SUMMARY
Mammals exhibit marked inter-individual variations in their gut microbiota, but it remains unclear if this is primarily driven by host genetics or by extrinsic factors like dietary intake. To address this, we examined the effect of dietary perturbations on the gut microbiota of five inbred mouse strains, mice deficient for genes relevant to host-microbial interactions (MyD88−/−, NOD2−/−, ob/ob, and Rag1−/−), and >200 outbred mice. In each experiment, consumption of a high-fat, high-sugar diet reproducibly altered the gut microbiota despite differences in host genotype. The gut microbiota exhibited a linear dose response to dietary perturbations, taking an average of 3.5 days for each diet-responsive bacterial groups to reach a new steady state. Repeated dietary shifts demonstrated that most changes to the gut microbiota are reversible, while also uncovering bacteria whose abundance depends on prior consumption. These results emphasize the dominant role that diet plays in shaping inter-individual variations in host-associated microbial communities.
INTRODUCTION
Although humans and other mammals exhibit many shared features of their resident gut microbial communities (Muegge et al., 2011), each individual harbors an idiosyncratic mixture of microbial strains and species (Faith et al., 2013). In healthy adults, the component members of the gut microbiota can be stable for years (Faith et al., 2013), whereas the relative abundance of each member, i.e. community structure, is highly dynamic (David et al., 2014). The underlying causes and consequences of these inter-individual and temporal variations remain poorly characterized.
Studies in animal models have led to the proposal that the gut microbiota might be considered a complex polygenic trait shaped by both environmental and host genetic factors (Benson et al., 2010). However, it remains unclear if host genotype or environment (e.g., diet) plays a more dominant role in shaping microbial ecology. Associations between genetic loci and the abundance of bacterial taxa have been described in mice fed a controlled diet (Benson et al., 2010; McKnite et al., 2012) and in humans (Li et al., 2012; Smeekens et al., 2014), the genetic distance between mouse strains was recently linked to the overall structure of the gut microbiota (Hildebrand et al., 2013), and numerous differences have been shown between transgenic animals and matched controls (Couturier-Maillard et al., 2013; Hashimoto et al., 2012; Spor et al., 2011).
Conversely, time series analyses of inbred mice have shown that the consumption of a high-fat, high-sugar diet dramatically alters the gut microbiota in a single day (Turnbaugh et al., 2008; Zhang et al., 2012). Endpoint analyses of multiple inbred mouse strains support the importance of current dietary intake (Parks et al., 2013), as does a recent study, which demonstrates that the microbial responses to the fucosylation of host glycans are diet-dependent (Kashyap et al., 2013). Finally, comparisons of human twins at various ages, from infants to adults, have failed to detect significantly more similar microbial communities in mono- versus dizygotic pairs, suggesting that environmental factors predominate over host genetics in shaping microbial ecology (Turnbaugh et al., 2009; Yatsunenko et al., 2012).
Here, we systematically test the relative impacts of dietary intake and host genetics on the gut microbiota, through the combined analysis of 5 inbred mouse strains, 4 transgenic lines, and a recently developed outbred mouse resource, the Diversity Outbred (DO) population (Churchill et al., 2012; Svenson et al., 2012). The DO population was derived from partially inbred lines of the Collaborative Cross (Threadgill and Churchill, 2012) that were outbred using a randomized breeding scheme with avoidance of sibling matings to obtain high genetic diversity, heterozygosity, and fine recombination block structure (Svenson et al., 2012). These traits make the DO population ideal for investigating the relative contributions of host and environmental factors in shaping complex traits.
We selected two diets that reflect distinctive macronutrient profiles, are widely used to study diet-induced obesity, and represent modern human dietary regimes: a low-fat, high-plant-polysaccharide diet (LFPP: 22.2% kcal protein, 16.0% fat, 61.7% carbohydrate) and a high-fat, high-sugar diet (HFHS: 14.8% kcal protein, 40.6% carbohydrate, 44.6% fat). Consecutive dietary shifts, longitudinal sampling, dietary mixtures, and computational modeling of microbial dynamics showed that inter-individual and inter-strain differences are rapidly and consistently reset by dietary perturbations. Our results also provide evidence that members of the gut microbiota undergo hysteresis in response to diet: their abundance is shaped by both current dietary intake and past exposures. Together, these findings have important implications for understanding host-microbial interactions during the consumption of a wide range of dietary regimes, and suggest that attempts to understand the genetic underpinnings of the human gut microbiota will require a careful consideration of our past and present dietary habits.
RESULTS
Consistent microbial responses to a high-fat, high-sugar diet in inbred mice
16S rRNA gene sequencing was used to profile the fecal microbiota of 73 mice representing 5 distinct inbred founder strains used in the Collaborative Cross: 129S1/SvImJ, A/J, C57BL/6J, NOD/LtJ, and NZO/HlLtJ (n=7–20 mice/genotype; 3–9 cages/genotype). For each genotype, we fed mice either a low-fat, high-plant-polysaccharide diet (LFPP) or a high-fat, high-sugar diet (HFHS) for ≥15 weeks (Figure S1a, Table S1a). 16S rRNA gene sequences were clustered into operational taxonomic units (OTUs) using a reference tree guided approach, prior to unsupervised ordination using Bray-Curtis dissimilarity-based principal coordinates analysis (PCoA).
