silicon oxide layer,33 the bilayer inner leaflet headgroups, the inner leaflet tails, the outer leaflet tails, and a thick layer (31.0 ± 1.0) predominantly composed of the LPS core oligosaccharide region facing the bulk solution. However, some related studies include an additional water layer between the oxide layer and the inner headgroup.34 Statistical analysis of these two models was performed by weighting the final χ2 value with the total number of free parameters, as described by Ihringer.35 This test revealed that the interfacial structure was optimally described as a five-layer structure across the silicon/water interface. The structural parameters obtained from model fitting are shown in Table 2. It is conceivable that, despite this, a small interfacial water does indeed exist; however, in the model it is then accounted for in the roughness of the adjacent layers.
Figure 3.
Neutron reflectometry profile and model data fits (A–C) and the scattering length density profiles these fits describe (D) for asymmetrically deposited DPPC (inner leaflet):RaLPS (outer leaflet) bilayer in the presence of 20 mM HEPES pH 7.2 buffer with 5 mM CaCl2. The six simultaneously fitted isotopic contrasts shown are (A) d-DPPC/RaLPS in D2O (red line), h-DPPC/RaLPS in D2O (blue line); (B) d-DPPC/RaLPS in SMW (black line), h-DPPC/RaLPS in SMW (gray line); and (C) d-DPPC/RaLPS in H2O (green line), h-DPPC/RaLPS in H2O (purple line).
Table 2. Structural Parameters Obtained for an Asymmetrically Deposited DPPC (inner leaflet):E. coli RaLPS (outer leaflet) Bilayer Deposited on a Silicon Surface in the Presence of 20 mM HEPES pH/D 7.2, 5 mM Ca2+ Solution.
| layer | thickness/Å | % DPPC | % RaLPS | % water | roughness/Å |
|---|---|---|---|---|---|
| silicon oxide | 14.6 ± 1.2 | N/A | N/A | 13 ± 2 | 3.0 ± 0.2 |
| inner headgroup | 13.0 ± 0.9 | – | – | – | bilayer roughness 5.5 ± 1.0 |
| inner tails | 17.0 ± 0.2 | 78 ± 4 | 18 ± 4 | 4 ± 4 | |
| outer tails | 14.5 ± 0.7 | 17 ± 4 | 79 ± 4 | 4 ± 4 | |
| core oligosaccharide (outer headgroup) | 31.0 ± 1.0 | – | – | – |
The difference in SLD between hydrogenous and deuterated lipid tails (SLDs of −0.39 × 10–6 and 7.45 × 10–6 Å–2, respectively) produces data sensitive to the leaflet structure within the tail region of the bilayer. When examined by NR without differential isotopic lipid/leaflet labeling, the hydrophobic tail region of lipid bilayers fits to a single layer of homogeneous SLD.34,36,37 However, isotopic labeling of the lipid components and significant asymmetry between the leaflets produces data sensitive to the differing inner and outer leaflet structure, which results in the hydrophobic tails region being modeled as two discrete layers to achieve the best fit.38,39 The isotopic labeling methodology described here allows us to accurately determine the asymmetry of this GNB-OM model by comparing the SLD obtained from the model fits of a given leaflet to the SLD of hydrogenous and deuterated lipid tails.19 Additionally, the reflectivity profiles obtained from the bilayer produced using hydrogenous DPPC and LPS are beneficial in the model fitting due to the sensitivity of these data sets to the overall bilayer structure and the core oligosaccharide region.
The bilayer structure was found to have lipid coverage of 96 ± 4% (% DPPC + % RaLPS) across the silicon surface and was asymmetric in its lipid composition, mirroring the condition under which it was deposited, with 78 ± 4% DPPC and 18 ± 4% LPS in the inner leaflet and 79 ± 4% LPS and 17 ± 4% DPPC in the outer leaflet. From these data it was clear that some mixing had occurred between inner and outer bilayer leaflets. This was likely a result of mechanical shock to the bilayer during the LS dipping stage of deposition.19,38 The bilayer roughness was 5.5 ± 1.0 Å, and this value is related to the in-plane height–height correlation function of the interface between described layers. The origin of this roughness is 2-fold: undulations in the bilayer due to headgroup size mismatch between the DPPC and LPS in inner headgroup region (which is directly next to the comparatively flat silicon oxide coating of the silicon crystal surface19) and a graded change in SLD between bilayer regions due to the shape/molecular complexity of the bilayer components.
