Abstract
Macromolecular nucleic acids such as DNA vaccines, siRNA, and splice-site switching oligomers (SSO) have vast chemotherapeutic potential. Nanoparticulate biomaterials hold promise for DNA and RNA delivery when a means for binding is identified that retains structure-function and provides stabilization by the nanoparticles. In order to provide these benefits of binding, we combined DNA and RNA with protamine— demonstrating association to gold microparticles by electrophoretic, gel shot, fluorescence, and dynamic laser light spectroscopy (DLLS). A pivotal finding in these studies is that the Au-protamine-DNA conjugates greatly stabilize the DNA; and DNA structure and vaccine activity are maintained even after exposure to physical, chemical, and temperature-accelerated degradation. Specifically, protamine formed nanoparticles when complexed to RNA. These complexes could be detected by gel shift and were probed by high throughput absorbance difference spectroscopy (HTADS). Biological activity of these RNA nanoparticles (RNPs) was demonstrated also by a human tumor cell splice-site switching assay and by siRNA delivery against B-Raf—a key cancer target. Finally, RNA:protamine particles inhibited growth of cultured human tumor cells and bacteria. These data provide new insights into DNA and RNA nanoparticles and prospects for their delivery and chemotherapeutic activity.
Keywords: Splice-site switching oligonucleotide, DNA nanoparticle, RNA nanoparticle, Protamine, DNA vaccine
Introduction
Despite the promise of nanotechnology for the delivery of therapeutic nucleic acids, a major current limitation is the inability to combine therapeutic DNA and RNA with nanomaterials so that structure and function can be maintained after bioprocessing and delivery. DNA vaccines [1], siRNA [2], and splice-site switching oligomers (SSO) [3] may have the ability to modulate gene expression in cells, thereby creating a new class of molecular chemotherapeutics. Given the emergent knowledge that alternative splicing plays an important role in the molecular basis of disease [4], there is an important impetus for SSO delivery, in particular, because nanoparticles delivering RNA may be able to elegantly target and correct the disease at the RNA level.
For binding to nanoparticle supports, the majority of work reported thus far has focused on DNA, not RNA. Typical modes of DNA binding involve (1) alkylthiol- or disulfide-terminated oligonucleotides on a metal nanoparticle surface, (2) covalent binding of oligonucleotides to a pre-activated nanoparticle surface, and (3) absorption of biotinylated oligonucleotides on surfaces coated with avidin or streptavidin [5–6]. Indeed, Seferos et al. [7] recently demonstrated that such oligoDNA-gold conjugates, termed “nanoflares,” can specifically detect mRNA. Thiol linkage to metal nanoparticles takes advantage of the strong coordination chemistry that exists between some metals and sulfur. Importantly, DNA-Au nanoparticles, functionalized with probe strands terminated in a triethylthiol group, were stable at higher temperatures and in the presence of reducing agents when compared to mono- or cyclic-dithiane species [5]. Furthermore, DNA-Au nanoparticles from the trithiol linker had higher DNA-probe surface coverages than those from mono-thiol or cyclic-dithiane species [6]. However, it is unclear currently whether or not these strategies can be applied to macromolecular DNA vaccine, siRNA, or SSO RNA analogs.
For DNA vaccines, one approach we have patented involves attaching DNA to the surface of gold microparticles via protamine for delivery by gene gun [8]. To enhance stability, several groups have reported that the attachment of proteins to gold nanoparticles affords the proteins resistance to temperature and protease degradation [9, 10]. In comparison to proteins, nucleic acids are considerably more susceptible to chemical and enzymatic degradation. Recently, we reported that protamine mediates attachment of DNA to gold microparticles and protects the DNA from nuclease degradation [11]. Herein our aim was to investigate whether or not Au-protamine-DNA conjugates enhance chemical stabilization of DNA as well; treating them for weeks to months against accelerated physical, chemical, and temperature degradation; and then testing their vaccine activity in mice.
In addition to its stabilization enhancing properties, protamine is well known for transporting macromolecules into cells. More importantly for this work, protamine is known to condense DNA into nano-structures [12–13] and to deliver antisense oligonucleotides as nanoparticles [14]. Protamine is known additionally to exhibit cell penetrating activity [15–16] and is an important component of several cancer targeting systems [17–18]. However protamine binding to siRNA or SSO RNA, and thus the formation of RNA:protamine nanoparticles and their delivery, has not been described to the best of our knowledge. Finally, we investigated the potential chemotherapeutic utility of these RNA and DNA nanoparticles and their effects on cultured human tumor cell and microbial cell growth.
Materials and Methods
Reagents and bio-molecules
Microparticle gold was obtained from DeGussa or from Bio-Rad. Protamine (clupeine) and all other reagents were obtained from Sigma-Aldrich. Plasmid DNA was obtained as previously reported [19]. Oligomer sequences (705 SSO) and siRNA (see below) were a generous gift from Professor Rudy Juliano, University of North Carolina-Chapel Hill, Chapel Hill, NC.
Nano-conjugation
Nanoparticles were prepared on a Cole-Parmer dual head Masterflex C/L (model #77120-62) pump at a constant rate with microfluidic inner diameter tygon tubing. Gold particles were included in some batches (3–35 mg batch sizes) mixed manually on a vortex at approximately 1 ml/sec by first adding protamine (0.01-2 mg/ml) followed sequentially by addition of DNA or RNA (0.01-0.1mg/ml). Thereafter, the particles were precipitated from 70% ethanol or stored in a 70% ethanol solution until use. In some cases, nanoparticles were separated by high speed density gradient centrifugation coupled to light scatter detection (University of North Carolina-Chapel Hill, Biophysical Instrumentation Core). Alternatively, nanoparticles were confirmed by dynamic laser light scatter spectroscopy (DLLS) in the laboratory of Professor Leaf Huang (University of North Carolina-Chapel Hill, Department of Molecular Therapeutics) or on a Malvern Zetasizer Nano-ZS90 DLLS from a suspension of water or phosphate-buffered saline.
Nano-characterization
UV/Vis and fluorescence spectroscopy of the nanoparticles and nucleic acids were conducted on Thermo Scientific NanoDrop™ instruments, either a NanoDrop 1000 Spectrophotometer or a NanoDrop 3300 Fluorospectrometer.