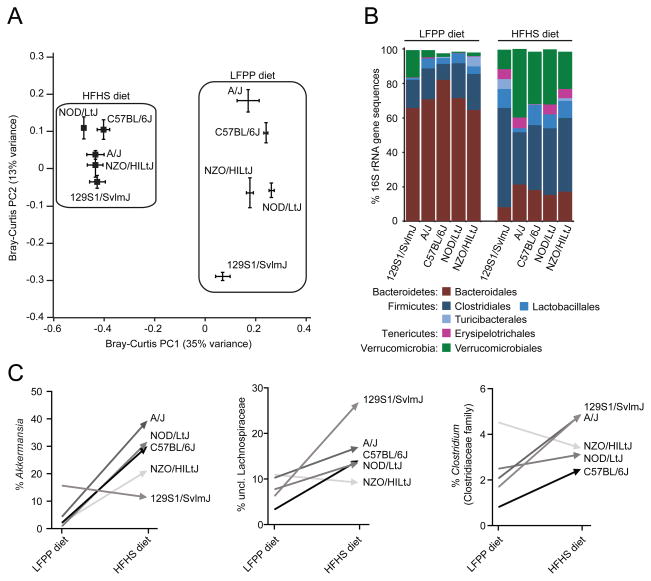
Consumption of the HFHS diet consistently modified the gut microbiota of all 5 strains such that all strains formed two distinct clusters defined by each diet [Figure 1a; p-value<0.001, permutational multivariate analysis of variance (PERMANOVA) of Bray-Curtis distances]. This shift was evident at the phylum level: the HFHS diet significantly increased the relative abundance of the Firmicutes (21.39±1.29% LFPP versus 52.92±4.45% HFHS), increased the Verrucomicrobia (5.90±1.20% LFPP versus 26.15±3.98% HFHS; see Supplemental Results), and decreased the Bacteroidetes (71.02±1.84% LFPP versus 16.35±1.48% HFHS) (all comparisons p-value<0.0001, Wilcoxon rank-sum test; Figure 1b). These trends were also detectable at finer taxonomic levels, ranging from class to genus (Table S2a).
Figure 1. Microbial responses to the high-fat, high-sugar diet in inbred mice.
(A) Microbial community structure is primarily determined by diet (see PC1; F=38.0, p-value<0.001, PERMANOVA on Bray-Curtis distances). Secondary clustering is by host genotype [see PC2; F=9.8, p-value<0.001 (LFPP) and F=2.9, p-value<0.001 (HFHS), PERMANOVA after splitting the datasets by diet]. Bray-Curtis dissimilarity-based principal coordinates analysis (PCoA) was performed on 16S rRNA gene sequencing data; the first two coordinates are shown (representing 48% of the total variance). Values are mean ± sem (n=2–13 animals/group). (B) Relative abundance of major taxonomic orders in 5 strains fed a LFPP or HFHS diet. Groups within the same bacterial phyla are indicated by different shades of the same color. Taxa with a mean relative abundance >1% are shown. (C) Diet-dependent bacterial genera with distinctive changes between genotypes. See Table S2a for the full set of taxa. Different genotypes are indicated by the shade of each line. Values in panels B,C are means (n=2–13 animals/group). See also Figure S1 and Tables S1,S2.
When considering each diet independently, we detected significant clustering by host genotype (p-value<0.001, PERMANOVA of Bray-Curtis distances). We also detected multiple taxa with significantly altered relative abundance between inbred strains; however, only 3 bacterial genera were significantly enriched in one mouse line relative to the other four (Table S3a). De novo clustering of our 16S rRNA gene sequences into OTUs confirmed these trends, while also identifying a member of the Erysipelotrichaceae family (OTU13400) that was consistently enriched in C57BL/6J mice regardless of diet (Table S3b). Interestingly, both the magnitude and direction of change for multiple bacterial genera were different between genotypes (Figure 1c, Table S2a), suggesting that the impact of the HFHS diet may depend on the broader host or microbial community context.
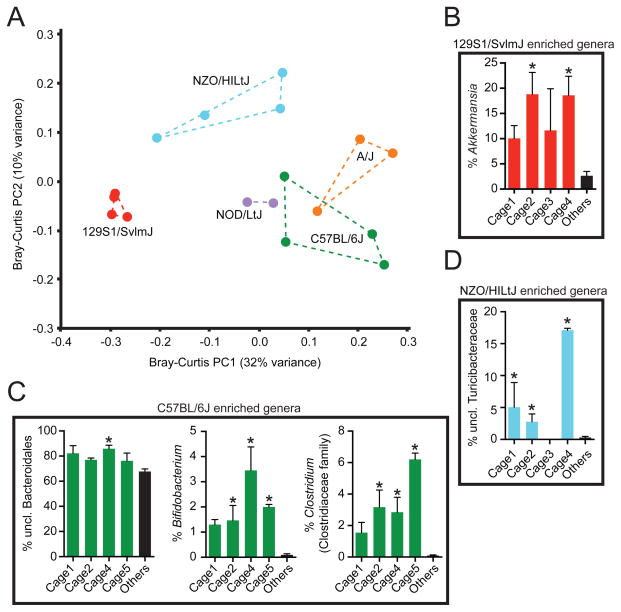
To test if these correlations were driven by host genotype or reflected a shared history of microbial exposures, we sampled multiple cages from each inbred line, each representing a unique set of littermates co-housed since birth and split according to sex at 3 weeks of age. Principal coordinates analysis confirmed that many of the differences between genotypes were consistently found in all cages (Figure 2a). Correspondingly, we identified bacterial genera that were significantly enriched in 129S1/SvlmJ, C57BL/6J, and NZO/HILtJ mice from multiple cages (Figures 2b–d). However, we also detected one genus that was only significantly altered in one cage (Figure 2c), and we did not detect any genera (or higher-level taxa) that were consistently enriched in the same host genotype on both diets (Table S3). Thus, our results imply that the effects of dietary intake overshadow any pre-existing differences between strains due to host genotype. Furthermore, we cannot fully exclude the possibility that some of the observed associations with host genotype may actually be the result of a shared history of environmental exposures.
Figure 2. Genotype-associated shifts in the gut microbiota are robust to cage effects.
