Abstract
SRT1720 is an activator of SIRT1, a NAD+ dependent protein and histone deacetylase that plays an important role in numerous biological processes. Several studies have illustrated that SRT1720 treatment could improve metabolic conditions in mouse models and a study in cancer where SRT1720 caused increased apoptosis of myeloma cells. However, the effect of SRT1720 on cancer may be complex, as some recent studies have demonstrated that SRT1720 may not directly activate SIRT1 and another study showed that SRT1720 treatment could promote lung metastasis. To further investigate the role of SRT1720 in breast cancer, we treated SIRT1 knockdown and control breast cancer cell lines with SRT1720 both in vitro and in vivo. We showed that SRT1720 more effectively decreased the viability of basal type MDA-MB-231 and BT20 cells as compared to luminal type MCF-7 breast cancer cells or non-tumorigenic MCF-10A cells. We demonstrated that SRT1720 induced lysosomal membrane permeabilization and necrosis, which could be blocked by lysosomal inhibitors. In contrast, SRT1720-induced cell death occurred in vitro irrespective of SIRT1 status, whereas in nude mice, SRT1720 exhibited a more profound effect in inhibiting the growth of allograft tumors of SIRT1 proficient cells as compared to tumors of SIRT1 deficient cells. Thus, SRT1720 causes lysosomal-dependent necrosis and may be used as a therapeutic agent for breast cancer treatment.
Keywords: SRT1720, breast cancer, necrosis, lysosomal membrane permeabilization
Introduction
Breast cancer is the most common type of cancer to affect American women. It is estimated that over 200,000 women will develop breast cancer in 2014 and about 40,000 will die from the disease (1). There are many treatment options for women with ER+/PR+/HER2+ luminal-type breast cancers, but for basal-type ER−/PR−/HER2− breast cancers (triple negative breast cancers, TNBCs) there are few options currently available (2, 3). Therefore, novel therapeutics targeting TNBCs and other breast cancers that are resistant to current therapies are desperately needed. In this study, we wanted to elucidate whether the small-molecule compound SRT1720 is a potential therapeutic agent for breast cancer. SRT1720 was initially identified as an activator of SIRT1, a NAD+ dependent deacetylase, which was shown to positively affect glucose and lipid homeostasis (4–7). However, more recently other researchers have demonstrated that SRT1720 has SIRT1-independent effects and does not directly activate SIRT1 (8).
Anti-cancer drugs may cause cell death either through the process of apoptosis or necrosis (9, 10). Apoptosis, also known as programmed cell death, results in the activation of caspases to cause membrane swelling and DNA fragmentation (11). Necrosis is another type of cell death that is not dependent on caspase activation but results from an alternative mechanism such as lysosomal membrane permeabilization, thus causing cell proteolysis and membrane disruption (12). In a previous study, it was shown that SRT1720 induced apoptosis of myeloma cells (13), while another study reported that SRT1720 could promote tumor cell migration and lung metastasis in mice (14). Neither study investigated whether such effects of SRT1720 were dependent on SIRT1. Therefore, it is inconclusive whether the effect of SRT1720 on cancer cells is dependent on SIRT1.
To determine the effect of SRT1720 in breast cancer and study its underlying mechanism, we treated SIRT1 deficient and proficient breast cancer cells with SRT1720. Our data demonstrated that SRT1720 caused lysosomal-dependent cell death in breast cancer cells in vitro irrespective of their SIRT1 status. SRT1720 could also inhibit the growth of allograft tumors in nude mice that was partially mediated by SIRT1. This data reveals that SRT1720 has both SIRT1-dependent and -independent functions and may potentially be a therapeutic agent for the treatment of breast cancer cells.
Materials and Methods
Cell lines and reagents
All human breast cancer cell lines (MCF-7, T47D, SKBR3, MDA-MB-231, SUM149, HS578T, BT-20) and the A549 lung adenocarcinoma cells were obtained from ATCC (Manassas, VA) and cultured with Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) (Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO) and 1% L-glutamine (Invitrogen). All cell lines from ATCC are authenticated by Short Tandem Repeat DNA profiling analysis. HCT116 colon adenocarcinoma cells were obtained from Bert Vogelstein (Johns Hopkins University, Baltimore, MD). These cells have not been authenticated. Mouse mammary tumor cells were from MMTV-neu mice (Neu) and from Brca1Co/Co;p53+/−;MMTV-Cre mice (69), respectively (15, 16). MCF10A immortalized mammary epithelial cells were obtained from ATCC and cultured with DMEM/F12 (1:1) (Invitrogen) supplemented with 5% horse serum (Invitrogen), hydrocortisone (0.5 µg/ml) (Sigma), epidermal growth factor (20 ng/ml) (Peprotech) (Rocky Hill, NJ), insulin (10 µg/ml) (Invitrogen), and cholera toxin (100 ng/ml) (Sigma). MEF cells were obtained from embryos of wild-type and SirT1−/− mice from our lab (17). MDA-MB-231/GFP-LC3 cells were generated by transfection and selection of stable cells with neomycin. Mixed cell clones were used for the experiments. SRT1720 was synthesized by Craig J. Thomas (National Cancer Institute, Bethesda, MD) and dissolved in dimethyl sulfoxide (DMSO) for cell culture experiments. Inhibitors of autophagolysosome function; chloroquine, ammonium chloride, and bafilomycin A1 were obtained from Sigma. The autophagy inhibitor 3-methyladenine (3-MA) was obtained from Sigma.
Preparation and transduction of lentiviral-delivered short-hairpin RNA (shRNA)
For transduction of lentiviral shRNA, pLKO.1 lentiviral vectors targeting SIRT1 were obtained from Sigma. The lentiviral SIRT1 shRNA clone, TRCN0000018979, targets the nucleotide sequence (5’- AAAGCCTTTCTGAATCTAT-3’) of SIRT1 mRNA. A lentiviral control shRNA, pLKO.1-Scrambled, was obtained through the plasmid repository Addgene (Cambridge, MA) (18). For production of lentiviral particles expressing SIRT1 shRNA, 293T cells (3 × 106) were seeded in 100 mm dishes. After the cells attached, the transfection complex was prepared as follows according to the manufacture’s instructions for X-tremeGENE9 (Roche Applied Science, Indiannapolis, IN). 3 µg of the pLKO.1-SIRT1 shRNA vector was added to 18 µl of X-tremeGENE9 in 500 µl DMEM along with 3 µg pCMV-dR8.2 dvpr packaging vector and 0.375 µg pCMV-VSV-G envelop vector. The packaging and envelop vectors were created by the lab of Robert Weinberg (19) and obtained through Addgene. The transfection complex was added to the cells for 24 hours of incubation, the cells were washed with medium, and 10 ml of fresh medium was added for another 24 hours. The medium containing lentiviral particles was then collected, centrifuged at 2,000 rpm for 5 minutes, filtered through a 0.45 µm Polyethersulfone syringe filter (EMD Millipore, Billerica, MA), and aliquots were stored at −80°C. For transduction of lentiviral particles, MDA-MB-231 (5 × 105) cells were seeded in 100 mm dishes and 1 ml of viral supernatant was added to 7 ml of medium after cell attachment. The cells were transduced for 24 hours in the presence of polybrene (8 µg/ml) (Sigma). Cells stably expressing SIRT1 shRNA were selected for 48 hours in the presence of puromycin (2 µg/ml) (Sigma) before plating for experiments.