Stabilization
Open circle (OC) DNA standard was generated by treating supercoiled DNA with citrate/DMED [20]. Linear (Lin) DNA standard was generated by restriction digestion with XMa1 following the vendor’s recommended protocol. Both OC and Lin nucleic acid species were precipitated from ethanol (Spectrum UV grade) (70% alcohol/water), re-dissolved in 1x Tris/NaCl/EDTA (TNE buffer), and stored frozen until use. Anion-exchange HPLC chromatography was performed as described by Molloy et al. [21], except with the employment of a Toso-Haas column and a sodium chloride/aqueous gradient on a Varian Prostar system with DAD detector. Oxidation was accomplished in 0.1% H2O2. Two week and one, two, or three month accelerated degradation treatments were conducted in standard cell culture incubators at 5% CO2 in the presence of dessicant at room temperature or 40°C or, in some cases, 60°C. Gamma irradiation treatment was executed by standard protocol (Sterigenics, Inc., NC). For autoclave treatments, particles enclosed in sealed Eppendorf microfuge tubes were exposed to 30 minute sterilization cycles “dry.” Particles (2–3 mg) exposed to accelerated degradation were then eluted in 1x TNE containing 0.1% SDS for 30 minutes at 37°C. Ten microliter aliquots were loaded in 1x load buffer (blue juice, Gibco-BRL) and electrophoresced through 1% agarose gels containing 0.5 µg/ml ethidium bromide for 45 minutes at 107 volts and 440 mA in a Horizon Life technologies gel system running in 1x TBE (Gibco BRL), and the gels were visualized on a gel documentation station (Syngene, Genetools).
Hela 705 splice-site shift assay, siRNA and DNA vaccine biological activity
HeLa 705 cells [32] were seeded into 96-well plates (5,000 cells/well) and allowed to attach overnight. Particles (100µL), containing 0.1–1 mg/ml protamine and 150 nM of either 705 sequence [3, 22] or anti-B-Raf siRNA sequence (5’-AUGAUCCAGAUCCAAUUCUdTdT-3’), were added to the well in a total volume of 200–300 µL Opti-Mem (Gibco-BRL). The medium was removed 6–8 hours later, 300 µL 10% FBS/DMEM (without indicator) was added, and the cells were allowed to grow for an additional 48 hours.
After this time the medium was aspirated, the cells washed twice with PBS buffer and then cell lysis buffer, luciferase kit reagents were added, and luminescence assayed (ProMega, Madison, WI). For B-Raf, A375 cultured melanoma cells were treated with anti-B-Raf siRNA sequence and B-Raf assayed by western blot according to our previously published procedure for MDR [23]. DNA vaccine studies were conducted as previously described [19, 24]. Briefly, HBV plasmid (pdp-SC18) was formulated onto gold particles with a 2 µg pDNA/mg gold target load. Mice were primed with a single dose of DNA vaccine by gene gun onto the shaved skin on the abdomen following PowderJect IACUC protocol under the direction of Dr. Lendon Payne. Two weeks later, blood was collected for hepatitis surface and core antigen ELISA. In some cases, Au-protamine-DNA or Au-spermidine-DNA controls were incubated at 4, 25, 40, or 60°C prior to gene gun administration. Student’s t-test was used for statistical analysis of the data; statistically significant differences were assumed at p < 0.05.
Microbiology, cell growth and morphology assays
From a slightly turbid culture of E. coli 10HB grown for approximately 6 hr at 30°C, a well-mixed 2 µL aliquot was transferred into a sterile glass tube containing 60 µL of nanosuspension formed just previously by the 1:1 vol/vol addition of protamine (5 mg/ml) to tRNA (1 mg/ml) or M9 media alone controls. The suspension of nanoparticles and E. coli cells or controls without nanoparticles were then brought up to 3 ml total volume with M9 minimal medium, mixed, and either 2 or 20 µL inocula volumes were placed onto sterile LB plates and grown overnight. Suspension cultures were grown for 5, 30, 60, 120, or 240 minutes in the presence of nanoparticles normalized to 10, 3, 1, or 0.3 mg/ml dosages or parallel controls of protamine, tRNA, or BSA. LB plates were then inoculated, grown overnight, and the colonies counted. A growth culture was prepared, 200 µL was spread onto an LB plate, and the plate was allowed to dry in a sterile air-flow hood for 10–15 minutes. A serial dilution series of tRNA:protamine nanoparticles, tRNA (1 mg/ml), protamine (5 mg/ml), or BSA (5 mg/ml) controls were spotted in 3 µL spots onto the pre-spread and dried LB plate and allowed to dry for an additional 15–20 min; subsequently, the cultures were allowed to incubate overnight. Live/dead cell assay was performed per manufacturer’s instructions (Invitrogen Corp.) with Nikon TE2000-E optics coupled to a Bio-Rad Radiance 2100 MP Confocal Laser. HeLa 705 cells were plated onto a 96-well plate in DMEM with 5% FBS and grown to 80% confluency. These cells were treated with tRNA (0.1 mg/ml), protamine (1 mg/ml), and the tRNA/protamine complexes (1: 1 v/v). The cells were incubated for 6 hours at 37°C. After 6 hours, the medium was removed from the wells. 100 µl of PBS were added to the wells with 100 µl of Trypan Blue. The cells were mixed and stained for 5–15 minutes. The stain was removed from the cells, and the cells were washed with PBS. Images of the cells were taken using an Olympus 1×70 inverted brightfield microscope. In some cases, yeast cells were treated and imaged similarly.
Results
Characterization of DNA binding to gold microparticles
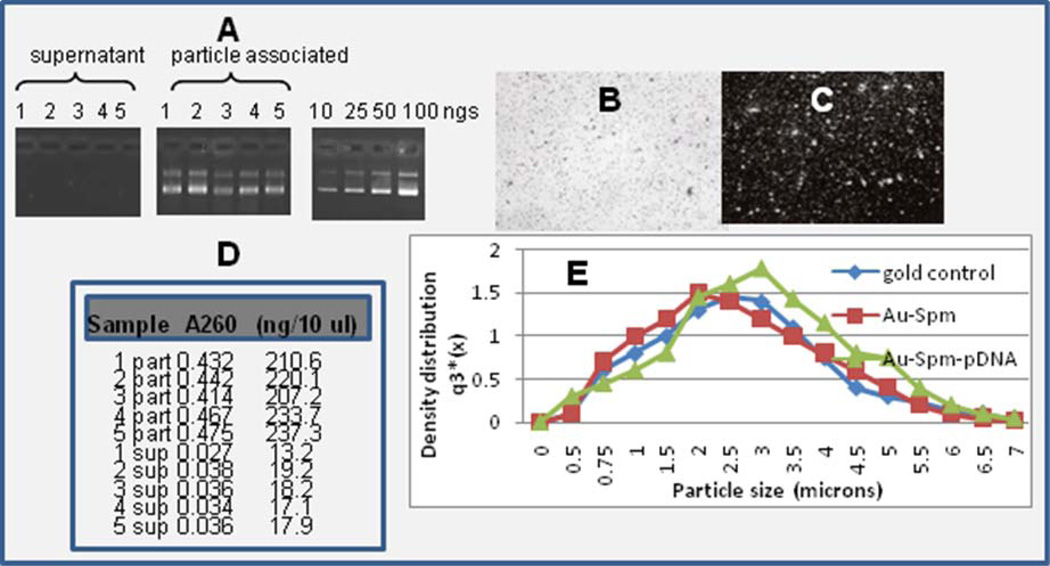
Proof of DNA attachment to gold is shown in Fig.1. Particles were prepared with DNA attached to gold [11, 19], and the supernatant or particle-associated fraction (eluted from the particles) was run by gel electrophoresis (panel A). In comparison to internal load standards, all five batches appeared to have 25 to 50 nanograms of DNA loaded per 0.5 mg of gold eluted. Particles were shot into agar gels by a gene gun in the absence (panel B) or presence (panel C) of ethidium dye and a non-fluorescence or fluorescence pattern photographed, revealing the particulate pattern of stained DNA (panel C). Alternatively, a series of five samples were prepared, and the supernatant (sup) or particle-associated fractions (part) were assayed by UV or fluorescence spectroscopy (panel D), indicating a loading efficiency of greater than 90%. Dynamic laser light scatter (DLLS) showed a slight right shift, indicative of larger particles, when DNA was bound by the standard spermidine approach [8, 11, 19].