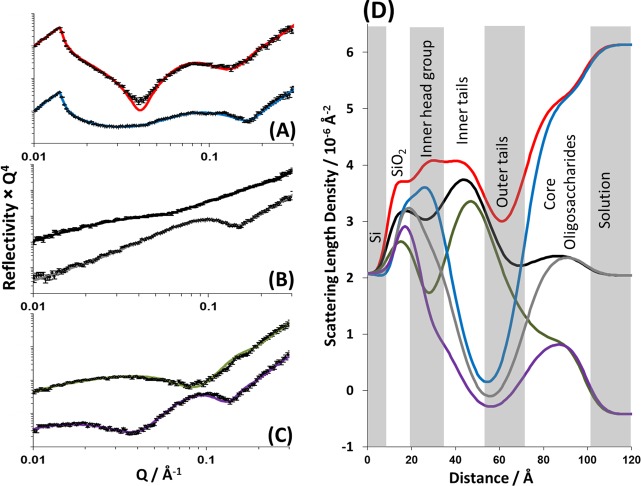
After NR data collection had taken place on bilayers in the presence of 5 mM CaCl2, the solution in the solid liquid flow cell was exchanged for a solution containing 3 mM EDTA, with 7.5 cell volumes of the buffer passed through the cell for complete buffer exchange. Ca2+ removal was assumed, as no further changes to the NR data were noted. Figure 4 shows the neutron reflectivity profiles obtained from the DPPC/RaLPS bilayer after Ca2+ sequestration by EDTA. Fitting of the reflectivity profiles revealed that the interfacial structure could be described by the same layer structure used for the bilayer in the presence of Ca2+. Figure 4 shows the experimental reflectivity profiles, model data fits, and the SLD profiles these fits describe, and Table 3 describes the structural parameters obtained from the fits to the data.
Figure 4.
Neutron reflectometry profile and model data fits (A–C) and the scattering length density profiles these fits describe (D) for asymmetrically deposited DPPC (inner leaflet):RaLPS (outer leaflet) bilayer in the presence of 20 mM HEPES pH 7.2 buffer with 3 mM EDTA. The six simultaneously fitted isotopic contrasts shown are (A) d-DPPC/RaLPS in D2O (red line), h-DPPC/RaLPS in D2O (blue line); (B) d-DPPC/RaLPS in SMW (black line), h-DPPC/RaLPS in SMW (gray line); and (C) d-DPPC/RaLPS in H2O (green line), h-DPPC/RaLPS in H2O (purple line).
Table 3. Structural Parameters Obtained for an Asymmetrically Deposited DPPC (inner leaflet):E. coli RaLPS (outer leaflet) Bilayer Deposited on a Silicon Surface in the Presence of a 20 mM HEPES pH/D 7.2, 3 mM EDTA Solution.