(A) Microbial community structure is consistent between cages. Bray-Curtis dissimilarity-based principal coordinates analysis (PCoA) was performed on 16S rRNA gene sequencing data collected during consumption of the LFPP diet. Each point represents a different cage; colored lines connect cages housing mice from the same genotype. (B–D) Relative abundance of bacterial genera that are associated with host genotype on the LFPP diet: (B) 129S1/SvlmJ (red), (C) C57BL/6J (green), and (D) NZO/HILtJ (blue) (also see Table S3a). Values are mean ± sem (n=2–5 mice/cage; 2–4 cages/genotype). Asterisks represent significant differences (p-value<0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). See also Figure S1 and Tables S1,S3.
Next, we sought to test if diet is capable of reproducibly shaping the gut microbiota in the context of more dramatic perturbations to host genotype and phenotype. Male C56BL/6J wild-type mice were fed a LFPP diet until they reached 7 weeks of age, at which point they were switched to the HFHS diet for one week. Fecal samples from multiple days prior to and after the diet shift were used for 16S rRNA gene sequencing (Figure S1b, Table S1b). The same procedure was performed for animals homozygous for mutations in four genes that have been previously shown to impact the gut microbiota (n=5 mice/genotype): MyD88−/− (Wen et al., 2008); NOD2−/− (Couturier-Maillard et al., 2013); ob/ob (Ley et al., 2005); and Rag1−/− (Scholz et al., 2014).
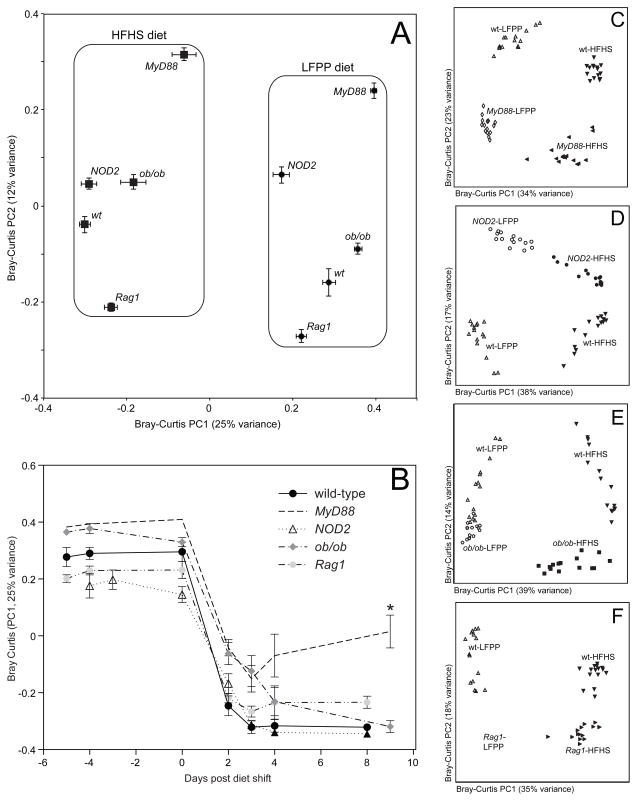
The HFHS diet consistently modified the gut microbiota of wild-type animals and all 4 transgenic lines (Figure 3a), with significant clustering by diet across the entire dataset (p-value<0.001, PERMANOVA of Bray-Curtis distances). Within each diet, there was significant clustering according to host genotype (p-value<0.001, PERMANOVA on the final time-point for each diet). Analysis of microbial community structure over time revealed that all genotypes responded within 2 days (Figure 3b); however, MyD88−/− animals exhibited a blunted response to the HFHS diet (p-value<0.05, Kruskal-Wallis test with Dunn’s correction for multiple comparisons using data from the final timepoint). In order to account for differences in sample number between groups, we re-analyzed the data for each transgenic mouse strain and the wild-type control (n=15 samples/group; the final timepoint on the HFHS diet was excluded to allow for matched comparisons on both diets). These analyses again identified diet as the primary factor driving the observed variations in microbial community structure, with genotype providing secondary clustering (Figures 3c–f). Pairwise statistical analysis of the final timepoint on each diet confirmed these trends, with diet explaining more of the observed variation than host genotype [R2 ranges: 0.08–0.23 (genotype) and 0.35–0.48 (diet); ADONIS test]. Diet also consistently altered community membership as indicated by the unweighted UniFrac metric (data not shown).
Figure 3. Microbial responses to the high-fat, high-sugar diet in transgenic mice.
(A) Microbial community structure is primarily determined by diet (see PC1; F=60.8, p-value<0.001, PERMANOVA on Bray-Curtis distances). Secondary clustering is by host genotype (see PC2; F=17.9, p-value<0.001). Bray-Curtis dissimilarity-based principal coordinates analysis (PCoA) was performed on 16S rRNA gene sequencing data; the first two coordinates are shown (representing 37% of the total variance). Values are mean ± sem (n=15–20 samples/group). (B) Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from the Bray-Curtis-based PCoA. Points and lines are labeled based on host genotype. Values are mean ± sem (n=5 mice/group). The asterisk represents significant differences at the final timepoint relative to wild-type controls (p-value<0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). (C–F) Bray-Curtis-based PCoA of the fecal microbiota of animals on a LFPP (white filled symbols) or HFHS (black filled symbols) diet (n=15 samples/group). Wild-type controls are included in all panels, indicated by white triangles (LFPP diet) and inverted black triangles (HFHS diet). Transgenic mice include: (C) MyD88−/− LFPP (white diamonds) and HFHS (black leftward triangles); (D) NOD2−/− LFPP (white circles) and HFHS (black circles); (E) ob/ob LFPP (white pentagons) and HFHS (black squares); and (F) Rag1−/− LFPP (white squares) and HFHS (black tilted triangles). See also Figure S1 and Tables S1,S2.
The HFHS diet significantly altered bacteria at taxonomic levels ranging from phylum to genus (Table S2a), confirming the overall trend of increased Firmicutes and decreased Bacteroidetes. Although not all diet-associated groups reached significance in both the transgenic and inbred strain experiments, the direction of change was consistent in all cases where a significant association was found in both datasets. The identified diet-dependent bacterial taxa were also generally consistent in their direction of change in each mouse strain following consumption of the HFHS diet; in all cases where there was a disagreement it was due to a single genotype (Table S2a).