Fig. 1.
Plasmid DNA isolated from the supernatant or particle-associated fractions (n=5) were assayed by gel electrophoresis relative to a set of load standards at 10, 25, 50, and 100 nanograms per lane (panel A). Gene gun delivery of 1 mg DNA-loaded gold into agar gels in the absence (panel B) or presence (panel C) of ethidium incorporation and UV visualization and photography. Five supernatant (sup) and five particle-associated (part) samples assayed by Hoechst fluorescence for DNA concentration in the nanogram range in comparison to A260 values (panel D). Dynamic laser light scatter analysis of gold microparticles with spermidine and spermidine-DNA (panel E).
Characterization of plasmid DNA vaccine delivery
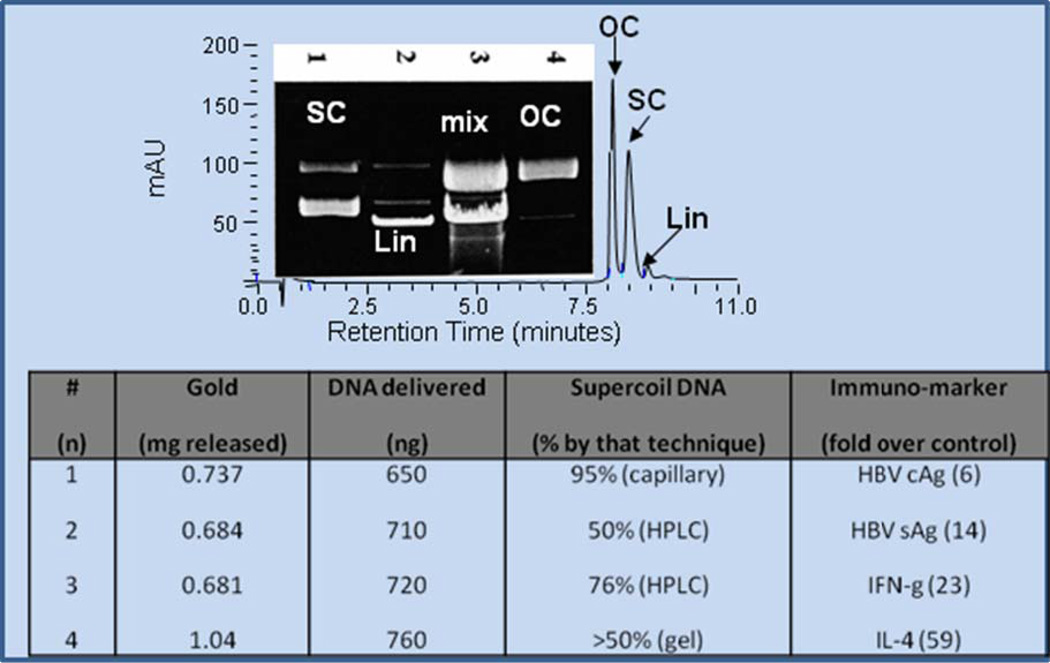
To generate standards, plasmid DNA in solution was degraded rapidly to open circle and linear forms on a time-scale of seconds to minutes after exposure to enzyme (lane 2), oxidative (lane 3), or hydrolytic treatments (lane 4) as shown in Fig. 2A. These degradation products could be base-line resolved with careful analysis by gel or anion exchange HPLC [31] (Fig. 2A).
Fig. 2.
Supercoiled DNA at approximately 95% purity was control-degraded with XMaI restriction endonuclease to generate linear forms (lane 2), oxidized in 0.1% H2O2 to generate a mixture of linear and open circle forms (lane 3), or treated with citrate/DMED[29] to generate stable open circles (lane 4). These samples were used as standards for electrophoresis or HPLC (panel A). Panel B represents four gene gun samples loaded with approximately one microgram of DNA bound onto gold (~1 mg) that were actuated into buffer, the amount of gold delivered determined by weighing the cartridge afterward, and DNA in the nano-range determined by Hoechst assay [24]. Percent supercoil of the DNA after expulsion from the gene gun was assayed by capillary, HPLC, and gel electrophoresis, and base-line immunogenicity determined in mice by assaying immuno-titer of HBV cAg, sAg, IFN-gamma, and IL-4 [8, 25–26].
Plasmid DNA vaccine encoding Hepatitis B core (cAg) and surface antigen (sAg) was bound to gold, and delivery was assayed by loss of gold from the gold cartridge after actuating the gene gun, and the DNA delivered was assayed by fluorescence spectroscopy [24], gel or capillary electrophoresis and HPLC as shown in Fig. 2B. After actuation from the gene gun, the DNA propelled into buffer retained 50–95% integrity, and when tested in mice, exhibited robust immunogenicity demonstrated by ELISA analysis of HBV cAg, sAg, interferon gamma, or interleukin-4 as standard measures [25–26].