| layer | thickness/Å | % DPPC | % RaLPS | % water | roughness/Å |
|---|---|---|---|---|---|
| silicon oxide | 13.9 ± 5.0 | N/A | N/A | 12 ± 2 | 2.9 ± 0.2 |
| inner headgroup | 15.4 ± 4.0 | – | – | – | bilayer roughness 8.4 ± 1.2 |
| inner tails | 15.9 ± 1.0 | 59 ± 4 | 37 ± 4 | 4 ± 4 | |
| outer tails | 11.0 ± 5.0 | 32 ± 5 | 63 ± 5 | 4 ± 5 | |
| core oligosaccharide (outer headgroup) | 28.4 ± 1.0 | – | – | – |
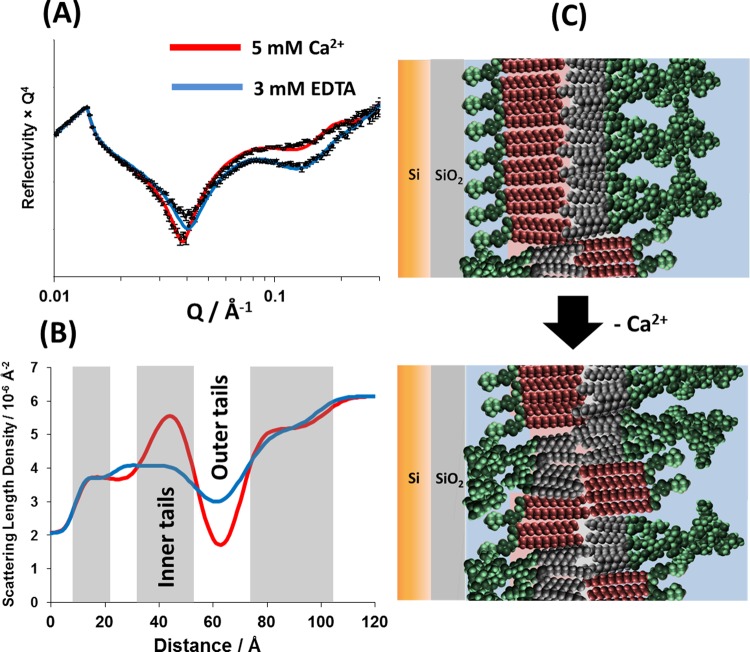
NR data revealed that the removal of Ca2+ from the bilayer was followed by both a decrease in the bilayer asymmetry (increased mixing) and an increase in roughness. The leaflet asymmetry was reduced to 59 ± 4% DPPC and 37 ± 4% RaLPS in the inner bilayer leaflet and 32 ± 5% DPPC and 63 ± 5% RaLPS in the outer leaflet (see Table 3). This is a loss of ∼20% of the DPPC from the inner leaflet and 20% LPS from the outer leaflet when compared with the same bilayer in the presence of Ca2+. Figure 5 gives a pictorial representation of these changes to the interfacial structure.
Figure 5.
A comparison of the neutron reflectometry profile and model data fits (A) and the scattering length density profiles these fits describe (B) for asymmetrically deposited d-DPPC (inner leaflet):RaLPS (outer leaflet) bilayer in the presence of 20 mM HEPES pD 7.2 D2O buffer with either 5 mM CaCl2 (red) or 3 mM EDTA (blue). A pictorial representation of the bilayer structure before and following Ca2+ sequestration by EDTA is shown (C).
The total lipid coverage at the interface remained unchanged with coverage found to be 96%. The roughness of the membrane was seen to increase in value following calcium sequestration with a value of 8.4 ± 1.2 Å found, an increase of 2.9 Å. This increase is likely caused by the increased amount of RaLPS, with its larger headgroup, in the inner leaflet of the bilayer. This size mismatch next to the relatively flat silicon oxide surface creates an increasingly undulating bilayer, which can only be expressed in the layer models used to fit the data as an increase in roughness, i.e., a smearing of the density profile.
The oligosaccharide region of the LPS outer membrane leaflet is a dense structure that concentrates negatively charged phosphate or carboxyl groups in close proximity near the bilayer interface. In the natural environment, this extraordinary buildup of repulsive forces is well-balanced by the presence of divalent cations such as Ca2+ or Mg2+, which not only screen these forces but also form salt bridges, which further strengthen the integrity of this complex lateral structure. The loss of asymmetry of the model silicon-supported DPPC/LPS bilayer observed upon removal of Ca2+ ions can therefore be attributed to the loss of sufficient electrostatic screening of the negative charges, which then provide enough energy to overcome the thermodynamic penalty of crossing the hydrophobic bilayer interior. The redistribution of LPS across the bilayer after mixing was not complete, suggesting that the localization of LPS on the inner leaflet is unfavorable and can be attributed to limited space between the bilayer and the silicon surface and repulsion between the weakly anionic SiO2 layer and the negatively charged LPS sugars.
