Abstract
Synpolydactyly 1 (SPD1; OMIM 186000), also known as type II syndactyly, is a dominantly inherited limb malformation that is characterized by an increased number of digits. SPD1 is most commonly caused by polyalanine repeat expansions in the coding region of the HOXD13 gene, which are believed to show a dominant-negative effect. In addition, missense and out-of-frame deletion mutations in the HOXD13 gene are also known to cause SPD, and the mechanism responsible for the phenotype appears to be haploinsufficiency. Here, we analyzed a large consanguineous family from Pakistan with SPD showing a wide variation in phenotype among affected individuals. We performed genetic linkage analysis, which identified a region on chromosome 2 containing the HOXD13 gene. Haplotype analysis with microsatellite markers suggested segregation of the phenotype with HOXD13 gene with incomplete penetrance. Direct sequencing analysis of HOXD13 gene revealed a nonsense mutation, designated Q248X. All affected individuals with the severe SPD phenotype are homozygous for the mutation, while those with the mild SPD phenotype are heterozygous for the mutation. Furthermore, some unaffected individuals also carry the mutation in the heterozygous state, showing incomplete penetrance. Our results demonstrate the first nonsense mutation in the HOXD13 gene underlying a severe form of SPD in the homozygous state, and a milder form of SPD with approximately 50% penetrance in the heterozygous state, most likely due to the production of 50% of protein compared to normal individuals..
Keywords: Synpolydactyly, haploinsufficiency, incomplete penetrance
Introduction
Trinucleotide repeat expansions in either the coding or non-coding sequences of genes are known to cause several hereditary diseases.1–3 One such example is the polyalanine repeat expansions in HOXD13 gene which have been shown to underlie synpolydactyly 1 (SPD1; OMIM 186000).2 In contrast to other types of nucleotide repeats, such as polyglutamine and polyglutamic acid repeats in Huntington disease and Friedreich’s ataxia respectively, polyalanine repeats in SPD1 are mitotically and meiotically stable, and thus polymorphisms tend to be rare.2
Synpolydactyly (SPD) is a rare autosomal dominant limb deformity, with a distinctive combination of syndactyly and polydactyly. The main features of SPD are webbing of the 3/4 fingers and 4/5 toes, with partial or complete digital duplication within the syndactylous web.4–6 Currently, SPD is classified into three types, SPD1, SPD2 and SPD3, yet mutations have been identified in only two types. SPD1 is caused mainly by polyalanine expansion repeats in the HOXD13 gene on chromosome 2.4 SPD2 (OMIM 608180) has been linked to chromosome 22 and mutations in fibulin 1 gene (FBLN1) have been reported.4 SPD3 (OMIM 610234) has been linked to chromosome 14, but no gene has yet been implicated.4
The HOX family of transcription factors are homeodomain-containing proteins that control cell fates and regional identities along the primary body and limb axes, through binding to cognate DNA sequences.7–9 HOXD13 is a member of the HOX family which encodes for a transcription factor with a crucial role in limb development. The expression of HOXD13, the most 5’ homeobox gene of the HOXD gene cluster, is involved in both the early and late phases of limb morphogenesis.10–12 The first phase is during the emergence of the limb buds, in which the limbs are endowed with AP polarity via the polarized expression of several genes.11,12 The second stage, which is involved in the distal outgrowth and specification of the most distal limb regions, depends on sonic hedgehog (Shh) expression.13–14 HOXD genes are critical in both phases, since they initiate Shh expression during the early phase and mediate the Shh morphogenic signal within the limb during the second phase. Therefore, mutations such as repeat expansions in HOXD13 are expected to result in limb malformation phenotypes.12,15,16 N-terminal polyalanine repeats of +7 to 14 polyalanine repeats are the most common mutations described in HOXD13 whereas the normal number of polyalanine repeats in HOXD13 is 15.12,15,16 To date, only a few missense and deletion mutations have been reported in the HOXD13 gene.17–22 Interestingly, nonsense mutations have not been previously reported in the HOXD13 gene.
In this study, we analyzed a large Pakistani family with SPD1 and identified a first nonsense mutation in HOXD13 gene. Our findings further underscore the crucial roles of HOXD13 in limb development in humans.
MATERIALS AND METHODS
Subjects and linkage analysis
After obtaining informed consent, we collected peripheral blood samples from the family members and 100 population-matched unrelated healthy control individuals in EDTA-containing tubes (under institutional approval and in adherence to the Declaration of Helsinki Principles). Genomic DNA was isolated from these samples according to standard techniques.