Stabilization of DNA vaccine by attachment to gold via protamine
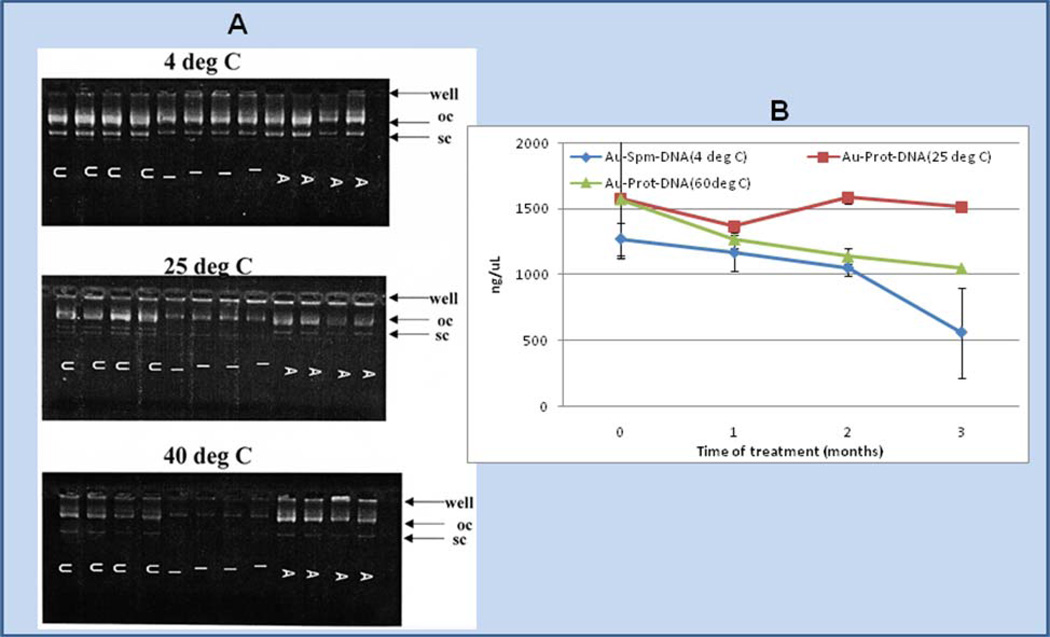
Nucleic acids are quite susceptible to both chemical and enzymatic degradation. During the process of preparing conjugates of DNA attached to microparticle gold, vortex and sonication steps physically contribute to DNA degradation. Thus (as shown in Fig. 3A), even untreated DNA, isolated from gold after attachment via spermidine, has lost a substantial amount of supercoil (shown in the wells labeled “U”). Further conversion from supercoiled to open circle forms occurs upon exposure of the particles to standard methods of sterilization, such as gamma irradiation (I) or autoclaving (A), prior to administration into cell cultures and to animals. Under these conditions exposure of the powders to higher temperatures for two weeks, prior to removing the DNA from the particles and assaying its integrity, accelerates degradation to the open circle form even though the Au-spermidine-DNA particles are dried from alcohol— presumably lessening hydrolysis by removing water. In contrast, for DNA conjugates prepared by protamine-ligation to gold, the DNA levels remained relatively constant after exposure to 25°C or 60°C assayed by Hoechst binding fluorescence analysis [24] for up to 2–3 months (Fig. 3B).
Fig. 3.
Au-spermidine-DNA or Au-protamine-DNA particles, prepared in a 35 mg standard amount [8, 25] were vortex-mixed, sonicated and dried from alcohol, and aliquots of the powders exposed to standard cycles of gamma irradiation (I) and autoclave sterilization (A) as described in the materials and methods; lanes containing untreated DNA are indicated by “U”. The particles were then subjected to exposure at 4 °C, 25 °C, 40 °C, and 60 °C for 2 weeks (panel A) or 1, 2, or 3 months (panel B). Afterwards, the DNA was isolated from 0.5 mg aliquots of the particles with elution buffer and samples were assayed by gel electrophoresis or Hoechst assay as shown above. Supercoiled DNA was completely lost and open circle band staining intensity diminished for Au-spermidine-DNA as shown by gel; however, more DNA was stained by Hoechst assay for Au-protamine-DNA at 25 °C and 60 °C in comparison to Au-spermidine-DNA stored at 4 °C (panel B).
Vaccine activity of Au-protamine-DNA exposed to accelerated degradation
Recently, we reported that attachment of DNA onto gold via protamine protected the DNA from nuclease degradation, prolonging its half-life in plant cell extracts and increasing its expression level and duration [11]. Maintenance of intact supercoil type plasmid is a hallmark stability indicator for pharmaceutical DNA [27]. Conjugation of proteins to nano-gold is similarly known to protect against proteases and, more importantly, guards against physico-chemical and temperature-accelerated degradation [9–10]. Clearly DNA structure is susceptible to heat, physical, and chemical degradation as well which is expected to effect the stability and activity of DNA vaccines. Notably, as shown above in Fig.3, temperature accelerates the physical and chemical degradation and the bands representing supercoil or open circle forms are lost or their fluorescence is greatly diminished.
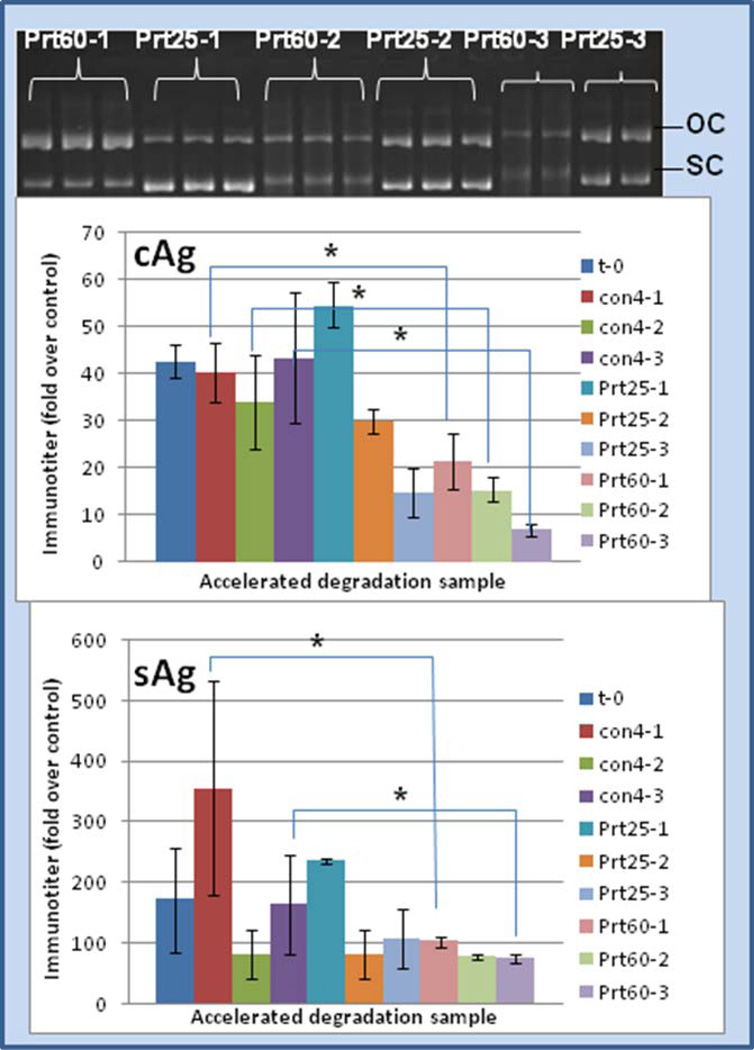
Here, in contrast and comparison to the above data, protamine greatly stabilized the DNA attached to gold. Thus, after exposing the Au:protamine:DNA ternary complexes to a 1, 2, or 3 month temperature-accelerated forced degradation at 25°C or 60°C, the DNA retained open circle and, to a lesser extent, supercoil form as shown by gel analysis (Fig. 4). More importantly, the DNA vaccine immunogenicity was retained even after exposure to 25 or 60 °C for several months as shown in Fig. 4. Thus, Au-protamine-DNA vaccine samples, aged for several months at 25°C or 60°C, retained Hepatitus B core (cAg) or surface antigen (sAg) activity comparable to a matched set of Au-spermidine-DNA samples stored at 4°C.