Western blotting
Cells were harvested from sub-confluent plates and whole cell lysates were prepared for immunoblot analysis. Cells were washed with cold phosphate buffered saline (PBS) and lysed with lysis buffer containing: 1% NP-40, 50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 10% glycerol, 50 mmol/L NaF, 2 mmol/L EGTA, 2 mmol/L EDTA, 1 µg/ml Pepstatin A, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 100 µg/ml AEBSF, and 1 mmol/L sodium orthovanadate. The lysate was centrifuged at 14,000 rpm at 4°C for 10 minutes and the protein concentration of the supernatant was quantified with the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA). Protein lysates (25–50 µg) were prepared in SDS-loading buffer containing the reducing agent β-mercaptoethanol (Sigma), heated at 95°C for 4 minutes, resolved by SDS-polyacrylamide gel electrophoresis on 4–12% Tris-glycine gels (Invitrogen), and then transferred to polyvinylidene diflouride (PVDF) Immobilon-P membranes (EMD Millipore). The membranes were blocked with 4% non-fat milk in PBS for 30 minute, incubated for 2 hours with primary antibodies (see Cell culture and reagents section) diluted in 1% milk in PBS, washed 3 times with PBS-T (PBS, 0.1% Tween-20), and then incubated with either an anti-rabbit or anti-mouse HRP-conjugated secondary antibody (GE Healthcare, Piscataway, NJ) for 45 minutes. The blots were washed 3 times with PBS-T and the protein bands were detected on enhanced chemiluminescence film (Denville Scientific, South Plainfield, NJ) with the Immobilon chemiluminescence detection reagent (EMD Millipore). Proteins were detected with the following antibodies: monoclonal anti-SIRT1 (1F3), LC3 (Cell Signaling, Beverly, MA), monoclonal anti-β-actin (Sigma),
Cell viability and apoptosis assays
For cell viability assays, MDA-MB-231 cells were plated at 15,000 cells/well in 24-well plates. The day after plating, SRT1720 was added to the cells for the designated time and concentration. Cell viability was assessed by methylthiazolydiphenyl-tetrazolium bromide (MTT) assay. A 0.5 mg/ml solution of Thiazolyl Blue (Sigma) in phenol-free DMEM was added to cells at 37° C for 1 hour. The substrate was then dissolved in isopropanol and absorbance was measured with a spectrophotometer at 570 nm. Apoptosis/necrosis was measured with the annexin V/propidium iodide staining kit from Sigma. For necrosis measurements alone, cells were trypsinized, washed in PBS, and incubated with 1 µg/ml of propdium iodide for 10 minutes. Both annexin V and propidium iodide staining were detected by FACS analysis with a FACSCalibur flow cytometer (BD Biosciences).
Acridine orange and LysoTracker Red staining
Cells were incubated at 37° C for 15 minutes with 1 µg/ml of acridine orange (Sigma). The cells were washed in PBS and observed with a FACSCalibur flow cytometer in both the red (FL-2) and green (FL-1) channel. For LysoTracker Red staining, cells were incubated with 100 nM LysoTracker Red (Invitrogen) at 37° C for 15 minutes.
In vivo tumor study
Neu cells (1×106) were injected into two sites of the mammary fat pads of five nude mice each for vehicle and SRT1720 treatment. There were a total of 10 tumor injection sites per treatment. SRT1720 (40 mg/kg/day) was delivered by intra-peritoneal injection every day for the duration of the treatment. SRT1720 was prepared in 40% PEG300 and 0.5% Tween 80 in water.
Statistical analysis
Data was analyzed using Prism GraphPad (GraphPad software, Inc.). Results are presented as the mean ± standard error of the mean (SEM) from independent triplicate samples. A P value of less than 0.05 was considered to be statistically significant and designated as P < 0.5 = *, P < 0.01 = **, or P < 0.001 = ***. For multiple comparisons, ANOVA followed by Bonferroni’s post tests was used. Otherwise the t-test was used for comparison of two treatments.
Results
SRT1720 inhibits the growth of breast cancer cells in-vitro in a SIRT1-independent manner
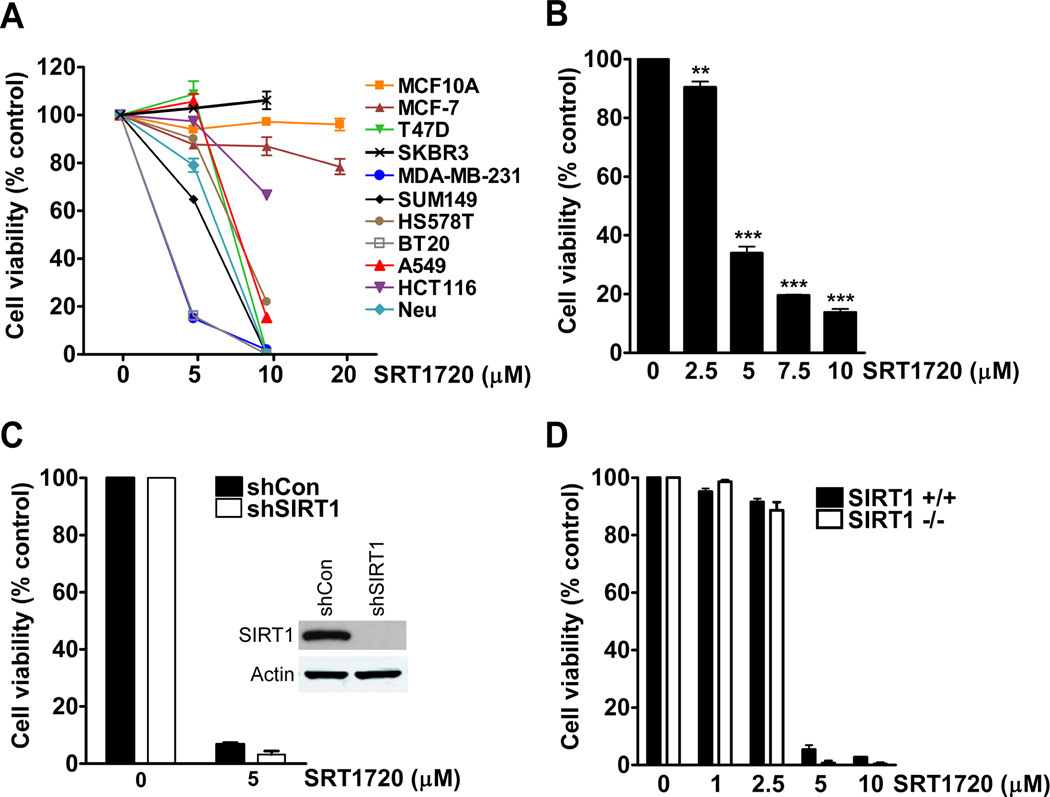
SRT1720 was previously shown to inhibit the proliferation of myeloma cell lines through the induction of apoptosis (13). In our study, we determined whether SRT1720 affected the viability of breast cancer cell lines. Human and mouse breast cancer cells, MCF10A immortalized human mammary epithelial cells, and A549 lung and HCT116 colon adenocarcinoma cell line were treated with various concentrations of SRT120 for 24 hours and assessed for cell viability by MTT assay. The viability of MDA-MB-231 and BT20 basal-type breast cancer cells decreased by more than 80% with 5 µmol/L of SRT1720, whereas the viability of MCF-7 luminal-type cells only decreased by 20% with 20 µmol/L of treatment (Fig. 1A). The viability of MCF10A immortalized mammary epithelial cells was not affected up to 20 µmol/L of SRT1720 treatment. Therefore, MDA-MB-231 cells were used for further experiments as they were more sensitive to SRT1720 treatment. Next we treated MDA-MB-231 cells with a dose range of SRT1720 from 2.5–10 µmol/L to determine the concentration, which begins to affect cell viability. The viability of MDA-MB-231 cells was decreased by 66–86% with a dose range from 5–10 µmol/L (Fig. 1B). Therefore, MDA-MB-231 cells treated with SRT1720 have a very narrow dose response. The sensitivity of MDA-MB-231 cells to 5 µmol/L of SRT1720 treatment was also affected by the plating density of the cells. If the cell density was doubled from 15,000 (low) to 30,000 (high) cells per well in a 24-well plate there was no effect on cell viability, however, 10 µmol/L of SRT1720 decreased the viability of cells plated at both low and high density (Supplementary Fig. 1A). To determine if a secreted factor could affect the sensitivity of MDA-MB-231 cells to SRT1720, we collected cell media from cells plated at a high density and added this media with or without SRT1720. Interestingly, MDA-MB-231 cells cultured in this media were resistant to SRT1720 treatment (Supplementary Fig. 1B). Therefore, a secreted factor from MDA-MB-231 cells grown at a high density could affect the sensitivity of cells to SRT1720 treatment although the identity of this secreted factor is currently unknown.