Genotyping and Data Analysis
The Affymetirx GeneChip Human Mapping 10K 2.0 array was used to genotype samples. Sample preparation followed the standard Affymetrix protocol, with minor alterations, as previously reported.23 Hybridization was performed by the Columbia University Gene Chip Facility. Genespring GT (Agilent Software) was used for quality control measures and to perform analyses. SNPs displaying Mendelian inheritance errors were removed so that the analyzed dataset contained 9833 variations. Haplotypes were inferred from the data by Genespring GT to mitigate the effect of linkage disequilibrium on multipoint linkage analysis, thus reducing type I error. Statistical analyses included nonparametric linkage analysis (NPL), parametic linkage analysis under assumptions of dominant mode of inheritance with reduced penetrance, and the haplotype-based haplotype relative risk (HHRR), which is a family-based test of association. Next genomic DNA from the family members was amplified by PC® using primers for microsatellite markers around the HOXD13 gene, D2S2188, D2S2314, HOXD13-MS, D2S148, D2S2261, D2S2273, and D2S1361. Of these, HOXD13-MS is a highly polymorphic marker which resides 0.35 Mb downstream of the HOXD13 gene, and the following primers were used for the amplification: forward (5’-CTCAGTGAATGTTCACAATCCA-3’) and reverse (5’-CAACCAACAAACAAGTGGCTC-3’). The amplification conditions for each PCR were 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, with a final extension at 72°C for 7 min. PCR products were run on 8% polyacrylamide gels and genotypes were assigned by visual inspection.
Mutation analysis of the HOXD13 gene
Using genomic DNA from the family members, all exons of the HOXD13 gene with adjacent sequences of exon-intron borders were amplified by PCR as previously described.24 The amplified PCR products were directly sequenced in an ABI Prism 310 Automated Sequencer, using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems).
The mutation c.742C>T in the HOXD13 gene generates a new restriction site for the endonuclease BfaI. A part of exon 1 and intron 1 of the HOXD13 gene was PCR-amplified using PlatinumR Taq DNA Polymerase High Fidelity (Invitrogen) and the following primers: forward (5’-GTGCCCGGCTATATCGACAT-3’) and reverse (5’-AGGCACAACTCCCACTCCCAAGTA-3’). The amplification conditions were 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 61°C for 30 sec, and 68°C for 30 sec, with a final extension at 68°C for 7 min. The amplified PCR products, 280 bp in size, were purified with Rapid PCR Purification System (Marligen), digested with BfaI at 37°C for 3 hours, and analyzed on 2.0% agarose gels.
Results
Patients
We studied a large consanguineous family from Pakistan with SPD. The inheritance pattern could be explained either by autosomal recessive or autosomal dominant with incomplete penetrance (Fig.1). The clinical presentation among the family members was highly variable. Of 16 affected individuals analyzed, the clinical features of 10 members showed a typical SPD1 phenotype, consisting of syndactyly between the 3/4 fingers with digital duplication, widely spaced 1st and 2nd toes, syndactyly involving toes 2/3/4/5 or 3/4/5, camptodactyly of the big toe and brachydactyly which mainly affected the second toe (Fig.2). On the other hand, remaining 6 affected individuals showed a milder phenotype which was restricted to the toes and consisted mainly of webbing of the 4th and the 5th digits or the 2nd and 3rd digits. In addition, a few affected individuals also had unilateral broad big toes, which is not a typical feature for SPD1 (Fig.2). None of the individuals had any urogenital abnormalities.
Figure 1.
Pedigree of a Pakistani family with with SPD consistent with an autosomal dominant pattern with incomplete penetrance. Individuals with
 and
and
 carry the mutation but are not affected. Individuals (a–i) are homozygous for the mutation and individuals (j–l) are heterozygous for the mutation.
carry the mutation but are not affected. Individuals (a–i) are homozygous for the mutation and individuals (j–l) are heterozygous for the mutation.
Figure 2.
Homozygous affected individuals from the Pakistani family with SPD, showed variable and severe phenotypes including syndactyly between the 3/4 fingers with digital duplication, widely spaced 1st and 2nd toes, syndactyly involving toes 2/3/4/5 or 3/4/5, camptodactyly of the big toe and brachydactyly which mainly affected the second toe. 50% of individuals carrying the heterozygous mutation had the disease which was characterized by a milder phenotype than those with the homozygous mutation and involved only the feet, consisting mainly of webbing of the 4th and the 5th digits or the 2nd and 3rd digits, in addition few of the patients had unilateral broad big toes.