Fig. 4.
Au-spermidine-DNA stored at 4°C or Au-protamine-DNA particles stored at 25°C or 60°C for 1, 2, or 3 months and then administered to mice as aged vaccine. Y-axis represents immuno-titer of Hepatitis B core antigen “cAg” (middle panel) or surface antigen “sAg” (bottom panel) relative to animals treated with control (standard HepB protein vaccine) [8, 25–26]. Protamine samples significantly different than their corresponding matched set of spermidine controls by Students t-test statistic are indicated with an asterisk (p < 0.050). Gel analysis (top panel) of these time and temperature treatments revealed retention of substantial intact supercoiled and open circle DNA forms for several months. Legend: OC = open coil DNA; SC = supercoiled DNA; t-0 = non-degraded sample; con4-1 = control at 4°C for 1 month; con4-2 = control at 4°C for 2 months; con4-3 = control at 4°C for 3 months; Prt25-1 = protamine at 25°C for 1 month; Prt25-2 = protamine at 25°C for 2 months; Prt25-3 = protamine at 25°C for 3 months; Prt60-1 = protamine at 60°C for 1 month; Prt60-2 = protamine at 60°C for 2 months; Prt60-3 = protamine at 60°C for 3 months.
Delivery of siRNA and SSO without gold and gene gun
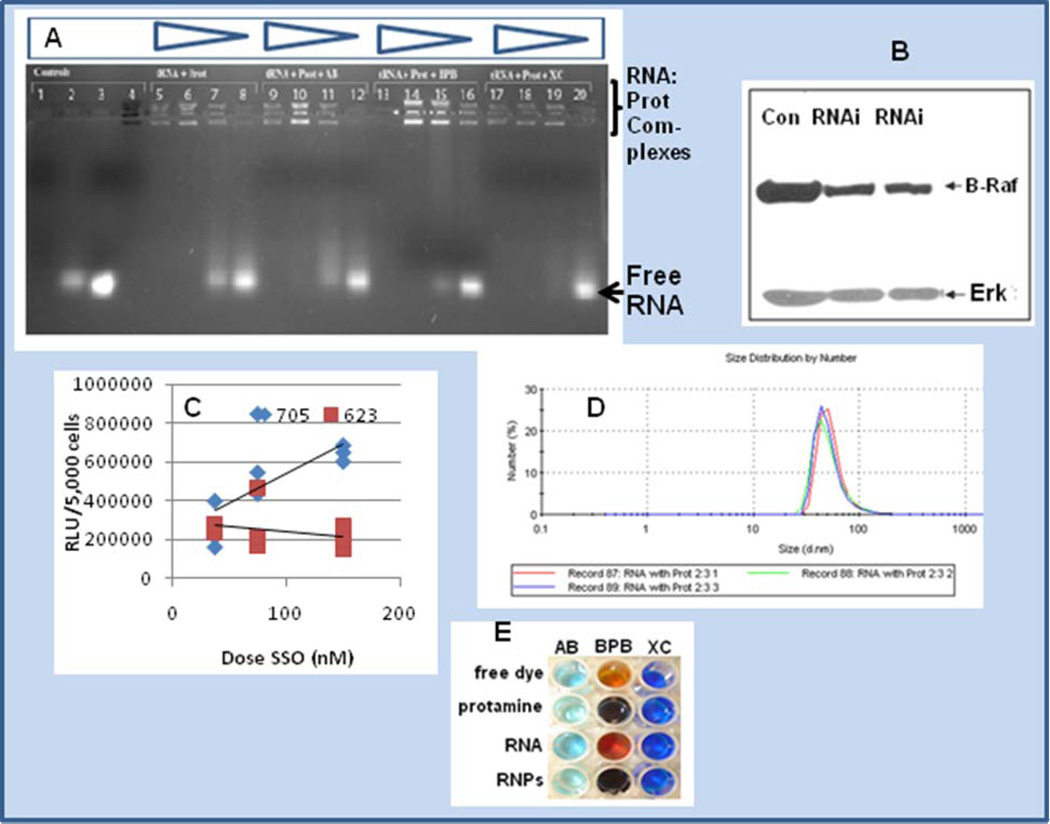
Despite its advantages as shown above, gene gun delivery of nucleic acids supported by microparticle gold has its limitations [28]. Protamine itself, on the other hand, has membrane translocating activity and is capable of macromolecular transport [15–16]. Further, protamine complexes oligonucleotides to form nanoparticles that have been shown to deliver antisense oligonucleotides into cells [14]. As shown in Fig. 5A, protamine binds RNA, producing a gel shift with RNA where the complex is retained in the well. Interestingly, multiple bands were observed in the well, perhaps indicative of isoforms of protamine [16]. The presence of amido black, which is known to stain protamine [29] as well as bromophenol blue and to a lesser extent xylene cyanol, appeared to impact the staining intensity of the complexes in the wells and putative RNA:protamine nanoparticles.
Fig. 5.
Panel A is a gel shift experiment of RNA:protamine in the presence of amido black, bromophenol blue and xylene cyanol. Staining of the multiple isoforms of protamine is shown in lane 4 containing amido black. Staining of the protamine:RNA complex in decreasing quantities of protamine is shown in lanes 5–8 (left to right, 0.5 mg/ml , 0.1 mg/ml, 0.05 mg/ml and 0.03 mg/ml). Lanes 9–12, 13–16, and 17–20 represent a matched set compared to lanes 5–8 with the inclusion of dyes amido black (lanes 9–12), bromophenol blue (lanes 13–16) or xylene cyanol (lanes 17–20), respectively. Panel B is a western blot of A375 human melanoma cells treated for 48 hrs with siRNA delivered as a protamine complex (RNAi lanes) or untreated controls (Con). Inhibition is shown, and Erk is blotted as a comparator. Panel C is a dose response experiment comparing SSO 705 and SSO 623 sequences for their ability to splice-site switch and up-regulate luciferase expression in the HeLa 705 pLuc system [3, 22, 33]. Panel D represents protamine-RNA complexes of approximate 50 nm size (n = 3 runs), where complexes were formed with vol/vol addition of protamine (1 mg/ml) and tRNA (0.1 mg/ml). Panel E represents chromophoric shift of amido black (AB), bromophenol blue (BPB) and xylene cyanol (XC) in the absence or presence of RNPs (RNA:protamine nanoparticles).