Figure 1.
SRT1720 decreases the viability of breast cancer cells independent of SIRT1 status. A, MTT viability assay of cancer cell lines treated with different doses of SRT1720 for 24 hours. B, MTT dose response of MDA-MB-231 breast cancer cells treated with SRT1720 for 24 hours. There was a significant decrease in cell viability with 5 (66%), 7.5 (80%), and 10 (86%) µmol/L of SRT1720 (P < 0.001, ***). C, Viability of control and SIRT1 shRNA knockdown MDA-MB-231 cells treated with 5 µmol/L of SRT1720 for 24 hours. There was no significant difference in cell viability in SRT1720 treated sh-control cells as compared with sh-SIRT1 knockdown cells. D, Viability of WT and SIRT1−/− MEF cells treated with SRT1720 for 24 hours. There was no significant difference in cell viability in SRT1720 treated WT as compared with SIRT1 −/− MEFs.
Previous reports have shown that SRT1720 exerts biological effects through the sirtuin deacetylase SIRT1 (4, 20). We determined whether the effect on cell viability by SRT1720 treatment was dependent on SIRT1 expression. We measured cell viability after SIRT1 knock-down with lentiviral-transduced SIRT1 shRNA in MDA-MB-231 cells or SIRT1 knockout mouse embryonic fibroblasts (MEFs). SRT1720 treatment of both MDA-MB-231 sh-control and sh-SIRT1 knockdown cells decreased cell viability by more than 90% which was also observed in immortalized wild-type (WT) and SIRT1−/− MEFs (Fig. 1C, D). Therefore, SRT1720 decreases the viability of MDA-MB-231 cells and MEF cells independently of SIRT1 expression. We also compared the effect of resveratrol, another reported activator of SIRT1 (21, 22), on the viability of MDA-MB-231 cells. There was a 30% decrease in the viability of sh-control MDA-MB-231 cells treated with 80 µmol/L of resveratrol as compared with a decrease of 47% for sh-SIRT1 knockdown cells, thus suggesting that SIRT1 is not required for resveratrol induced cell death (Supplementary Fig. 2). We next determined whether the effect of SRT1720 on breast cancer cells was dependent on other members of the sirtuin family by co-treatment with the pan-sirtuin inhibitor nicotinamide. Our results showed that other sirtuin members also do not have a role in SIRT1720-induced cell death (Supplementary Fig. 3).
SRT1720 decreases the viability of breast cancer cells by increased necrosis
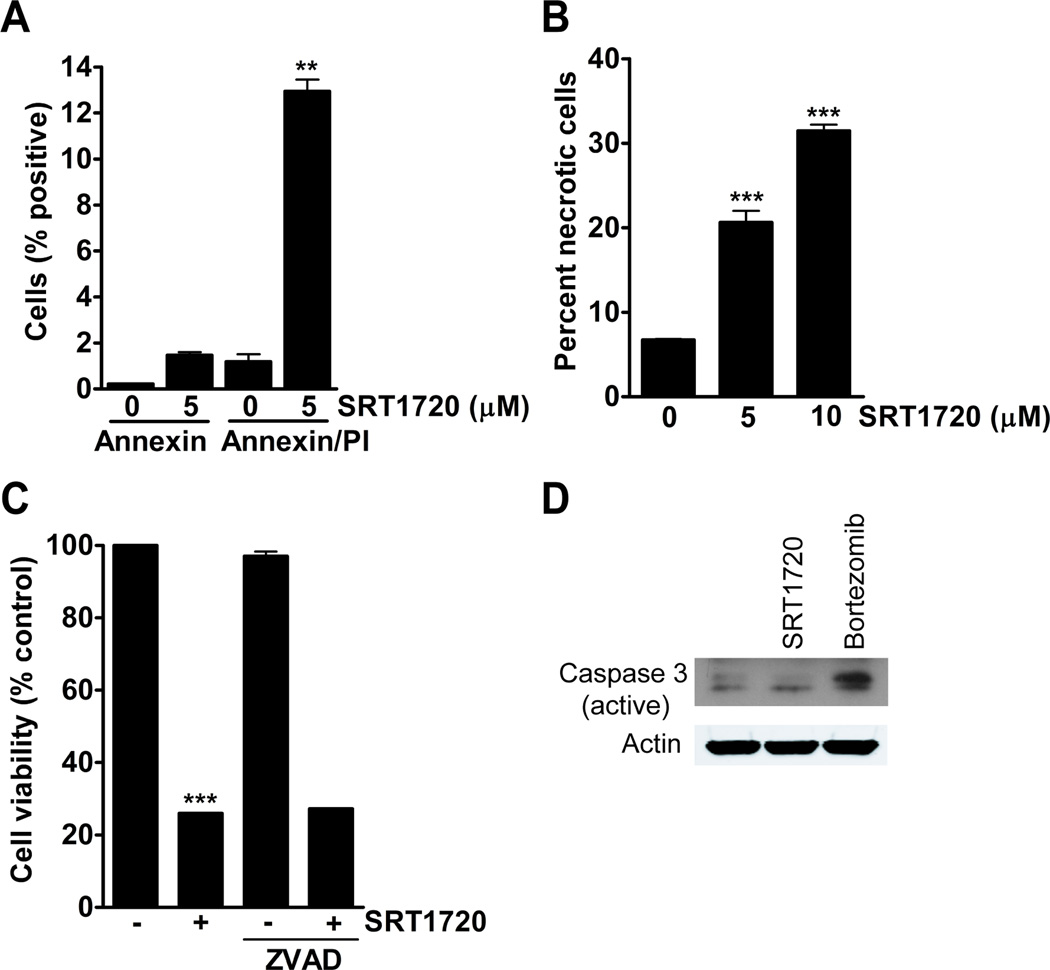
A decrease in cell viability may result from decreased cell cycle progression or through increased cell death. The cause of a reduction in cell viability was determined by measuring apoptosis and necrosis. Cells were treated with SRT1720 for 8 hours and apoptosis/necrosis was measured by annexin V/propidium iodide staining, respectively. After 5 µmol/L of SRT1720 treatment, 1% percent of the cells were positive for early apoptosis (only annexin V positive) and 12% of the cells were positive for late apoptosis/necrosis (double positive for annexin V and propidium iodide) (Fig. 2A). Measurement of necrosis with prodium iodide alone showed a 13% and 24% increase in necrotic cells with 5 and 10 µmol/L SRT1720, respectively, after 8 hours of SRT1720 treatment (Fig. 2B). The pan-caspase inhibitor ZVAD was used to determine if apoptosis had any role in SRT1720-induced cell death. There was a 74% in cell viability in SRT1720 treated cells, however, co-treatment of MDA-MB-231 cells with ZVAD did not reduce cell death caused by SRT1720 treatment (Fig. 2C). Also, SRT1720 did not cause caspase-3 cleavage as was observed with the proteosome inhibitor bortezomib (Fig. 2D).