The in vivo effects of chelating agents on the GNB-OM include the release of LPS into the bulk solution9 and the appearance of phospholipids in the outer leaflet of the bilayer.10 A loss of LPS from the bilayer as a result of calcium sequestration was not observed here, probably because the RaLPS is too hydrophobic to form solution soluble aggregates as might occur with longer, smooth LPS. Indeed, vesicle studies on rough LPS types generally involve the preparation of samples in similar fashion to phospholipids40 and these truncated LPS types can be deposited as insoluble monolayers,15,23 whereas smooth LPS is readily able to form micelles in solution in a similar way as surfactants.41,42 The electrostatic repulsion between neighboring molecules, which are suggested to drive the bilayer leaflet mixing observed on insoluble, rough LPS types studied here, are likely to also be the same forces driving the partial release of smooth LPS into solution when Gram-negative bacteria are treated with EDTA.9,43 The leaflet mixing observed in the GNB-OM models upon EDTA treatment produced an increase in phospholipids in the bilayers outer leaflet. Therefore, we present here a previously unknown effect of removal of divalent cations on the structure of the OM, LPS–LPS repulsion driven OM leaflet mixing.
Previously, we have described the production of GNB-OM models without Ca2+ present during the deposition of the individual bilayer leaflets.19 In an effort to improve interfacial coverages and bilayer asymmetry, the solution subphase was cooled below room temperature (10 °C) and 5 mM CaCl2 was present in the solution subphase during the deposition of the bilayer as well as during initial NR measurements. These changes to the fabrication procedure increased bilayer coverage at the interface from ∼80% to ∼95% and leaflet asymmetry from ∼65 to ∼80%. Interestingly, the DPPC/RaLPS bilayer following EDTA treatment showed a closer resemblance to the bilayers deposited without divalent cations in terms of the asymmetry of the inner and outer leaflets. Indeed, an asymmetrical DPPC/RaLPS bilayer deposited in the absence of calcium was found to have an outer leaflet composed of 67 ± 7% LPS and 22 ± 5% DPPC, which is close to the 63 ± 5% LPS and 32 ± 5% found for the EDTA-treated bilayer described here. This suggests the important role divalent cations play in the fabrication of these OM models, enabling high coverage and asymmetric bilayers to be fabricated by reducing electrostatic repulsion between the polyanionic LPS.
Conclusions
Using XRR and NR techniques, we reveal the intricate molecular details of the divalent-cation-driven stabilization of the outer membranes in Gram-negative bacteria. We examine the effects that take place as a result of calcium removal leading to the destabilization of the bilayer asymmetry and mixing of LPS molecules between the inner and outer leaflets. The results highlight the importance of salt bridges formed by divalent cations such as Ca2+ and Mg2+ with negatively charged sugars in LPS core oligosaccharide. These interactions appear to be crucial to the structural integrity of the outer membrane, with the disruptive nature of the electrostatic repulsive forces between adjacent LPS molecules revealed when charge screening is removed.
Acknowledgments
This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work was supported by ISIS beam-time award 1410126. J.H.L. wishes to thank the Wellcome Trust for support (Grant Nos. 080342 and 093581). A.P.L.B. thanks the Australian Research Council for a Discovery Early Career Researcher Award (DE140101788).
Glossary
Abbreviations Used
- AA-1
acrylamide foldamer
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- d-DPPC
tail-deuterated DPPC
- h-DPPC
tail-hydrogenated DPPC
- GNB
Gram-negative bacteria
- LPS
lipopolysaccharide
- NR
neutron reflectivity
- OM
outer membrane
- RaLPS
Ra chemo type rough mutant lipopolysaccharide
- RcLPS
Rc chemo type rough mutant lipopolysaccharide
- ReLPS
Re chemo type rough mutant lipopolysaccharide
- SLD
scattering length density
- SMW
silicon scattering length density matched water.