To facilitate siRNA entry into cancer cells, investigators have fused protamine to Fab antibody [17]. Furthermore, LPD nanoparticles containing protamine have been shown to target siRNA to tumors [18]. Although cancer evolution is complicated, and it is difficult to identify a specific single gene to target with siRNA-bearing RNPs, B-Raf and the MEK-Erk pathway it regulates have received recent attention for their importance as a molecular target in cancer [30–31]. B-Raf is alternatively spliced [32] and is involved in the regulation of cell growth, cell-cycle control, and programmed cell death. Therefore, we reasoned that it was logical to target B-Raf with RNA-bearing nanoparticles (RNP) bearing anti-B-Raf siRNA, and we were able to achieve B-Raf knock-down relative to untreated cells as shown in Fig. 5B. RNPs were also tested in combination with splice-site switching oligonucleotides (SSO) in the well-described HeLa tumor cell 705 system; delivery of an SSO overlapping an aberrant splice-site corrects splicing and switches-on luciferase [3, 22]. Two known SSO sequences, 705 and 623, were compared in a dose-response study, and these data are shown in Fig. 5C. As recently reported, treatment with the SSO 705 sequence versus the alternative SSO sequence 623 [33] yielded a higher dose response. Protamine complexation to RNA forms nanoparticles as detected directly by dynamic laser light scatter spectroscopy as shown in Fig. 5D. The RNA:protamine nanoparticles (RNPs) were probed by chromophoric shift assay in the presence of amido black, bromophenol blue, and xylene cyanol. In each case the presence of the RNP provided a color change indicative of nanoparticle staining (Fig. 5E).
Microbial and tumor cell growth inhibition
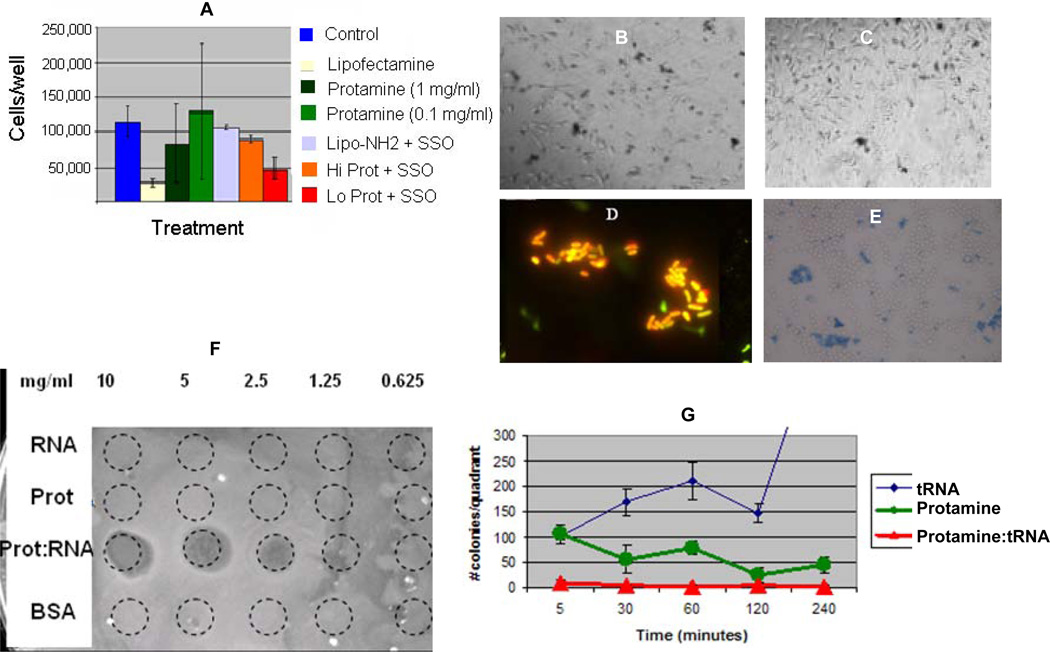
Consistent with protamine’s well-known membrane traversal [15–16], the data above suggest that protamine is capable of conveying RNA into the cells, and that the RNA exhibits functionality afterwards, either by siRNA activity or splice-site switching. Thus, we examined next the effect of the RNPs on cell growth and cytotoxicity. Little growth inhibition and almost no killing were evident for treatment of HeLa cells with equimolar concentrations of RNPs or controls (Fig. 6A-6C). In marked contrast however, trypan blue viability assays demonstrated approximately 30–40% cell killing, when these RNPs were incubated with yeast, and almost 100% killing of E. coli as shown by light or fluorescence microscopy (Fig. 6D, 6E). Plaque or zone of killing and colony formation assays revealed the antibacterial activity of the RNPs (Fig. F, G). Protamine itself belongs to a class of cationic antimicrobial peptides [34–35], yet the nanoparticles themselves appear to exhibit antimicrobial properties as has been shown for other nanoparticles [36–37].
Fig. 6.
Panel A shows the effects of lipofectamine, protamine (Hi = 1 mg/ml, Lo = 0.1 mg/ml), or their complexes with SSO 705 sequence on the number of HeLa cells after a 48-hr incubation as counted by Coulter counting in a 48 well plate assay. Panels B and C represent comparable treatments of HeLa cells with protamine:RNA complexes at the high or low levels and stained afterwards with trypan blue; dark cells represent dead or dying cells indicating low toxicity. In contrast, Panels D and E represent E. coli bacteria or yeast treated with RNA:protamine RNPs stained with live/dead fluorescent stain (Invitrogen) or trypan blue, respectively, and imaged by light or fluorescence microscopy; yellow or blue cells reveal dead or dying cells, indicating more overt toxicity for bacteria or yeast. Panel F represents a plaque formation assay wherein cell death was induced by RNP treatment at the various mass/volume inocula versus protamine, tRNA, or BSA controls. Panel G is a colony formation assay of E. coli colonies counted after treatment with RNPs or controls as described in the methods section, demonstrating that colony formation was completely inhibited by protamine:RNA nanoparticles (green line) or partially inhibited by protamine (red line) versus untreated bacteria (blue line).
Discussion
In summary, we have formed and characterized DNA- and RNA-based nanoparticles for stabilization and delivery of DNA vaccine, siRNA, and splice-site switching oligomers (SSOs) targeted towards cancer and infectious disease. Attachment of DNA vaccine to gold via protamine appears to stabilize the DNA. These Au-protamine-DNA conjugates provide the DNA resistance to physico-chemical and temperature-accelerated degradation and prolong the DNA vaccine activity. Simple protamine complexed with siRNA and SSO provide for RNA delivery activity, and these RNA nanoparticles (RNPs) appear to have differential cell growth and inhibitory effects on human and microbial cells, thus supporting their chemotherapeutic potential.
In our studies, attachment of DNA to gold by protamine enhanced DNA resistance to physico-chemical degradation. Poly L-lysine (PLL) has been similarly shown to condense and stabilize plasmid DNA [38]. Interaction of protamine with tRNA was first reported by X-ray diffraction and circular dichroism [39]; however, here we use the more novel DLLS and nano-probe UV/Vis approaches, in addition to gel shift, as a means to confirm these RNA:protamine nanoparticles. Gold is relatively inert and as such has been the focus of much work to date; however, other inorganic nanoparticles may be better substrates for nucleic acid binding [28,40]. Furthermore, although our nanoparticles are biologically-based (formed with RNA and protamine), other nanomaterials, such as iron oxide [41] or silica-based materials [42], may potentially be useful for the binding, stabilization, and delivery of DNA and RNA.