Figure 2.
The viability of MDA-MB-231 cells treated with SRT1720 is decreased due to necrosis but not apoptosis. A, Apoptosis/necrosis measurement by annexin V/propidium iodide analysis of MDA-MB-231 cells treated with 5 µmol/L of SRT1720 for 8 hours. There was a 1 % increase in early apoptosis and a 12% increase in late apoptosis/necrotic cells (P < 0.01, **). B, Propidium iodide positive MDA-MB-231 cells treated with SRT1720 for 8 hours as a measurement for necrosis. There was a significant increase in necrosis with 5 (13%) and 10 (24%) µmol/L of SRT1720 treatment (P < 0.001, ***). C, Viability of MDA-MB-231 cells treated with SRT1720 and 20 µmol/L of the pan-caspase inhibitor ZVAD for 24 hours. SRT1720 caused a 74% reduction in cell viability (P < 0.001, ***). There was no significant difference between SRT1720 treatment alone and in combination with ZVAD. D, Expression of active caspase-3 in MDA-MB-231 cells treated with either 5 or 10 µmol/L SRT1720 or 10 µmol/L of the proteosome inhibitor bortezomib as a control for caspase-3 activation. There was increased caspase 3 cleavage with bortezomib but not SRT1720 (upper band on immunoblot).
SRT1720 caused the formation of acidic vacuoles and an increase in autophagy markers
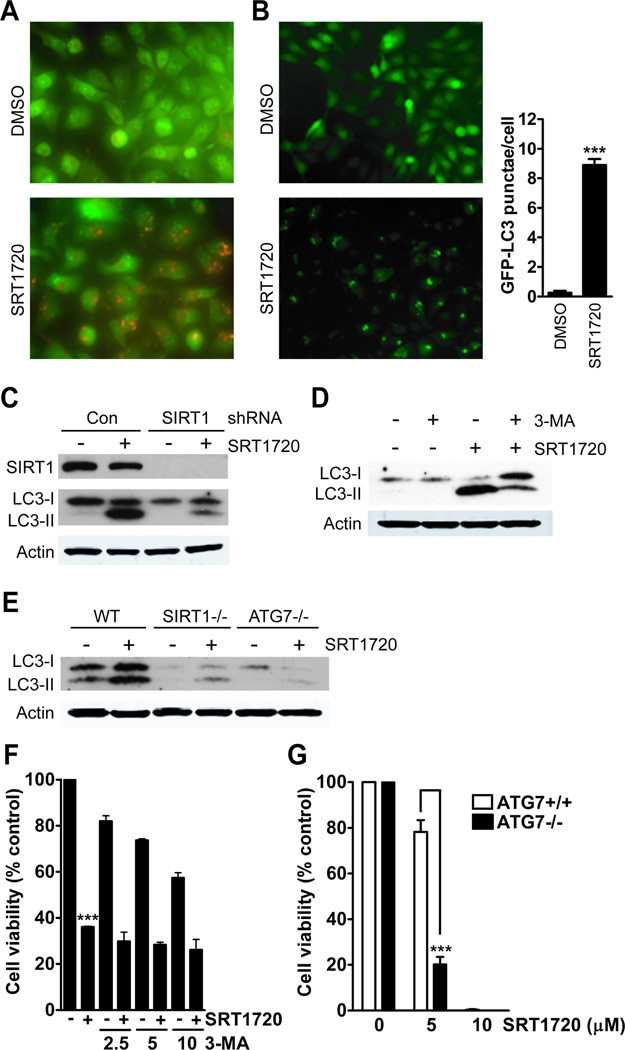
Treatment of MDA-MB-231 breast cancer cells with SRT1720 for 8 hours resulted in the formation of acidic vacuoles (Fig. 3A) as detected by acridine orange staining. Acridine orange will emit red fluorescence in a low pH environment, which can be used to measure acidic vacuoles. Autophagy is a cellular process by which the cell catabolizes its own cytoplasmic organelles during nutritional deprivation. It is mainly a survival pathway, which may result in cell death during prolonged nutrient deprivation. One of the markers for autophagy is the autophagosome membrane marker LC3 that is converted from LC3-I to LC3-II through lipidation involving both ATG7 and ATG3, which leads to LC3 fusion with autophagosome membranes (23). We generated MDA-MB-231 cells with stable expression of LC3-GFP to measure the affect of SRT1720 on autophagy by observation of GFP punctae formation. Treatment with SRT1720 resulted in an increase in the number of cells with an average of 8 GFP punctae per cell (Fig. 3B). An immunoblot of LC3 will show an increase in the lower LC3-II band during autophagy. We observed that treatment with SRT1720 caused an increase in the autophagy marker LC3-II, and this was decreased in sh-SIRT1 knockdown MDA-MB-231 cells (Fig. 3C). This validates a previous study demonstrating that SIRT1 is involved in the regulation of autophagy (24). Next, we determined whether blocking the PI3K pathway responsible for initiation of autophagy would affect autophagy induction by SRT1720. Co-treatment of MDA-MB-231 cells with the type-III PI3K inhibitor, 3-MA, resulted in decreased LC3-II formation caused by SRT1720 treatment (Fig 3D). Also, LC3-II formation was reduced in SIRT1 and ATG7 knockout MEFs (Fig 3E). Therefore, SRT1720 increases the formation of the autophagy marker, LC3-II, through SIRT1 and ATG7. Then we determined whether blocking autophagy induction by SRT1720 with 3-MA or ATG7 knockout cells could affect cell death. MDA-MB-231 cells treated with either SRT1720 or 2.5 mM 3-MA resulted in a 65% and 18% decrease in cell viability, respectively. However, co-treatment of 3-MA with SRT1720 did not reduce SRT1720-induced cell death (Fig. 3F). In addition, treatment of wild-type MEFs with 5 µmol/L of SRT1720 resulted in a 22% decrease in cell viability, whereas, the number of ATG7 knockout cells decreased by 80% (Fig. 3G). Therefore, these results suggest that autophagy is increased during SRT1720 treatment due to cellular stress as a mechanism to promote cell survival.