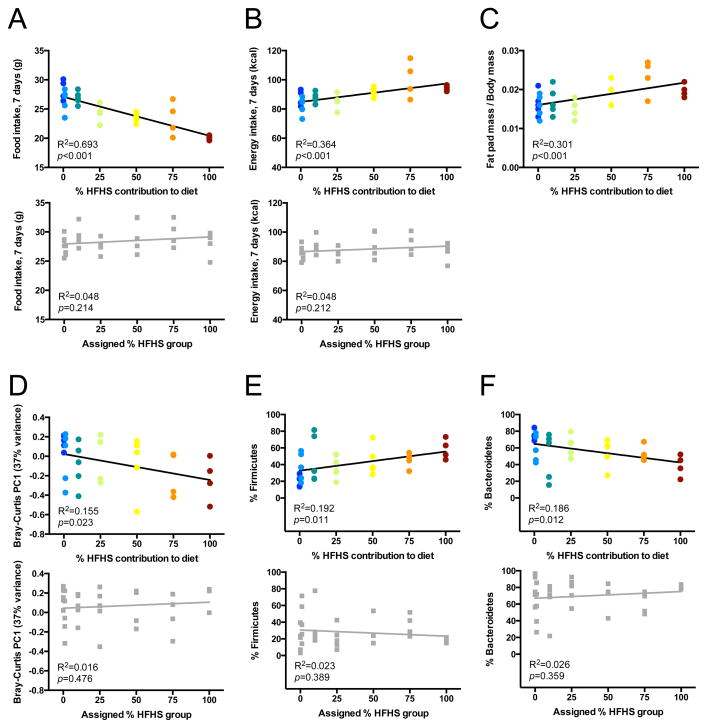
One explanation for the marked impact of the HFHS diet across multiple inbred strains, including transgenic animals, is that this represents a relatively strong dietary shift: 16.0% to 40.6% fat accompanied by a shift from plant polysaccharides to more readily digestible carbohydrates. Ecological theory predicts that the gut microbiota may be able to tolerate more subtle dietary interventions (Costello et al., 2012). To test if the magnitude of microbial response is directly proportional to the degree of dietary perturbation, we fed adult male C57BL/6J wild-type mice re-pelleted mixtures of the LFPP and HFHS diet (0, 1, 10, 25, 50, 75, and 100% HFHS by weight; n=4–5 mice/diet). Fecal samples were collected prior to and 7 days after each dietary intervention and analyzed by 16S rRNA gene sequencing (Figure S1c, Table S1c). Dietary HFHS content influenced host adiposity over the 7 days of gradient feeding. Despite reducing food intake as the proportion of the HFHS diet increased (Figure 4a), mice consumed more calories overall on HFHS-rich diets (Figure 4b). Correspondingly, we observed HFHS-dependent increases in epididymal fat pad mass, whether measured on an absolute basis (R2=0.272; F=11.93; p-value=0.002) or as a fraction of body mass (Figure 4c).
Figure 4. Microbial responses are proportional to the degree of dietary perturbation.
(A–C) Physiological responses of mice to diets with differing HFHS contents: (A) food intake decreases as dietary HFHS content increases; nevertheless both (B) caloric intake and (C) body fat increase on HFHS-rich diets. (D) Dose-dependent relationship between dietary HFHS content and the first principal coordinate from a Bray-Curtis dissimilarity-based PCoA of microbial community composition. (E, F) Dose-dependent relationships between dietary HFHS content and the two most abundant diet-associated bacterial phyla: (E) Firmicutes increase with HFHS content; (F) Bacteroidetes decrease with HFHS content. Within each panel, the upper graph (colored circles) represents data collected during gradient feeding, whereas the lower graph (grey squares) represents data collected during the baseline week, when mice had been assigned to a diet group but had not yet initiated gradient feeding. R2 and p-values reflect linear regression (n=4–5 animals/group). See also Figure S1 and Tables S1,S2.
HFHS content was also a significant predictor of gut microbial community structure. Bray-Curtis dissimilarity-based PCoA revealed significant clustering of the gut microbiota by diet group (p-value<0.01; PERMANOVA of Bray-Curtis distances), with diet separating microbial communities along the first principal coordinate (PC1, 37% of variance). PC1 values decreased linearly as HFHS content increased (Figure 4d). We also observed correlations between HFHS content and the relative abundance of microbial taxa previously associated with the HFHS diet, including an increased proportion of Firmicutes (Figure 4e) and a decreased proportion of Bacteroidetes (Figure 4f). Verrucomicrobia, a phylum associated with the HFHS diet in the earlier experiments, was either below the limit of detection or present at low abundance in all samples from this experiment, regardless of diet (mean % of 16S sequences: 0.0013±0.0001). These linear associations were also detectable at the genus level (Table S2b), confirming many of the previously observed differences between the LFPP and HFHS diet (Table S2a). Together, these data suggest that the gut microbiota responds to diet in a dose-dependent manner with even subtle perturbations detectably changing microbial community structure.
A rapid and sustained microbial response to the HFHS diet in outbred mice
To more systematically vary host genetics in mice with a common history of environmental exposures, we obtained 60 Diversity Outbred (DO) mice (30 females, 30 males). All mice were from generation 6 of outbreeding, each representing a unique combination of 8 founder alleles from the 5 strains surveyed previously, in addition to CAST/EiJ, PWK/PhJ, and WSB/EiJ (Churchill et al., 2012; Svenson et al., 2012). Genome-wide analyses of our animals confirmed that we successfully recaptured nearly all of the genetic diversity in the DO population (FST=0.05 for the 60 animals in the DO time series experiment; FST=0.03 for the expanded set of 203 animals). Consecutive dietary shifts were employed to assess the speed, reproducibility, and reversibility of the microbial responses to dietary perturbations. Individually housed mice were fed a LFPP diet from weaning until 7 weeks of age, at which time they were fed a HFHS for one week, returned to the LFPP diet for one week, and then maintained on the HFHS diet for 2 months. Fecal pellets were analyzed from the 60 mice at 18 timepoints, spanning 14 weeks (n=977 samples; Figure S1d, Table S1d).