Protamine is an incorporated component in tumor targeting [17–18] and is known to facilitate membrane transversal [15–16], observations that are consistent with the data shown here for protamine:RNA nanoparticles associated with delivery of siRNA and splice-site switching oligomers (SSO) and up-regulating luciferase gene expression in the HeLa-705 engineered splicing system. Furthermore, we show that protamine mediates the plasmid DNA vaccine attachment to gold microparticles. Recently, a similar approach has demonstrated that conjugation of DNA vaccine to nanoparticle gold by chitosan increases serum antibody titer and cytotoxic T cell response [26]. Polylactide co-glycolide (PLGA) nanoparticles have been shown to incorporate DNA and increase uptake into macrophages [43]; therefore, it may be of some interest to combine these systems into the nano-range in the near future.
In terms of the biochemical effects of RNPs, we observed that the dyes in the gel shift, particularly bromophenol blue (BPB), displace the RNA:protamine complexes. Although this method is indirect, it constitutes a putatively novel approach for probing these types of nanoparticles and perhaps others. For example, BPB and Coomassie Blue can probe hydrophobic sites in proteins [44]; therefore, other chromophoric and fluorometric characterizations of the RNPs may be possible [45–46].
Biologically, the RNPs appeared to elicit distinctive cytotoxic effects in bacteria and yeast in comparison to the HeLa cells. In this case, the RNA:protamine particles were simply formed by complexation of protamine to transfer RNA (tRNAphe). Neither the protamine itself nor the tRNA gave these results, suggesting that there are unique properties of these nanoparticles. RNA processing, such as splicing, is quite different in eukaryotic versus prokaryotic organisms. Therefore, it is possible that RNPs bearing RNA molecules designed for RNA-level intervention may exhibit mechanistic differences that could be crucial for chemotherapeutic selectivity.
Conclusions
DNA and RNA nanoparticles may be useful in the binding, stabilization, and delivery of DNA vaccines, siRNA, and splice-site switching oligomers. As shown here, one such approach is through formation of DNA or RNA nanoparticles by complexation of the nucleic acid with protamine and its ensuing attachment to gold. We have demonstrated that these Au-protamine-DNA conjugates greatly resist physico-chemical destruction and retain DNA vaccine structure and function. In addition to DNA processing, stability, and delivery advantage, RNA:protamine nanoparticles (RNPs) provide for delivery of siRNA and inhibit B-Raf, an important cancer target. The RNPs are capable of delivering splice-site switching oligomers into human tumor cells, with subsequent up-regulation of luciferase marker gene expression under splice-site switching control. The RNPs do not appear to inhibit HeLa cell growth significantly, yet appear to be selectively inhibitory to bacteria and, to a lesser extent, yeast. These observations imply promising chemotherapeutic potential.
Acknowledgements
We are grateful to Dr. Rudy Juliano for his mentorship and support over the years and for the provision of the siRNA and 705 SSO sequences, to Chris R. Lively for his insights on measuring DNA bound onto gold, and to S. Grizzard, D. Davis and K. Grozinger for their preliminary experiments on RNA nanoparticles. HeLa 705 cells were kindly provided by Ryszard Kole. We thank Ms. Nancy Maxfield for critically reviewing and editing this manuscript.
Funding sources for this work: This work was supported by a research initiation grant to R.K.DeLong and an Howard Hughes Medical Institute Summer Undergraduate Research Fellowship to R.K. DeLong and E. Engstrom.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9(10):776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13(24):3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abes R, Arzumanov A, Moulton H, Abes S, Ivanova G, Gait MJ, et al. Arginine-rich cell penetrating peptides: design, structure-activity, and applications to alter pre-mRNA splicing by steric-block oligonucleotides. J Pept Sci. 2008;14(4):455–460. doi: 10.1002/psc.979. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22(5):535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Jin R, Merkin CA, Letsinger RL. Multiple thiol-anchor capped DNA–gold nanoparticle conjugates. Nucleic Acids Res. 2002;Vol. 30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R, et al. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Analytical Chemistry. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 7.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lively CR, DeLong RK. Inventors: Europe. Nucleic acid coated particles. Applicant: POWDERJECT RES LTD (GB), EC:A61K9/16K2; A61K48/00B IPC: A61K9/16; A61K48/00; A61K9/16 (+4) Publication info: EP1545593 A1 - 2005-06-29. [Google Scholar]
- 9.Phadtare S, Vinod VP, Mukhopadhyay K, Kumar A, Rao M, Chaudhari RV, et al. Immobilization and biocatalytic activity of fungal protease on gold nanoparticle-loaded zeolite microspheres. Biotech and Bioengineering. 2004;85:629–637. doi: 10.1002/bit.10856. [DOI] [PubMed] [Google Scholar]
- 10.Asuri P, Karajanagi SS, Vertegel AA, Dordick JS, Kane RS. Enhanced stability of enzymes adsorbed onto nanoparticles. J Nanosci Nanotechnol. 2007;7(4–5):1675–1678. doi: 10.1166/jnn.2007.453. [DOI] [PubMed] [Google Scholar]
- 11.Sivamani E, DeLong RK, Qu R. Protamine-mediated DNA coating remarkably improves bombardment transformation efficiency in plant cells. Plant Cell Rep. 2009;28(2):213–221. doi: 10.1007/s00299-008-0636-4. Epub 2008, Nov 18. [DOI] [PubMed] [Google Scholar]
- 12.Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286(5437):120–123. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- 13.Brewer L, Corzett M, Balhorn R. Condensation of DNA by spermatid basic nuclear proteins. J Biol Chem. 2002;277(41):38895–38900. doi: 10.1074/jbc.M204755200. [DOI] [PubMed] [Google Scholar]
- 14.Mayer G, Vogel V, Weyermann J, Lochmann D, van den Broek JA, Tziatzios C, et al. Oligonucleotide-protamine-albumin nanoparticles: Protamine sulfate causes drastic size reduction. J Control Release. 2005;106(1–2):181–187. doi: 10.1016/j.jconrel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Rhee M, Davis P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J Biol Chem. 2006;281(2):1233–1240. doi: 10.1074/jbc.M509813200. Epub 2005, Nov 4. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds F, Weissleder R, Josephson L. Protamine as an efficient membrane-translocating peptide. Bioconjug Chem. 2005;16(5):1240–1245. doi: 10.1021/bc0501451. [DOI] [PubMed] [Google Scholar]