Figure 3.
Autophagy is induced in MDA-MB-231 cells treated with SRT1720. A, SRT1720 treatment for 8 hours increases the number of acidic vesicular organelles in MDA-MB-231 cells. B, SRT1720 treatment of MDA-MB-231 cells increases the number of GFP-LC3 punctae. GFP-LC3 was assessed from counting the number of GFP-LC3 punctae per cell from 100 cells as shown by representative microscopy images. The bar graph shows an average of eight punctae per cell in SRT1720 treated cells as compared with one for DMSO treated cells (P < 0.0001, ***). C, The autophagy marker, LC3-II, increases after 8 hours of treatment with 5 µmol/L of SRT1720 in control but not in sh-SIRT1 knockdown MDA-MB-231 cells. D, The class III PI3K inhibitor 3-MA reduces LC3-II formation caused by SRT1720 treatment. E, LC3-II formation after treatment with SRT1720 for 8 hours in WT, SIRT1−/−, and ATG7−/− MEFs. F, Viability of MDA-MB-231 cells after treatment with 2.5, 5, or 10 mmol/L 3-MA and SRT1720 for 24 hours. There was no significant difference in cell viability with SRT1720 alone or in combination with 2.5, 5, or 10 mM 3-MA. G, Viability of WT and ATG7−/− MEFs after treatment with SRT1720 for 24 hours. There was a significant decrease in cell viability in ATG7−/− MEFs treated with 5 µmol/L SRT1720 as compared with WT MEFs (P < 0.001, ***), whereas there was no difference with 10 µmol/L.
SRT1720 induces lysosomal membrane permeabilization
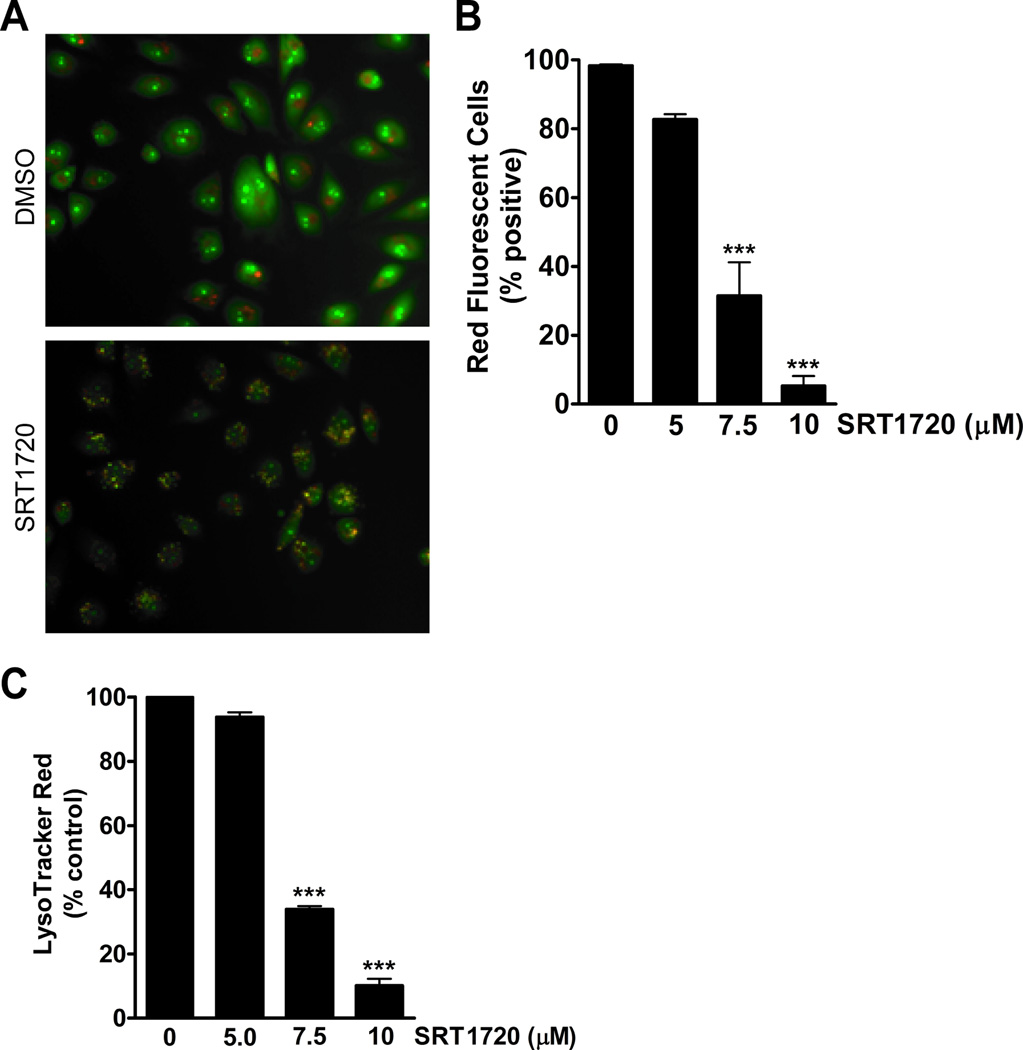
Necrotic cell death may be caused by permeabilization of the lysosomal membrane (12). Lysosomal membrane permeabilization (LMP) causes the release of cathepsins and other hydrolases from the lysosomal lumen to the cytosol (25). To determine if the death of MDA-MB-231 cells was caused by permeabilization of the lysosomal membrane, cells were incubated with either acridine orange or LysoTracker Red. Acridine orange and LysoTracker Red normally concentrate in the lysosomal compartment and emit red fluorescence, but upon permeabilization these dyes will diffuse out of the lysosome (12). In the case of acridine orange, the lysosomal vacuoles become yellow as a result of an increase in the pH. After treatment of MDA-MB-231 cells with SRT1720 for 16 hours, the cells were incubated with acridine orange and observed by microscopy. SRT1720 caused an increase in yellow stained cells, therefore indicating an increase in lysosomal membrane permeability (Fig. 4A). An analysis of the cells by FACS showed that there is a dose-dependent decrease in the percentage of red fluorescent cells with SRT1720 treatment The percentage of red cells decreased by 68% with 7.5 µmol/L and 95% with 10 µmol/L of SRT1720 (Fig. 4B). We also incubated the cells with LysoTracker Red and observed a 66% and 90% decrease in LysoTracker Red staining by FACS analysis after treatment with 7.5 and 10 µmol/L of SRT1720, respectively (Fig 4C).
Figure 4.
Lysosomal membrane permeabilization increases in MDA-MB-231 cells treated with SRT1720. A, Representative microscopy images of acridine orange stained MDA-MB-231 cells with or without 5 µmol/L SRT1720 treatment for 16 hours. B, SRT1720 decreases the percentage of red fluorescent cells in MDA-MB-231 cells stained with acridine orange as measured by FACS analysis. There was a decrease in red fluorescent cells with 7.5 (68%) and 10 (95%) µmol/L of SRT1720 treatment (P < 0.001, ***). C, SRT1720 decreased LysoTracker Red uptake in MDA-MB-231 cells as measured by FACS analysis. There was a decrease in LysoTracker Red positive cells with 7.5 (66%) and 10 (90%) µmol/L of SRT1720 treatment (P < 0.001, ***).