Consistent with our analysis of inbred strains and transgenic mice, the HFHS diet rapidly altered microbial community membership (Figure S2a) and structure (Figures 5a, b) despite substantial variations in host genotype (Svenson et al., 2012). The diet shifts also reset inter-individual variability in community structure (i.e. β-diversity patterns). Mantel tests were performed for all pairwise comparisons of Bray-Curtis distance matrices representing differences in gut community structure between animals at each timepoint. Although within each diet β-diversity was stable over time (median p-value<0.001), microbial community structure was rapidly altered by the HFHS diet such that the differences between mice could not be accurately predicted using samples collected on different diets (median p-value>0.1; Figure S2b).
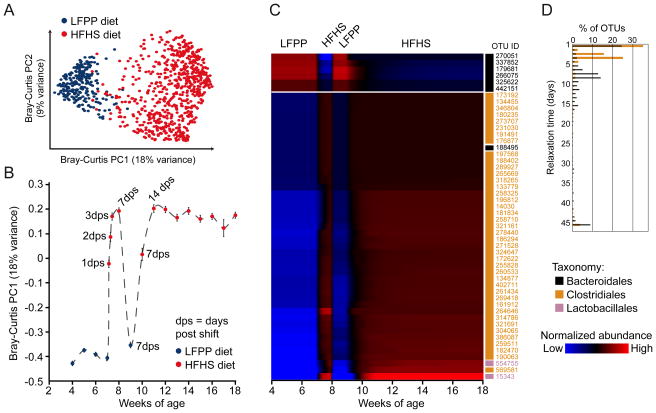
Figure 5. A rapid and reproducible microbial response to the high-fat, high-sugar diet in outbred mice.
(A) Bray-Curtis-based PCoA analysis of the fecal microbiota of animals on a LFPP (blue) or HFHS (red) diet. The first two principal coordinates are shown (representing 27% of the total variance), which clearly separate the 977 fecal samples by diet. (B) Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from the Bray-Curtis-based PCoA. Points are labeled based on the current diet: LFPP (blue) or HFHS (red). Samples were collected weekly, with daily sampling during the first week of the HFHS diet (indicated by the number of days post shift, dps). On average, 52 mice were sampled at each timepoint. Values are mean ± sem. (C) Time-map of consistently responsive species-level bacterial operational taxonomic units (OTUs) in outbred mice. The selected OTUs were present, responsive, and had consistent temporal patterns in ≥50% of mice. Each row represents a consensus temporal signature (aggregated model estimates across mice) for an OTU, ordered by agglomerative clustering of signatures. Blue indicates relative abundances below the mean abundance for the entire signature, and red indicates relative abundances above the mean. Values represent model estimates, in units of log transformed and standardized relative abundances. The taxonomic assignment for each OTU is indicated on the right of the heatmap: Bacteroidales (black), Clostridiales (orange), and Lactobacillales (pink). (D) Relaxation time constant distributions on the second HFHS diet regimen, for OTUs belonging to the bacterial orders Clostridiales and Bacteroidales (data from all OTUs are shown, including those with inconsistent behavior across mice). Relaxation time characterizes how quickly an OTU’s relative abundance reaches an equilibrium level, with shorter times indicating more rapid equilibration. See also Figures S1–4 and Tables S1,S2,S4.
As with the inbred mice, consumption of the HFHS diet by outbred mice led to a significant increase in the Firmicutes phylum (39.10±1.17% LFPP versus 64.65±0.73% HFHS), and a decrease in the Bacteroidetes (57.81±1.25% LFPP versus 32.58±0.73% HFHS; both comparisons p-value<0.0001, Wilcoxon rank-sum test; Figure S2c). The Verrucomicrobia were consistently found at low abundance in the outbred mice (<1% maximum abundance across the entire time series), but were still significantly increased on the HFHS diet (2.4e−4±9.3e−5% LFPP versus 5.1e−3±1.8e−3% HFHS; p-value<0.0001, Wilcoxon rank-sum test; see Supplemental Results). We also detected Lactococcus lactis on the HFHS diet, likely due to the presence of free nucleic acids in fermented casein (see Supplemental Results). These trends were detectable at multiple taxonomic levels, and were confirmed on an independent set of 143 outbred mice sampled after 21 weeks on either the LFPP or HFHS diet (Figure S3 and Tables S1e, S2a).
To assess the time-dependent responses of individual species-level bacterial operational taxonomic units (OTUs), we implemented the Microbial Counts Trajectories Infinite Mixture Model Engine (MC-TIMME) model (Gerber et al., 2012) (Figure S4a). MC-TIMME uses nonparametric Bayesian methods to infer patterns of change in OTU relative abundances over time, referred to as signatures (see Supplemental Experimental Procedures for model validation). We focused on the 81 OTUs that were present and responsive to dietary changes in >50% of mice. An additional 389 OTUs were responsive to diet in <50% of the animals, due to genotypic, environmental, or stochastic effects. We also found 38 OTUs that were present in >50% of the mice but non-responsive to diet (Table S2c). Non-responsive OTUs were significantly enriched for members of the Ruminococcaceae family (55% relative to 17% of the diet-responsive OTUs; p-value<10−4, Fisher’s Exact Test), and found at a significantly higher median relative abundance as compared to the diet-responsive OTUs (2.5×10−2±4.7×10−3 versus 8.3×10−3±3.8×10−3; p-value<10−10, Wilcoxon rank-sum test). Our model found that 62% of the 81 diet-responsive OTUs exhibited consistent patterns of change across ≥50% of the mice (Figure 5c, Table S4a). Almost all (98%) of the analyzed OTUs that increased on the HFHS diet were assigned to the Firmicutes phylum, whereas all of the OTUs that increased on the LFPP diet were Bacteroidetes.