- 17.Williams BR. Targeting specific cell types with silencing RNA. N Engl J Med. 2005;353(13):1410–1411. doi: 10.1056/NEJMcibr052492. [DOI] [PubMed] [Google Scholar]
- 18.Li SD, Huang L. Surface-modified LPD nanoparticles for tumor targeting. Ann N Y Acad Sci. 2006;1082:1–8. doi: 10.1196/annals.1348.001. [DOI] [PubMed] [Google Scholar]
- 19.Medberry P, Dennis S, Van Hecke T, DeLong RK. pDNA bioparticles: comparative heterogeneity, surface, binding, and activity analyses. Biochem Biophys Res Commun. 2004;319(2):426–432. doi: 10.1016/j.bbrc.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 20.Poxon SW, Hughes JA. A biofunctional assay to study pRL-CMV plasmid DNA formulation stability. PDA J Pharm Sci Technol. 1999;53(6):314–317. [PubMed] [Google Scholar]
- 21.Molloy MJ, Hall VS, Bailey SI, Griffin KJ, Faulkner J, Uden M. Effective and robust plasmid topology analysis and the subsequent characterization of the plasmid isoforms thereby observed. Nucleic Acids Res. 2004;32(16):e129. doi: 10.1093/nar/gnh124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang SH, Cho MJ, Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37(18):6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- 23.Alahari SK, DeLong R, Fisher MH, Dean NM, Viliet P, Juliano RL. Novel chemically modified oligonucleotides provide potent inhibition of P-glycoprotein expression. J Pharmacol Exp Ther. 1998;286(1):419–428. [PubMed] [Google Scholar]
- 24.Latt SA. Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proc Natl Acad Sci U S A. 1973;70(12):3395–3399. doi: 10.1073/pnas.70.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowle R, Werner A, DeLong RK. R4 peptide-pDNA nanoparticle coated HepB vaccine microparticles: sedimentation, partitioning, and spray freeze dry bioprocesses. J Nanosci Nanotechnol. 2006;6(9–10):2783–2789. doi: 10.1166/jnn.2006.427. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Zhang X, Yu X, Zha X, Fu Q, Liu B, et al. The effect of conjugation to gold nanoparticles on the ability of low molecular weight chitosan to transfer DNA vaccine. Biomaterials. 2008;29(1):111–117. doi: 10.1016/j.biomaterials.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Middaugh CR, Evans RK, Montgomery DL, Casimiro DR. Analysis of plasmid DNA from a pharmaceutical perspective. J Pharm Sci. 1998;87(2):130–146. doi: 10.1021/js970367a. Review. [DOI] [PubMed] [Google Scholar]
- 28.Sokolova V, Epple M. Inorganic nanoparticles as carriers of nucleic acids into cells. Angew Chem Int Ed Engl. 2008;47(8):1382–1395. doi: 10.1002/anie.200703039. Review. [DOI] [PubMed] [Google Scholar]
- 29.Tsai JT, Furstoss KJ, Michnick T, Sloane DL, Paul RW. Quantitative physical characterization of lipid-polycation-DNA lipopolplexes. Biotechnol Appl Biochem. 2002;36(Pt 1):13–20. doi: 10.1042/ba20020010. [DOI] [PubMed] [Google Scholar]
- 30.King AJ, Patrick DR, Batorsky RS, Ho ML, Do HT, Zhang SY, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66(23):11100–11105. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 31.Madhunapantula SV, Robertson GP. Is B-Raf a good therapeutic target for melanoma and other malignancies? Cancer Res. 2008;68(1):5–8. doi: 10.1158/0008-5472.CAN-07-2038. Review. [DOI] [PubMed] [Google Scholar]
- 32.Hmitou I, Druillennec S, Valluet A, Peyssonnaux C, Eychène A. Differential regulation of B-raf isoforms by phosphorylation and autoinhibitory mechanisms. Mol Cell Biol. 2007;27(1):31–43. doi: 10.1128/MCB.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alam MR, Dixit V, Kang H, Li ZB, Chen X, Trejo J, et al. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36(8):2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen LT, Gill TA. Solubility and antimicrobial efficacy of protamine on Listeria monocytogenes and Escherichia coli as influenced by pH. J Appl Microbiol. 2000;88(6):1049–1055. doi: 10.1046/j.1365-2672.2000.01074.x. [DOI] [PubMed] [Google Scholar]
- 35.Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. 1998;180(15):4002–4006. doi: 10.1128/jb.180.15.4002-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Z, Neoh KG, Kang ET, Wang W. Antibacterial and mechanical properties of bone cement impregnated with chitosan nanoparticles. Biomaterials. 2006;27(11):2440–2449. doi: 10.1016/j.biomaterials.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 37.Kim JS, Kuk E, Yu KN, Kim JH, Park SJ, Lee HJ, et al. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;1:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. Stability of peptide-condensed plasmid DNA formulations. J Pharm Sci. 1998;87(6):678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 39.Warrant RW, Kim SH. alpha-Helix-double helix interaction shown in the structure of a protamine-transfer RNA complex and a nucleoprotamine model. Nature. 1978;271(5641):130–135. doi: 10.1038/271130a0. 298, 982-6. [DOI] [PubMed] [Google Scholar]
- 40.Kim SH, Shin WC, Warrant RW. Heavy metal ion-nucleic acid interaction. Methods Enzymol. 1985;114:156–167. doi: 10.1016/0076-6879(85)14016-4. [DOI] [PubMed] [Google Scholar]
- 41.Butoescu N, Jordan O, Petri-Fink A, Hofmann H, Doelker E. Co-encapsulation of dexamethasone 21-acetate and SPIONs into biodegradable polymeric microparticles designed for intra-articular delivery. J Microencapsul. 2008;25(5):339–350. doi: 10.1080/02652040801999551. [DOI] [PubMed] [Google Scholar]
- 42.Thomas JC, Pacholski C, Sailor MJ. Delivery of nanogram payloads using magnetic porous silicon microcarriers. Lab Chip. 2006;6(6):782–787. doi: 10.1039/b516121e. [DOI] [PubMed] [Google Scholar]
- 43.Mok H, Park TG. Direct plasmid DNA encapsulation within PLGA nanospheres by single oil-in-water emulsion method. Eur J Pharm Biopharm. 2008 Jan;68(1):105–111. doi: 10.1016/j.ejpb.2007.04.022. Epub 2007 Jul 18. [DOI] [PubMed] [Google Scholar]
- 44.Bertsch M, Mayburd AL, Kassner RJ. The identification of hydrophobic sites on the surface of proteins using absorption difference spectroscopy of bromophenol blue. Anal Biochem. 2003;313(2):187–195. doi: 10.1016/s0003-2697(02)00590-0. [DOI] [PubMed] [Google Scholar]
- 45.Shcharbin D, Klajnert B, Mazhul V, Bryszewska M. Dendrimer interactions with hydrophobic fluorescent probes and human serum albumin. J Fluoresc. 2005;15(1):21–28. doi: 10.1007/s10895-005-0209-7. [DOI] [PubMed] [Google Scholar]
- 46.Sharma A, Rao M, Miller R, Desai A. Fluorometric assay for detection and quantitation of polyamidoamine dendrimers. Anal Biochem. 2005;344(1):70–75. doi: 10.1016/j.ab.2005.06.025. [DOI] [PubMed] [Google Scholar]