Linkage analysis
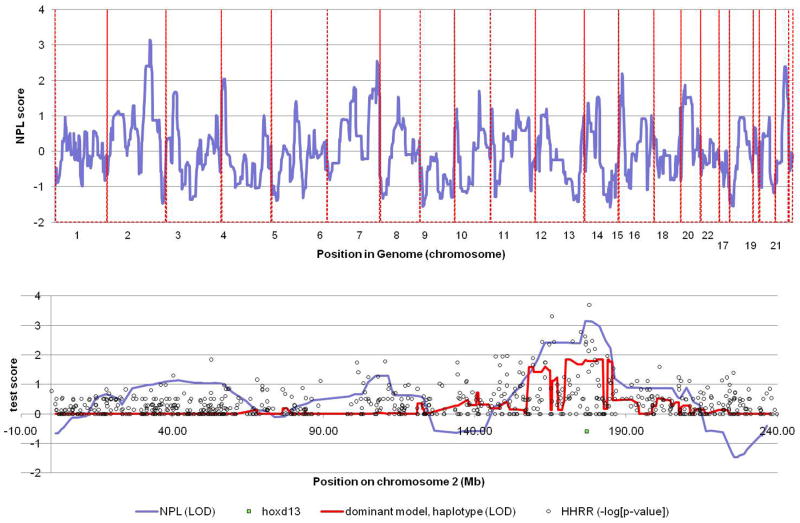
SPD is characterized as an autosomal dominant disorder with wide variability in the phenotypic consequence of particular mutations. In addition to reduced penetrance, a number of investigators have observed that carriers of two mutations often have a more severe phenotype than heterozygous carriers. We therefore initially performed parametric linkage analysis under the assumption of a dominant mode of transmission with reduced penetrance. This analysis failed to reveal any regions of linkage. Because of the extensive consanguinity in our pedigree, we considered that a majority of affected individuals may be homozygous carriers, so we performed parametric linkage analysis under a recessive mode of inheritance. This approach also failed to reveal and regions of linkage. Finally, we performed nonparametric linkage analysis which does not rely on a prior assumption of a mode of inheritance and we discovered a significant NPL score at chromosome 2q22.3–34 (Z=3.15) (Fig.3).
Figure 3.
Non parametric linkage analysis reveals a maximum LOD score on chromosome 2q22.3–34 (Z=3.15). This region contains the HOXD13 gene, the cause of SPD.
Haplotype analysis with markers near the HOXD gene cluster
We analyzed a total of 27 members (16 affected and 11 unaffected) of the SPD family. We performed microsatellite marker analysis using seven makers that span the region of linkage. Ten of the sixteen affected individuals were homozygous for a common haplotype, designated haplotype A, between the markers D2S2188 and D2S148 (Fig.1). This region overlapped with HOXD13 gene. Six of the affected individuals, as well as six of the eleven unaffected individuals, carried the haplotype A in the heterozygous state.
Identification of a nonsense mutation in the HOXD13 gene
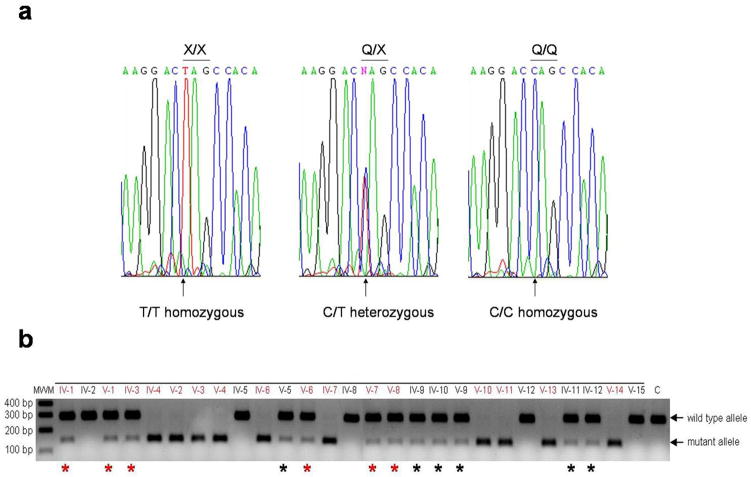
Based on the results of hapolotype analysis, we postulated that affected individuals might carry a mutation in the HOXD13 gene with an incomplete penetrance. We next performed direct sequencing analysis of the HOXD13 gene of this family. Affected individuals, who are homozygous for haplotype A, carry a homozygous nonsense mutation c.742C>T (p.Q248X) in exon 1 of the HOXD13 gene (Fig.4a). Affected and some unaffected individuals, who are heterozygous for haplotype A, carry the mutation heterozygously. Unaffected individuals without haplotype A do not carry the mutation. Restriction enzyme digestion analysis confirmed the result, and excluded the mutation from 100 population-matched unaffected control individuals (Fig.4b; data not shown).