We next used our model to calculate relaxation times, representing the time constant (half-life) for each OTU to reach a new steady state after a dietary switch. OTUs with consistent patterns of change in response to the dietary shift generally exhibited short relaxation times (3.57±0.33 days; Table S4a). Overall, members of the Bacteroidales order had significantly longer relaxation times than the Clostridiales (Figure 5d; median of 8.6 versus 3.4 days, respectively; p-value<10−16, Wilcoxon rank sum test). These results are consistent with recent evidence that Clostridiales may be more active than the Bacteroidales (Maurice et al., 2013), allowing them to respond more rapidly to a dietary perturbation. Thus, despite marked host genetic diversity the gut microbiota exhibits a rapid and stereotypical microbial response to consumption of the HFHS diet.
Microbial responses to sequential diet shifts
Given the rapid and reproducible microbial response to HFHS intake, we next sought to test whether this phenomenon could be attributed fully to current dietary intake or whether there were lingering effects of past dietary history. Analysis of the consecutive dietary shifts in outbred mice suggested that both community membership and structure were markedly altered after 3–7 days of HFHS diet consumption in naïve mice, whereas the gut microbiota of mice previously exposed to the HFHS diet took 1–2 weeks to respond (Figure 5b). To more rigorously address this question, we collected daily fecal samples from inbred C57BL/6J mice (n=15) oscillating between LFPP and HFHS diets every 3 days. All animals were maintained on a LFPP diet in individual cages prior to the beginning of the experiment at 7 weeks of age. In total, we analyzed 4 groups, representing 2 sets of oscillating mice staggered by 3 days, and 2 control groups maintained on either a continuous LFPP or a continuous HFHS diet. All animals were switched to the HFHS diet for the final week. Daily fecal samples were subjected to 16S rRNA gene sequencing, representing 536 total samples across the 38-day experiment (Figure S1e, Table S1f).
Mouse physiology and chow consumption were rapidly altered by diet. Body weight consistently increased during consumption of the HFHS diet, while decreasing on the LFPP diet (Figure S5a). Similarly, control mice continuously fed a HFHS diet had an elevated weight relative to LFPP controls (Figure S5b). On average, each 3-day diet period resulted in 3.40±0.54% weight loss on the LFPP diet, and 5.95±0.56% weight gain on the HFHS diet (Figure S5c). Weight gain during the 5-week experiment was significantly higher in control mice continuously fed the HFHS diet, as compared to the mice on oscillating diets or LFPP-fed controls (Figure S5d). The mice on oscillating dietary regimens displayed excess caloric intake on the HFHS diet and diminished intake on the LFPP diet during the first day following each shift (Figures S5e, f).
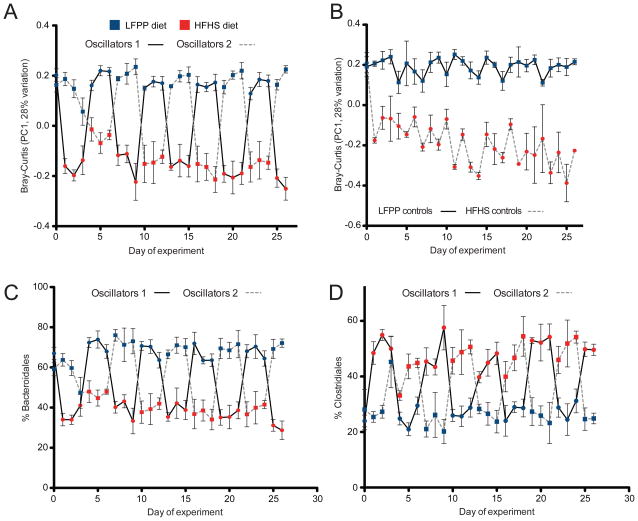
Analysis of microbial community membership (Figure S6a) and structure (Figures 6a, S6b) over time revealed that the gut microbiota was rapidly and consistently re-shaped by both diets. Control mice continuously fed the LFPP or HFHS diets were clearly distinguishable (Figure 6b). The observed changes to overall microbial community structure corresponded to rapid shifts in the relative abundance of the two major bacterial orders in the distal gut, the Bacteroidales (phylum: Bacteroidetes) and Clostridiales (phylum: Firmicutes) (Figures 6c, d). These high-level changes occurred consistently during each successive diet shift and were detectable after only a single day. These trends were consistent with our previous experiments and significant at multiple taxonomic levels (Table S2a).
Figure 6. Impact of successive dietary shifts on the gut microbiota.
(A) Analysis of the microbial response to the LFPP (blue) and HFHS (red) diet over time, using the first principal coordinate from the Bray-Curtis-based PCoA. The two oscillating groups are indicated by a solid line (group 1) or a dashed line (group 2). Timepoints are colored based on the diet consumed over the prior 24 hours; i.e. oscillator group 1 was switched onto the HFHS diet on day zero. (B) Results from control mice continuously fed a LFPP (solid line) or HFHS diet (dashed line). The full time series, including additional baseline and maintenance samples, is shown in Figure S6a,b. (C,D) The abundance of (C) the Bacteroidales (phylum: Bacteroidetes) and (D) Clostridiales (phylum: Firmicutes) is shown over time. The two oscillating groups are indicated by a solid line (group 1) or a dashed line (group 2). Values are mean ± sem (n=3–5 mice per group). See also Figures S1,S5,S6 and Tables S1,S2.