Supporting Information Available
Surface pressure vs area isotherms for RaLPS monolayers deposited on HEPES buffer containing either 5 mM CaCl2 or 3 mM EDTA, tables of parameters obtained from model fits to the experimental NR data, NR profiles obtained from a d-DPPC/RaLPS bilayer after flushing with 22.5 and 90 mL of 20 mM HEPES 5 mM CaCl2 D2O buffer at a pD of 7.2, and a description of the calculation of Ca2+ ions per LPS molecules in the monolayers. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Pagès J. M.; James C. E.; Winterhalter M. The Porin and the Permeating Antibiotic: A Selective Diffusion Barrier in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [DOI] [PubMed] [Google Scholar]
- Stavenger R. A.; Winterhalter M. TRANSLOCATION Project: How to Get Good Drugs into Bad Bugs. Sci. Transl. Med. 2014, 6, 228ed7. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C.; Bennett-Guerrero E.; Poxton I. R. Structure and Function of Lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [DOI] [PubMed] [Google Scholar]
- Nilsson C.; Skoglund A.; Moran A. P.; Annuk H.; Engstrand L.; Normark S. Lipopolysaccharide Diversity Evolving in Helicobacter pylori Communities through Genetic Modifications in Fucosyltransferases. PLoS One 2008, 3, e3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroff M.; Karibian D. Structure of Bacterial Lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [DOI] [PubMed] [Google Scholar]
- Vaara M. Agents That Increase the Permeability of the Outer Membrane. Microbiol. Rev. 1992, 56, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneck E.; Schubert T.; Konovalov O. V.; Quinn B. E.; Gutsmann T.; Brandenburg K.; Oliveira R. G.; Pink D. A.; Tanaka M. Quantitative Determination of Ion Distributions in Bacterial Lipopolysaccharide Membranes by Grazing-Incidence X-ray Fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 9147–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leive L. Release of Lipopolysaccharide by EDTA Treatment of E. coli. Biochem. Biophys. Res. Commun. 1965, 21, 290–296. [DOI] [PubMed] [Google Scholar]
- Nikaido H.; Vaara M. Molecular Basis of Bacterial Outer Membrane Permeability. Microbiol. Rev. 1985, 49, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin H. J. P.; Witholt B. A Highly Efficient Procedure for the Qautitative Formation of Intact and Viable Lysozyme Spheroplasts from Escherichia coli. Anal. Biochem. 1987, 164, 320–330. [DOI] [PubMed] [Google Scholar]
- Wooley R. E.; Blue J. L.. In-Vitro Effect of EDTA–Tris–Lysozyme Solutions on Selected Pathogenic Bacteria. J. Med. Microbiol. 1975, 8. [DOI] [PubMed] [Google Scholar]
- Schneck E.; Papp-Szabo E.; Quinn B. E.; Konovalov O. V.; Beveridge T. J.; Pink D. A.; Tanaka M. Calcium Ions Induce Collapse of Charged O-Side Chains of Lipopolysaccharides from Pseudomonas aeruginosa. J. R. Soc. Interface 2009, 6, S671–S678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham T.; Schooling S. R.; Beveridge T. J.; Katsaras J. Monolayer Film Behavior of Lipopolysaccharide from Pseudomonas aeruginosa at the Air–Water Interface. Biomacromolecules 2008, 9, 2799–2804. [DOI] [PubMed] [Google Scholar]