Figure 4.
a) Affected individuals, who are homozygous for the haplotype A, carry a homozygous nonsense mutation c.742C>T (p.Q248X) in exon 1 of the HOXD13 gene. Affected and unaffected individuals are heterozygous for haplotype A. b) The mutation c.742C>T in the HOXD13 gene generates a restriction site for the endonuclease enzyme, thus leading to the digestion of a PCR product from the mutant allele into two fragments of 144bp and 136bp in size. The PCR fragment from the wild type allele is 280bp and is not digested.
Discussion
SPD is a rare genetic malformation of the limb. SPD is clinically heterogeneous, and in addition to the typical clinical presentation of syndactyly involving the 3/4 fingers and 4/5 toes and digital duplication within the syndactylous web, many patients demonstrate a wide range of other clinical manifestations including clinodactyly, camptodactyly, brachydactyly, duplicated metatarsals in different digits, broad hallux, extra phalangeal creases, widely spaced first and second toes and pre/postaxial digital duplication.4 Variations in clinical presentation occur not only within different families, but even within members of the same family.4 Atypical clinical features have been more commonly reported in the setting of deletion and missense mutations than with polyalanine repeat expansions.4,17–22
The role of HOXD13 during embryogenesis is crucial in limb development.13,25 It is postulated that the pathophysiology of the abnormal HOXD13 gene is such that mutant HOXD13 proteins with additional polyalanine repeats are predicted to destabilize the normal protein conformation, leading to its aggregation. This impedes the protein translocation from the cytoplasm to the nucleus where it acts as a transcription factor.5,26 The mutant proteins also show a dominant-negative activity against the wild type protein.6 By contrast, it was recently demonstrated that the consequence of a missense mutation in the HOXD13 gene was likely to be 50% reduction in protein levels, although the underlying mechanism remains unclear.21
Here, we studied a family from Pakistan with clinically heterogeneous SPD. Because of the high rate of consanguinity, the inheritance pattern was most likely autosomal recessive, while there was also the possibility of autosomal dominant inheritance with incomplete penetrance. We initially performed parametric linkage analysis, which was non-revealing. We then performed non parametric linkage analysis and identified linkage to a region on chromosome 2 containing the HOXD13 gene, under an autosomal dominant inheritance model.
Direct sequencing analysis led to the identification of a nonsense mutation Q248X in the HOXD13 gene. All ten individuals with severe SPD were homozygous for the mutation, while twelve individuals were heterozygous for the mutation, among which six were affected and showed a mild SPD phenotype.
It is well-known that the size of the polyalanine expansion repeats in HOXD13 correlates with the severity of the disease. Individuals with short repeats (i.e. 7 or less repeats) are associated with lower penetrance and expressivity, while those with longer repeats (+10 or more) are associated with higher penetrance and more severe disease.27 Moreover, individuals who are homozygous for the polyalanine repeat expansion show a more severe phenotype than those who are heterozygous within members of the same family 24,25,28 Much less is known about HOXD13 mutations other than those associated with polyalanine repeat expansions, since very few families were reported with missense or deletion mutations, and none previously with nonsense mutations. The few reports of families with in-frame deletions and missense mutations presented affected individuals with only heterozygous mutations and complete penetrance of the disease,17–22 though frameshift mutations could be associated with incomplete penetrace.18 The phenotype was generally mild and the clinical features were not consistent with the typical features known for SPD. The mild and atypical phenotypes in non-polyalanine repeat expansion mutations suggested considerable residual activity of the HOXD13 gene, which implicated haploinsufficiency rather than a dominant-negative mechanism. Functional studies on a missense mutation designated G220V in the HOXD13 gene in a Greek family with synpolydactyly, showed that haploinsufficiency is likely to be the mechanism leading to the phenotype, although aggregation of the mutant protein within the cytoplasm still occurred, but to a lesser extent than that observed in polyalanine expansions.21
In the Pakistani family presented here, we observed highly variable phenotypes among affected individuals. All affected individuals who were homozygous for the mutation had very severe phenotypes. In contrast, affected individuals carrying the heterozygous mutation had mild and more atypical features. Interestingly, 50% of individuals heterozygous for the mutation were clinically unaffected, suggesting incomplete penetrance of SPD in the setting of nonsense mutations in HOXD13.