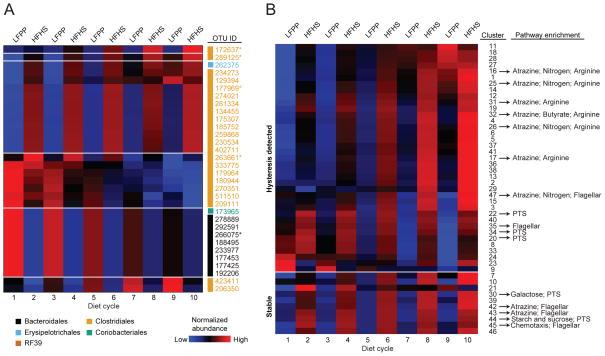
To analyze the impact of successive dietary perturbations on the time-dependent responses of species-level bacterial OTUs, we modified our MC-TIMME algorithm to model temporal signatures using simple linear models with constant levels for each dietary regimen (LFPP and HFHS) and linear increases or decreases with subsequent dietary switches (oscillation number) (Figure S4b). Our model merged data from the staggered (counter-oscillatory) experiments to produce consensus signatures for each OTU (see Supplemental Experimental Procedures). 125 OTUs were consistently responsive (present in >50% of mice, with significant changes on the first or last dietary shifts), and exhibited patterns of change in response to diet that were consistent in >50% of mice. 32 OTUs exhibited dependence of their levels over time on the serial dietary switches (Figures 7a, S7a–e, Table S4b), whereas 93 had a consistent difference in abundance on the LFPP versus HFHS diets but displayed no detectable change in abundance after each sequential shift (Figure S6c, S7f, g, Table S4c).
Figure 7. Identification of bacterial species and genes dependent on prior dietary intake.
(A) Relative abundance of species-level OTUs that were consistently present, responsive to diet, had consistent temporal patterns, and exhibited dependence of levels on serial dietary changes (see Experimental Procedures for thresholds used). Each row represents a temporal signature for an OTU (model estimate from combined data from the staggered dietary oscillation groups). Blue indicates relative abundances below the mean abundance for the entire signature, and red indicates relative abundances above the mean. Values represent model estimates, in units of log transformed and standardized relative abundances. The taxonomic assignments for each OTU are labeled on the right of each heatmap: Bacteroidales (black), Clostridiales (orange), Erysipelotrichales (blue), Coriobacteriales (green), and RF39 (brown). *OTUs with detailed graphs shown in Figure S7a–e. (B) Bacterial gene content (KEGG orthologous groups) was inferred using an ancestral state reconstruction method (Langille et al., 2013). The MC-TIMME algorithm identified 47 clusters of orthologous groups (mean of 68 orthologous groups per cluster) showing consistent differences in abundance on the LFPP versus HFHS diets. Each row in the time-map represents a consensus temporal signature for the indicated cluster. Blue indicates relative abundances below the mean abundance for the entire signature, and red indicates relative abundances above the mean. Values represent model estimates, in units of log transformed and standardized relative abundances. The top 37 clusters (above the white line) exhibited dependence of their levels over time on the serial dietary switches (hysteresis). See also Figures S1,S4,S6,S7 and Tables S1,S4.
We performed three analyses to assess whether these trends were primarily dependent on the dietary oscillations, rather than being dependent on temporal drift caused by other host or environmental factors. First, we found no bias in the temporal consistency of the behavior of the detected OTUs between the two groups of mice subjected to staggered dietary oscillations (p-value=0.94, paired t test corrected for differing group sizes). Second, we tested if the 32 OTUs exhibiting dependence on the serial dietary shifts were also altered over time in control mice maintained on a constant LFPP or HFHS diet. Of the 32 OTUs, only eight demonstrated any significant change in abundance over time on either the LFPP or HFHS diet (Table S4d; q-value<0.05, F-test and slope≥0.1).
Finally, we tested if the observed hysteresis patterns of species-level OTUs were accompanied by a change in the abundance of groups of functionally coherent bacterial genes. 16S rRNA gene sequencing data was used to predict the abundance of enzyme-level orthologous groups, which were then filtered and clustered according to their temporal dynamics with MC-TIMME (see Experimental Procedures). We identified 47 clusters of orthologous groups (containing on average 68 orthologous groups each) with a consistent difference in abundance on the LFPP versus HFHS diets (Figure 7b). 37 of these clusters exhibited dependence of their levels over time on the serial dietary switches (hysteresis, using the same criteria as for OTUs). The clusters that increased on the HFHS diet were significantly enriched for orthologous groups from pathways for the metabolism of sucrose, the dominant carbohydrate in the HFHS diet, including a phosphotransferase system for sucrose import (K02808/K02809) and a key enzyme for sucrose catabolism (levansucrase, K00692) (Table S4e). Of note, on the HFHS diet we also found a steady increase in the abundance of orthologous groups for urea metabolism (found within the “arginine metabolism” pathway), including urease(K01428-30), allophanate hydrolase (K01457), and urea carboxylase (K01941). We also observed significant enrichments for orthologous groups in the sucrose and arginine metabolism pathways in our outbred mouse time series experiment (data not shown).
Taken together, these results suggest that gut microbial community structure and metabolic activity are, at least in part, determined by prior dietary history (i.e. oscillation number) and not simply by current dietary intake. Our use of a relatively simple linear model provides a conservative estimate of these effects, which could conceivably include changes in equilibration time with serial dietary shifts or nonlinear dependencies on oscillation number that our model did not capture.
DISCUSSION
A recent endpoint analysis of 52 matched inbred strains of mice fed a comparable LFPP or HFHS diet reported a significant association between host genotype and microbial community structure (Parks et al., 2013). However, our current results, based on both endpoint and extensive time series analyses, emphasize that the microbial response to the consumption of the HFHS diet consistently overshadows pre-existing genetic associations. We found that (i) the gut microbiotas of inbred, transgenic, and outbred mice were consistently shaped by diet despite widely divergent genotypes, (ii) inter-strain and inter-individual differences in community structure were reset after dietary perturbation, and (iii) the changes in microbial community structure were rapid, reproducible, and reversible. These results suggest that environmental factors, namely host diet, play a dominant role in shaping gut microbial ecology.