- Jeworrek C.; Evers F.; Howe J.; Brandenburg K.; Tolan M.; Winter R. Effects of Specific versus Nonspecific Ionic Interactions on the Structure and Lateral Organization of Lipopolysaccharides. Biophys. J. 2011, 100, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E. L.; Engström O.; Jo S.; Stuhlsatz D.; Yeom M. S.; Klauda J. B.; Widmalm G.; Im W. Molecular Dynamics and NMR Spectroscopy Studies of E. coli Lipopolysaccharide Structure and Dynamics. Biophys. J. 2013, 105, 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam N. H.; Ma Z.; Ha B.-Y. Electrostatic Modification of the Lipopolysaccharide Layer: Competing Effects of Divalent Cations and Polycationic or Polyanionic Molecules. Soft Matter 2014, 10, 7528–7544. [DOI] [PubMed] [Google Scholar]
- Pink D. A.; Hansen L. T.; Gill T. A.; Quinn B. E.; Jericho M. H.; Beveridge T. J. Divalent Calcium Ions Inhibit the Penetration of Protamine through the Polysaccharide Brush of the Outer Membrane of Gram-Negative Bacteria. Langmuir 2003, 19, 8852–8858. [Google Scholar]
- Clifton L. A.; Skoda M. W. A.; Daulton E. L.; Hughes A. V.; Le Brun A. P.; Lakey J. H.; Holt S. A.; Hughes V. Gram-Negative Bacterial Outer Membrane Mimic Asymmetric Phospholipid: Lipopolysaccharide Bilayers; a Gram-Negative Bacterial Outer Membrane Mimic. J. R. Soc. Interface 2013, 10, 20130810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. V.; Howse J. R.; Dabkowska A.; Jones R. A. L. .; Lawrence M. J.; Roser S. J. Floating Lipid Bilayers Deposited on Chemically Grafted Phosphatidylcholine Surfaces. Langmuir 2008, 24, 1989–1999. [DOI] [PubMed] [Google Scholar]
- Hughes A. V.; Holt S. A.; Daulton E.; Soliakov A.; Charlton T. R.; Steven J.; Lakey J. H.; Roser S. J.. High Coverage Fluid-Phase Floating Lipid Bilayers Supported by v-Thiolipid Self-Assembled Monolayers. J. R. Soc. Interface 2014, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm L. K.; McConnell H. M. Supported Phospholipid Bilayers. Biophys. J. 1985, 47, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Brun A. P.; Clifton L. A.; Halbert C. E.; Lin B.; Meron M.; Holden P. J.; Lakey J. H.; Holt S. A. Structural Characterization of a Model Gram-Negative Bacterial Surface Using Lipopolysaccharides from Rough Strains of Escherichia coli. Biomacromolecules 2013, 14, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenson T. R.; Kiaer K.. Structural Properties and Interactions of Thin Films at the Air–Liquid Interface Explored by Synchrotron X-ray Scattering. In Novel Methods in the Study of Interfacial Layers; Moebius D., Miller R., Eds.; Elsevier: New York, 2001; pp 206–252. [Google Scholar]
- Webster J.; Holt S.; Dalgliesh R. INTER the Chemical Interfaces Reflectometer on Target Station 2 at ISIS. Phys. B Condens. Matter 2006, 385-386, 1164–1166. [Google Scholar]
- Born M.; Wolf E.. Principles of Optics; Pergamon Press: Oxford, 1970. [Google Scholar]
- Nevot L.; Croce P. Caractérisation Des Surfaces Par Réflexion Rasante de Rayons X. Application À L’étude Du Polissage de Quelques Verres Silicates. Rev. Phys. Appl. 1980, 761–779. [Google Scholar]
- Coughlin R. T.; Tonsager S.; Mcgroarty E. J. Quantitation of Metal Cations Bound to Membranes and Extracted Lipopolysaccharide of Escherichia coli. Biochemistry 1983, 22, 2002–2007. [DOI] [PubMed] [Google Scholar]
- Als-Nielsen J.; Jacquemain D.; Kj K.; Leveiller F.; Lahav M.; Leiserowitz L. Principles and Applications of Grazing Incidence X-ray and Neutron Scattering from Ordered Molecular Monolayers at the Air–Water Interface. Phys. Rep. 1994, 246, 251–313. [Google Scholar]