It is likely that aberrant HOXD13 transcripts with the nonsense mutation Q248X undergo nonsense-mediated mRNA decay, and thus no protein will be synthesized from the mutant allele, explaining why affected individuals homozygous for the mutation show a severe SPD phenotype. Likewise, the heterozygous mutation is predicted to result in production of 50% of the protein, leading to either a mild SPD phenotype or a clinically normal appearance. A similar example of incomplete penetrance was previously reported in families with SPD1 carrying out-of-frame deletion mutations in the HOXD13 gene.18 Taken together, these data suggest that haploinsufficiency of HOXD13 expression resulting from premature termination codon mutations can show incomplete penetrance with some frequency.
It is estimated that the penetrance of HOXD13 mutations is approximately 95% and this is mainly due to the length variation in polyalanine repeats, while it is possible that missense and in-frame deletion mutations are maybe completely penetrant.4 Here, we observed that the penetrance of the heterozygous nonsense mutation, Q248X, is around 50%, while it is 100% penetrant in the homozygous state. Incomplete penetrance could be due to genetic background such as the presence of modifier genes and/or due to environmental variability. Transcription factors with major roles in development, such as HOXD13, are required to be active at specific times and at certain dosages during development to initiate the correct morphogenic process, therefore disturbances in any of these features could result in abnormal development. Recently, it has been shown that Hoxd13 may cause polydactyly in SPD by inducing extraneous interdigital chondrogenesis, both directly and indirectly, via a reduction in RA levels.29 Interestingly, even within the same family, gene expression may be different, and these differences may be sufficient to modify the development of disease.30 It was recently shown in C.elegans that mutations in critical developmental networks can lead to variability in gene expression and thus variable penetrance and expressivity of phenotypes.31 Although abnormal HOXD13-mRNA with the mutation Q248X is most likely degraded via nonsense-mediated mRNA decay, it is possible that a truncated HOXD13 protein may be generated and behave in a dominant-negative manner. Furthermore, the expression levels of the mutant protein may be variable between affected individuals, leading to phenotypic variability in heterozygous mutation carriers from mild SPD to clinically normal appearance.
In conclusion, our results expand the spectrum of mutations in the HOXD13 gene and enhance our understanding of the molecular basis of SPD1.
Acknowledgments
We gratefully acknowledge the families for having participated in this study. We also thank Helen Lam for expert technical assistance. This study was supported in part by NIH grant from USPHS, NIH/NIAMS RO1 AR44924 (to A.M.C.). Y.S. is a recipient of a Research Career Development Award from the Dermatology Foundation.
Footnotes
Conflict of interest : None.
References
- 1.Ranum LP, Day JW. Dominantly inherited, non-coding microsatellite expansion disorders. Curr Opin Genet Dev. 2002;12(3):266–271. doi: 10.1016/s0959-437x(02)00297-6. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht A, Mundlos S. The other trinucleotide repeat polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15(3):285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Cummings CJ, Zoghbi HY. Fourteen and counting, unraveling trinucleotide repeat diseases. Hum Mol Genet. 2000;6:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 4.Malik S, Grzeschik KH. Synpolydactyly, clinical and molecular advances. Clin Genet. 2008;73(2):113–120. doi: 10.1111/j.1399-0004.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- 5.Albrecht AN, Kornak U, Böddrich A, Süring K, Robinson PN, Stiege AC, Lurz R, Stricker S, Wanker EE, Mundlos S. A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet. 2004;13(20):2351–2359. doi: 10.1093/hmg/ddh277. [DOI] [PubMed] [Google Scholar]
- 6.Brown LY, Brown SA. Alanine tracts, the expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20(1):51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 8.Favier P, Dolle B. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- 9.Salsi V, Vigano MA, Cocchiarella F, Mantovani R, Zappavigna V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev Biol. 2008;317(2):497–507. doi: 10.1016/j.ydbio.2008.02.048. [DOI] [PubMed] [Google Scholar]