Lysosomal inhibitors block SRT1720-induced cell death
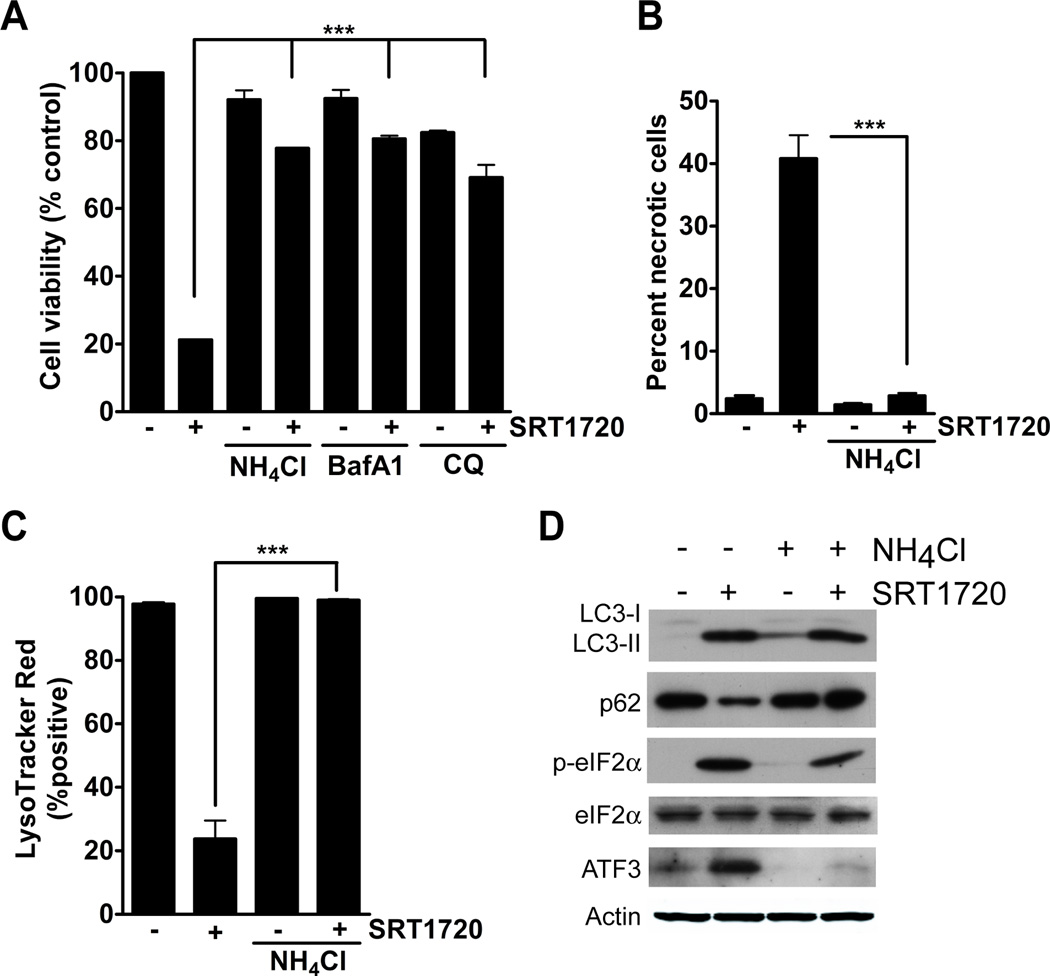
As a result of our observation that SRT1720 increases lysosomal membrane permeabilization, we next determined whether inhibitors of lysosomal acidification could affect SRT1720-induced cell death. The lysosomotropic alkalinazation agents, ammonium chloride, bafilomycin A1, and chloroquine affect lysosomal acidification through different mechanisms. Ammonium chloride directly neutralizes lysosomal pH, bafilomycin A1 is an inhibitor of the V-ATPase that pumps protons into the lysosome, and chloroquine blocks the fusion of the lysosome and autophagosome (23, 26, 27). We assessed the viability of MDA-MB-231 cells treated with SRT1720 and lysosomal inhibitors. We observed that ammonium chloride, bafilomycin A1, and chloroquine could significantly reduce SRT1720-induced cell death. Treatment with SRT1720 alone resulted in a 79% decrease in viable cells, whereas, co-treatment with ammonium chloride, bafilomycin A1, and chloroquine resulted in only a 23%, 20%, and 30% decrease in cell viability, respectively (Fig. 5A). We also observed that ammonium chloride could significantly reduce the death of Hs578T breast cancer cells caused by SRT1720 treatment (Supplementary Fig. 4). There was an 84% decrease in cell viability with 10 µmol/L of SRT1720 and only a 36% decrease with ammonium chloride co-treatment. Ammonium chloride could also inhibit SRT1720-induced cell necrosis (Fig. 5B) and lysosomal membrane permeabilization of MDA-MB-231 cells (Fig. 5C). There was a 41% increase in necrotic cells with SRT1720 and only 3% in combination with ammonium chloride. LysoTracker Red positive cells decreased 76% with SRT1720 treatment and only 1% in combination with ammonium chloride. We then determined whether ammonium chloride treatment could affect SRT1720-induced changes in autophagy and protein homeostasis. MDA-MB-231 cells were treated with SRT1720 and/or ammonium chloride, and markers for autophagy and cellular protein stress were assessed. LC3-II induction by SRT1720 could not be blocked by ammonium chloride (Fig. 5D). An increase in LC3-II may be caused by increased autophagy or inhibition of autophagy. The protein p62/SQSTM1 is an ubiquitin binding protein that binds to LC3-II and during increased autophagy is normally degraded after fusion of the autophagosome and lysosome (23). Therefore, p62 has been used as a marker for autophagic flux. Treatment of MDA-MB-231 cells with SRT1720 decreases the level of p62 protein, which indicates an increase in the rate of autophagy by SRT1720, whereas ammonium chloride inhibits SRT1720-induced p62 degradation by inhibiting lysosome function (Fig. 5D). We also determined whether SRT1720 caused activation of the endoplasmic reticulum (ER) stress pathway. ER stress is caused by a malfunction in protein processing in the ER lumen, which leads to an increase in unfolded/misfolded proteins (28). This will cause a block in protein translation by stress-kinase induced phosphorylation of the eukaryotic initiation factor 2 alpha (eIF2α) and an induction of genes necessary for the proper folding of proteins including activating transcription factor 3 (ATF3) (29). As a result of SRT1720 treatment, the phosphorylation of eIF2α and protein level of ATF3 was increased (Fig. 5D). Also, the cells were co-treated with ammonium chloride to determine if the inhibition of SRT1720-induced death could be through the ER stress pathway. There was a slight reduction in eIF2α phosphorylation with ammonium chloride co-treatment, however, the induction of ATF3 with SRT1720 was completely inhibited. This observation suggests that the induction of eIF2α phosphorylation and enhanced ATF3 expression level by SRT1720 treatment may be through a different mechanism. We then determined whether SRT1720-induced cell death was dependent on ATF3. The viability of WT and ATF3−/− MEFs was measured after treatment with SRT1720, however, both cells were equally affected by SRT1720 treatment (Supplementary Fig. 5).