- Jensen T. R.; Balashev K.; Bjornholm T.; Kjaer K. Novel Methods for Studying Lipids and Lipases and Their Mutual Interaction at Interfaces. Part II. Surface Sensitive Synchrotron X-ray Scattering. Biochimie 2001, 83, 399–408. [DOI] [PubMed] [Google Scholar]
- Neville F.; Hodges C. S.; Liu C.; Konovalov O.; Gidalevitz D. In Situ Characterization of Lipid A Interaction with Antimicrobial Peptides Using Surface X-ray Scattering. Biochim. Biophys. Acta, Biomembr. 2006, 1758, 232–240. [DOI] [PubMed] [Google Scholar]
- Ivankin A.; Livne L.; Mor A.; Caputo G. a; Degrado W. F.; Meron M.; Lin B.; Gidalevitz D. Role of the Conformational Rigidity in the Design of Biomimetic Antimicrobial Compounds. Angew. Chem., Int. Ed. 2010, 49, 8462–8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold J.; Staples E. J.; Tucker I.; Thompson L. J. Adsorption of Mixed Cationic and Nonionic Surfactants at the Hydrophilic Silicon Surface from Aqueous Solution: Studied by Specular Neutron Reflection. Langmuir 1997, 13, 6638–6643. [Google Scholar]
- Fragneto G.; Graner F.; Charitat T.; Dubos P.; Magiste M. Interaction of the Third Helix of Antennapedia Homeodomain with a Deposited Phospholipid Bilayer: A Neutron Reflectivity Structural Study. Langmuir 2000, 1, 4581–4588. [Google Scholar]
- Ihringer J. A Quantitative Measure for the Goodness of Fit in Profile Refinements with More than 20 Degrees of Freedom. J. Appl. Crystallogr. 1995, 28, 618–619. [Google Scholar]
- Fernandez D. I.; Le Brun A. P.; Whitwell T. C.; Sani M.-A.; James M.; Separovic F. The Antimicrobial Peptide Aurein 1.2 Disrupts Model Membranes via the Carpet Mechanism. Phys. Chem. Chem. Phys. 2012, 14, 15739–15751. [DOI] [PubMed] [Google Scholar]
- Callow P.; Fragneto G.; Cubitt R.; Barlow D. J.; Lawrence M. J. Interaction of Cationic Lipid/DNA Complexes with Model Membranes As Determined by Neutron Reflectivity. Langmuir 2009, 25, 4181–4189. [DOI] [PubMed] [Google Scholar]
- Gerelli Y.; Porcar L.; Fragneto G. Lipid Rearrangement in DSPC/DMPC Bilayers: A Neutron Reflectometry Study. Langmuir 2012, 28, 15922–15928. [DOI] [PubMed] [Google Scholar]
- Wacklin H. P. Composition and Asymmetry in Supported Membranes Formed by Vesicle Fusion. Langmuir 2011, 27, 7698–7707. [DOI] [PubMed] [Google Scholar]
- D’Errico G.; Silipo A.; Mangiapia G.; Molinaro A.; Paduano L.; Lanzetta R. Mesoscopic and Microstructural Characterization of Liposomes Formed by the Lipooligosaccharide from Salmonella Minnesota Strain 595 (Re Mutant). Phys. Chem. Chem. Phys. 2009, 11, 2314–2322. [DOI] [PubMed] [Google Scholar]
- Aurell C. A.; Wistrom A. O. Critical Aggregation Concentrations of Gram-Negative Bacterial Lipopolysaccharides (LPS). Biochem. Biophys. Res. Commun. 1998, 253, 119–123. [DOI] [PubMed] [Google Scholar]
- Santos N. C.; Silva A. C.; Castanho M. A.; Martins-Silva J.; Saldanha C. Evaluation of Lipopolysaccharide Aggregation by Light Scattering Spectroscopy. ChemBioChem 2003, 4, 96–100. [DOI] [PubMed] [Google Scholar]
- Van Alphen L.; Verkleij A.; Leunissen-Bijvelt J.; Lugtenberg B. Architecture of the Outer Membrane of Escherichia coli Complexes, I I I Protein-Lipopolysaccharide Complexs in Intramembranous Particles. J. Bacteriol. 1978, 134, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.