- 10.Dolle P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell. 1993;75(3):431–441. doi: 10.1016/0092-8674(93)90378-4. [DOI] [PubMed] [Google Scholar]
- 11.Te Welscher P, Zuniga A, Fernandez-Teran M, Ros M, Kuijper S, Drenth T, Goedemans H, Meijlink F, Zeller R. Patterning the limb before and after SHH. J Anat. 2002;201:417. doi: 10.1046/j.1469-7580.2002.01021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zákány J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior- posterior asymmetry. Science. 2004;304(5677):1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 13.Deschamps J. Developmental biology. Hox genes in the limb, a play in two acts. Science. 2004;304(5677):1610–1611. doi: 10.1126/science.1099162. [DOI] [PubMed] [Google Scholar]
- 14.Zakany J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- 15.Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- 16.Tarchini B, Duboule D, Kmita M. Regulatory constraints in the evolution of The tetrapod limb anterior-posterior polarity. Nature. 2006;443(7114):985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- 17.Calabrese O, Bigoni S, Gualandi F. A new mutation in HOXD13 associated with foot pre-postaxial polydactyly. Eur J Hum Genet. 2000;8:140. [Google Scholar]
- 18.Goodman F, Giovannucci-Uzielli ML, Hall C, Reardon W, Winter R, Scambler P. Deletions in HOXD13 segregate with anidentical, novel foot malformation in two unrelated families. Am J Hum Genet. 1998;63:992–1000. doi: 10.1086/302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debeer P, Bacchelli C, Scambler P, De Smet L, Fryns JP, Goodman FR. Mutations in HOXD13 and HOXA13 act synergistically to cause severe digital abnormalities. J Med Genet. 2002;39:852–856. doi: 10.1136/jmg.39.11.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan SH, Johnson D, Giele H, Wilkie AO. An acceptor splice site mutation in HOXD13 results in variable hand, but consistent foot malformations. Am J Med Genet A. 2003;121A:69–74. doi: 10.1002/ajmg.a.20103. [DOI] [PubMed] [Google Scholar]
- 21.Fantini S, Vaccari G, Brison N, Debeer P, Tylzanowski P, Zappavigna V. G220V substitution within the N-terminal transcription regulating domain of HOXD13 causes a variant synpolydactyly phenotype. Hum Mol Genet. 2009;18(5):847–860. doi: 10.1093/hmg/ddn410. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Sun M, Zhao J, Leyva JA, Zhu H, Yang W, Zeng X, Ao Y, Liu Q, Liu G, Lo WH, Jabs EW, Amzel LM, Shan X, Zhang X. Mutations in HOXD13 underlie syndactyly type V and a novel brachydactyly-syndactyly syndrome. Am J Hum Genet. 2007;80(2):361–371. doi: 10.1086/511387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petukhova L, Sousa EC, Martinez-Mir A, Vitebsky A, Dos Santos LG, Shapiro L, Haynes C, Gordon D, Shimomura Y, Christiano AM. Genome- wide linkage analysis of an autosomal recessive hypotrichosis identifies a novel P2RY5 mutation. Genomics. 2008;92(5):273–278. doi: 10.1016/j.ygeno.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wajid M, Ishii Y, Kurban M, Dua-Awereh MB, Shimomura Y, Christiano AM. Polyalanine repeat expansion mutations in the HOXD13 gene in Pakistani families with synpolydactyly. Clin Genet. 2009;76(3):300–302. doi: 10.1111/j.1399-0004.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- 25.Muragaki Y, Mundlos S, Upton J, Olsen BR. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science. 1996;272(5261):548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 26.Graba Y, Aragnol D, Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays. 1997;19:379–388. doi: 10.1002/bies.950190505. [DOI] [PubMed] [Google Scholar]
- 27.Goodman F, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, Upton J, Winter RM, Olsen BR, Scambler PJ. Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci U S A. 1997;94(14):7458–7463. doi: 10.1073/pnas.94.14.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsnell K, Ali M, Malik S, Wilson L, Hall C, Debeer P, Crow Y. Clinical phenotype associated with homozygosity for a HOXD13 7-residue polyalanine tract expansion. Eur J Med Genet. 2006;49(5):396–401. doi: 10.1016/j.ejmg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Kuss P, Villavicencio-Lorini P, Witte F, Klose J, Albrecht AN, Seemann P, Hecht J, Mundlos S. Mutant Hoxd13 induces extra digits in a mouse model of synpolydactyly directly and by decreasing retinoic acid synthesis. J Clin Invest. 2009;119(1):146–156. doi: 10.1172/JCI36851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression. Implications for haploinsufficiency. Proc Natl Acad Sci U S A. 1998;95(26):15641–15646. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]