Figure 5.
Lysosomal inhibitors block SRT1720-induced death of MDA-MB-231 cells. A, Viability of MDA-MB-231 cells treated with SRT1720 in the presence of the lysosomal inhibitors, 10 mmol/L ammonium chloride (NH4Cl), 1 nmol/L Bafilomycin A1 (BafA1), and 50 µmol/L chloroquine (CQ). Lysosomal inhibitors significantly reduced SRT1720-induced cell death (P < 0.001). B, Percentage of necrotic cells after treatment with SRT1720 and the lysosomal inhibitor NH4Cl. Co-treatment with NH4Cl significantly decreased the percentage of necrotic cells after SRT1720 treatment (P < 0.001, ***). C, Percentage of LysoTracker Red positive cells after treatment with SRT1720 and NH4Cl. Co-treatment with NH4Cl significantly reduced the loss of LysoTracker Red positive cells (P < 0.001, ***). D, Immunoblot of autophagy and endoplasmic reticulum (ER) stress markers after treatment with SRT1720 and NH4Cl for 8 hours.
SRT1720 inhibits breast cancer cell growth in vivo
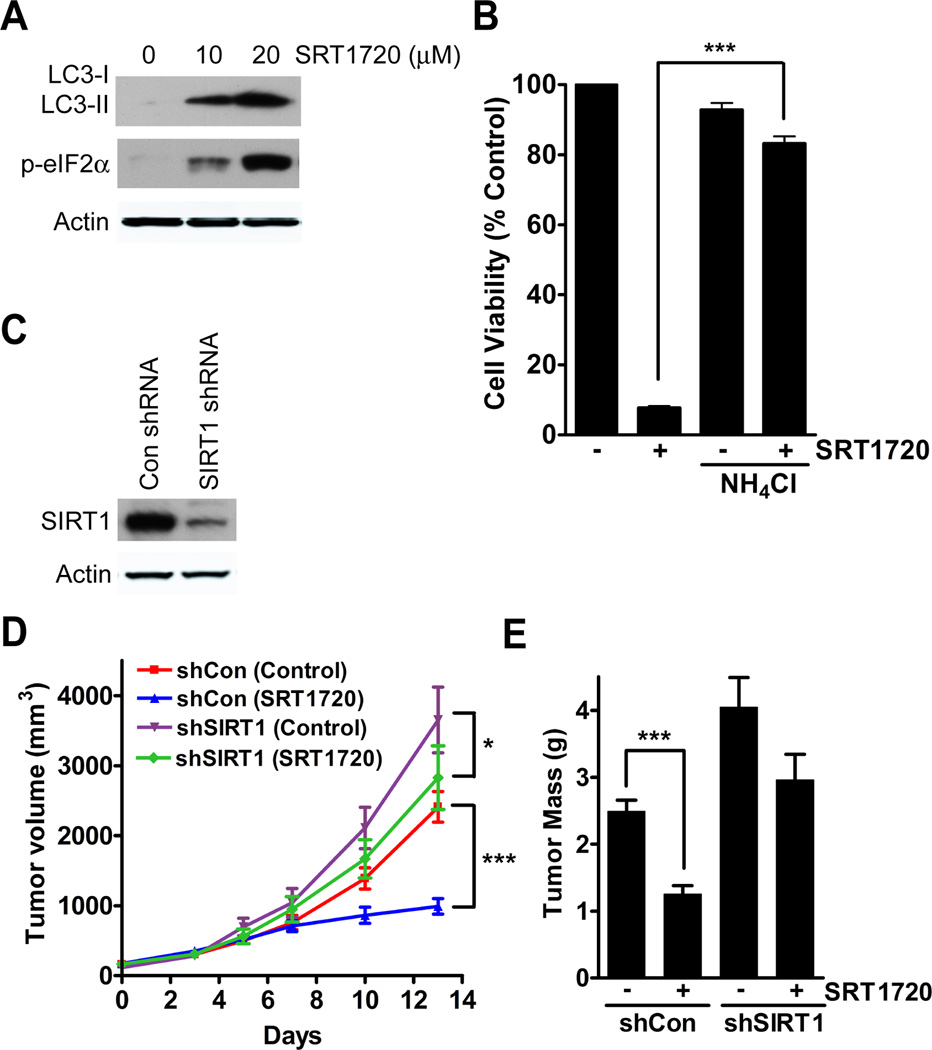
We have shown that SRT1720 inhibits the growth of both human and mouse breast cancer cells in vitro, so we next tested the effect of SRT1720 on tumor growth in mice. We were limited in the availability of SRT1720 so we used neu cells, a tumor cell line derived from a mammary tumor of MMTV-neu transgenic mouse (15), which grow faster as tumor xenografts than MDA-MB-231 and so require less SRT1720 for treatment. To verify that SRT1720 also induces cellular stress in neu cells we determined by immunoblot the expression of LC3-II and p-eIF2α after SRT1720 treatment, which showed an increase in both (Fig. 6A). We had also observed that the viability of neu cells in vitro was decreased by SRT1720 treatment (Fig. 1A). We then determined whether lysosomal inhibition could also decrease cell death caused by SRT1720 as we previously observed in MDA-MB-231 cells. As a result of co-treatment of ammonium chloride with SRT170, there was a decrease in cell death of neu cells (Fig. 6B). For the in vivo study with SRT1720, neu cells were initially infected with either control or SIRT1 shRNA (Fig. 6C). Then the cells were injected into to the mammary fat of nude mice and SRT1720 was administered daily by intra-peritoneal injection for 14 days. Tumor volumes were measured with a caliper during the length of the experiment and the data showed a significant difference between neu-sh-control tumor xenografts treated with vehicle and SRT1720 (59% decrease, P < 0.001), whereas there was a less significant difference in SRT1720 treated neu-sh-SIRT1 tumors (23%, P < 0.05) (Fig. 6D). We also detected enhanced growth of neu tumor xenografts carrying shSIRT1 knockdown than sh-control tumors (66%, P < 0.001) (Fig. 6C), which is consistent with the tumor suppressor function of SIRT1 as demonstrated previously (17, 30). The tumors were also weighed at the end of 14 days, which showed a significant decrease in tumor mass of the neu-control tumors treated with SRT1720 (50%, P < 0.001) but not of neu-SIRT1 knockdown tumors (27%, P = 0.0545) (Fig. 6E). This observation suggests that the inhibitory effect of SRT1720 is at least partially mediated by SIRT1. There was no effect of SRT1720 treatment on the body weight of the mice (Data not shown).
Figure 6.
SRT1720 reduces tumor xenograft growth in a SIRT1-dependent and -independent manner. A, SRT1720 increases autophagy and ER stress markers in neu-expressing mouse cancer cells after 8 hours of treatment. B, Percentage of viable neu cells after treatment of SRT1720 and NH4Cl. Co-treatment with NH4Cl significantly reduced the death of neu cells treated with SRT1720 (P < 0.001, ***). C, Immunoblot showing knockdown of SIRT1 in neu cells. D, Graph showing neu tumor volume of control (n=10) and SIRT1 (n=10) knockdown tumors in mice treated with SRT1720. There was a 69% decrease in tumor volume of sh-control tumors (P < 0.001, ***) after SRT1720 treatment as compared to a 23% decrease in sh-SIRT1 tumors (P < 0.05, *) after SRT1720 treatment. E, Graph of neu tumor mass from control (n=10) and SIRT1 (n=10) knockdown tumors after SRT1720 treatment. There was a significant decrease in tumor mass in sh-control tumors (P < 0.0001, ***) but not in sh-SIRT1 tumors (P = 0.0545)
Discussion
SIRT1 is the initial member of the 7-member sirtuin family of protein and histone deacetylases (7, 31, 32). Numerous studies have revealed important functions of SIRT1 in many biological processes including cell proliferation, differentiation, apoptosis, senescence, metabolism, calorie restriction, lifespan regulation, and tumorigenesis (5, 6, 33–39). Although SIRT1 was initially considered as a tumor promoter in some studies, recent work has uncovered a tumor suppressor function of SIRT1, especially from analyzing SIRT1 mutant mouse models (17, 30, 40). While SIRT1 might have both tumor promoter and tumor suppressor activity depending on the tissue context (41–43), a few studies have tested the effectiveness of a SIRT1 activator, SRT1720, for cancer therapy. These studies demonstrated that SRT1720 could serve as an inhibitor of myeloma (13), however, it could also promote tumor cell migration and lung metastasis in mice (14). In addition, there has been conflicting data in the use of SRT1720 as a direct activator of SIRT1 (8).
In the present study we sought to determine if SRT1720 could affect breast cancer cells in vitro and in vivo in a SIRT1-dependent manner. We observed that SRT1720 caused increased cell death in multiple breast cancer cell lines and other tumor types. In the panel of breast cancer cell lines that we tested, the basal type cells including MDA-MB-231 were more sensitive to SRT1720 than luminal-type MCF-7 or non-transformed cells. However, unexpectedly the increased cell death with SRT1720 treatment in vitro was independent of SIRT1 levels. We also treated mice with SRT1720 in which neu mouse mammary tumor cells were grown in the mammary fat pad. SRT1720 also inhibited the growth of neu tumors, but in contrast to in vitro, this effect was partially dependent on SIRT1. It is unclear why there was a difference but it may be due to the function of SIRT1 in the stromal environment. It will be important to explore the role of SIRT1 and the tumor extracellular environment. From our in vitro experiments, we determined that SRT1720 caused cell death by necrosis but not apoptosis. We observed that SRT1720 activated several cellular stress pathways including autophagy and the unfolded protein response. However, the effect of SRT1720 was independent of the activation of these pathways, because inhibition of either autophagy or the unfolded protein response through either chemical or genetic means did not inhibit SRT1720-induced cell death. Interestingly, we found that MDA-MB-231 cells are more resistant to SRT1720 treatment when cultured with conditioned media from cells plated at a high density. This observation suggests that a secreted factor from MDA-MB-231 cells may attenuate the effect of SRT1720. Future studies should be directed to identify this secreted factor as it may help to further understand the mechanism of action of SRT1720.
We identified lysosomal membrane permeabilization as the cause of SRT170-induced cell death. This could be verified with three different chemical modifiers of lysosomal function including bafilomycin A1, chloroquine, and ammonium chloride, which could all inhibit lysosomal membrane permeabilization and cell death caused by SRT1720. It has been established that cancer cells exhibit structural and functional changes in their lysosomes as compared with normal cells that allow cancer cells to survive (44). Therefore, targeting lysosomes to induce cell death has emerged as a possible target for cancer therapy to overcome apoptosis resistant cells and drug resistance to chemotherapy (31). Many therapeutic agents have been reported to effect the death of cancer cells through modulation of lysosome function (25). Therefore, SRT1720 may serve as a useful agent for studying lysosomal-dependent cell death in cancers, which may eventually lead to the development of effective therapeutic approaches for cancer prevention and treatment.
Supplementary Material
Acknowledgements
We thank Ms. Cuiling Li for technical assistance and Dr. Craig J. Thomas for synthesis of SRT1720. We are grateful for the critical reading and helpful discussion of the manuscript by members of the Deng laboratory. This work was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
We declare that there are no conflicts of interest for this study.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Hudis CA, Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16(Suppl 1):1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- 3.Schmadeka R, Harmon BE, Singh M. Triple-negative breast carcinoma: current and emerging concepts. American journal of clinical pathology. 2014;141:462–477. doi: 10.1309/AJCPQN8GZ8SILKGN. [DOI] [PubMed] [Google Scholar]
- 4.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. The Journal of clinical investigation. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang RH, Xu X, Kim HS, Xiao Z, Deng CX. SIRT1 deacetylates FOXA2 and is critical for Pdx1 transcription and beta-cell formation. International journal of biological sciences. 2013;9:934–946. doi: 10.7150/ijbs.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. The Journal of biological chemistry. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:7271–7279. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann SH, Vaux DL. Alterations in the apoptotic machinery and their potential role in anticancer drug resistance. Oncogene. 2003;22:7414–7430. doi: 10.1038/sj.onc.1206945. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan D, Bandi M, Singh AV, Ray A, Raje N, Richardson P, et al. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. British journal of haematology. 2011;155:588–598. doi: 10.1111/j.1365-2141.2011.08888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Hayashi R, Ichikawa T, Imanishi S, Yamada T, Inomata M, et al. SRT1720, a SIRT1 activator, promotes tumor cell migration, and lung metastasis of breast cancer in mice. Oncology reports. 2012;27:1726–1732. doi: 10.3892/or.2012.1750. [DOI] [PubMed] [Google Scholar]
- 15.Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–7523. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [see comments]. [DOI] [PubMed] [Google Scholar]
- 17.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 19.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 Activation Mimics Low Energy Levels and Protects against Diet-Induced Metabolic Disorders by Enhancing Fat Oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell metabolism. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aits S, Jaattela M. Lysosomal cell death at a glance. Journal of cell science. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 26.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. European journal of pharmacology. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. The Journal of biological chemistry. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 28.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. The Journal of cell biology. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nature reviews Drug discovery. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 30.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groth-Pedersen L, Jaattela M. Combating apoptosis and multidrug resistant cancers by targeting lysosomes. Cancer letters. 2013;332:265–274. doi: 10.1016/j.canlet.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Guarante LP. Regulation of Aging by SIR2. Ann N Y Acad Sci. 2005;1055:222. [Google Scholar]
- 33.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 34.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 35.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 36.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free radical biology & medicine. 2013;56:133–171. doi: 10.1016/j.freeradbiomed.2012.10.525. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Kazgan N. Mammalian sirtuins and energy metabolism. International journal of biological sciences. 2011;7:575–587. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herranz D, Maraver A, Canamero M, Gomez-Lopez G, Inglada-Perez L, Robledo M, et al. SIRT1 promotes thyroid carcinogenesis driven by PTEN deficiency. Oncogene. 2013;32:4052–4056. doi: 10.1038/onc.2012.407. [DOI] [PubMed] [Google Scholar]
- 43.Fang Y, Nicholl MB. Sirtuin 1 in malignant transformation: friend or foe? Cancer letters. 2011;306:10–14. doi: 10.1016/j.canlet.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.