The robust linkage between dietary intake and gut microbial community structure is exemplified by our analysis of C57BL/6J mice alternating between the LFPP and HFHS diet every 3 days. Remarkably, each diet shift rapidly and reproducibly reshaped the gut microbiota, detectable after a single day. Additional work is necessary to identify the mechanisms responsible for the observed changes to the gut microbiota during the consumption of the HFHS diet. Although total caloric intake has been associated with changes to the human gut microbiota (Jumpertz et al., 2011), studies in both mice and humans suggest that these effects are minimal relative to that of dietary composition (David et al., 2014; Ravussin et al., 2012). But what components of the LFPP and HFHS diets matter most? The answer will likely require systematic comparisons of defined mixtures that span the nutritional space encompassed by these two diets (Faith et al., 2011). Whether changes in the microbial community arise directly from the altered nutrient environment in the gastrointestinal tract, or indirectly due to effects of altered diets on host physiology that are consistent across genotypes, remains a fascinating area for future inquiry.
These studies also provide a basis from which to consider the many functional consequences of a gut microbiota altered by diet. Rapid re-structuring of the gut microbiota may contribute to the beneficial effects of time-restricted feeding (Hatori et al., 2012), and could broadly alter the microbial contributions toward metabolism and immunity. It will be important to determine if similar shifts occur in the context of humans undergoing analogous “yo-yo” diets reflecting alternating periods of increased and reduced caloric intake (Atkinson et al., 1994). More broadly, it is interesting to consider whether or not the observed microbial plasticity may be a selective trait that helped our ancestors maintain energy balance given a volatile diet that was dependent on season and foraging success (Hawkes et al., 1991). Diet-induced changes in the gut microbiota could also alter key metabolic activities, as shown recently for the interaction between dietary protein and the bacterial inactivation of cardiac drugs (Haiser et al., 2013).
Our data also support a role for host genetics and/or other environmental factors in mediating variations in community structure within each dietary regime. Controlling for diet, microbial communities clustered by genotype among inbred strains and transgenic mice. We identified 12 outbred mice with divergent community structure (2 that resembled LFPP during consumption of the HFHS diet, and 10 with the opposite response; Figure S3b). Our computational modeling also highlighted species-level bacterial OTUs that behaved inconsistently between mice. The range of relative abundances found within each diet group was substantial, especially for the Akkermansia genus (phylum: Verrucomicrobia), which dominated the gut microbiota of some mice and was nearly undetectable in many others (Figure S3c, d and Supplemental Results). Additional studies are warranted to determine what other host and/or environmental factors may drive variations in microbial community structure within and between animal facilities (Ivanov et al., 2009).
These results also demonstrate that during dietary perturbations the gut microbiota can exhibit hysteresis: the dependence of a complex system not only on its current environment but also on past exposures. These effects have been described previously in a wide range of fields, including ecology (Scheffer et al., 2001), immunology (Das et al., 2009), developmental biology (Sha et al., 2003), economics (Blanchard and Summers, 1987), and physics (Jiles and Atherton, 1986). There is also preliminary evidence for hysteresis in the human gut microbiota following repeated exposure to broad-spectrum antibiotics (Gerber et al., 2012). Additional work is needed to test the underlying source of this effect, whether it is due to a dynamic lag between the dietary input and the change in bacterial abundance (i.e. rate-dependent hysteresis) or to another as-of-yet unknown mechanism that allows the gut microbiota to remember its past dietary exposures (e.g., through bacterial interaction networks) (Levy and Borenstein, 2013).
Finally, this study provides a cautionary note for ongoing efforts to link host genetics to the composition and function of the human gut microbiota. Perhaps more so than any other complex trait, the gut microbiota is shaped by a wide range of environmental factors, including diet (David et al., 2014), antibiotics and other drugs (Maurice et al., 2013), and geography (Yatsunenko et al., 2012). It will be difficult to identify host factors that are reproducibly associated with alterations to the gut microbiota without adequately controlling for these environmental factors. We anticipate that studies of large cohorts of outbred mice, fed a wide range of diets, coupled to experiments with inbred, transgenic, and gnotobiotic animal models, will be an important strategy to elucidate the complex interrelationships between diet, host genetics, and the gut microbiota.
EXPERIMENTAL PROCEDURES
Animal husbandry
Animal procedures were approved by the Jackson Lab and Harvard University. The LFPP diet (LabDiet, Richmond, IN; 5K52) contained 16.0% kcal from fat (primarily soybean oil) and 61.7% from carbohydrates (primarily whole grains). The HFHS diet (Harlan Laboratories Inc, Madison, Wisconsin USA; TD.08811) contained 44.6% kcal from fat (primarily milk-derived saturated fat) and 40.6% from carbohydrates (sucrose, maltodextrin, and corn starch). Mice in the dietary gradient experiment were fed custom diets representing defined mixtures of TD.08811 and TD.96338, a Harlan product that is the nutritional equivalent of LabDiet 5K52.
16S rRNA gene sequencing and analysis
DNA was extracted using the PowerSoil bacterial DNA extraction kit (MoBio, Carlsbad CA) and PCR-amplified using barcoded universal bacterial primers targeting variable region 4 of the 16S rRNA gene. Barcoded amplicons from multiple samples were pooled and sequenced using the Illumina HiSeq platform. 16S rRNA gene sequences were analyzed on the Harvard Odyssey cluster using the QIIME (Quantitative Insights Into Microbial Ecology) software package (Caporaso et al., 2010) along with custom Perl scripts. LefSe was used to identify taxonomic groups associated with each group (Segata et al., 2011).
Data pre-processing for dynamical modeling
An operational taxonomic unit (OTU) was defined as present in a given mouse if there were ≥10 sequence counts at ≥5 timepoints. For the outbred mouse time series, we defined each OTU as responsive in a given mouse if its relative abundance on the first LFPP diet interval was significantly different from that on the final HFHS diet interval (q-value<0.05, Wilcoxon rank sum test) (Benjamini and Hochberg, 1995). For the dietary oscillation experiment, we defined each OTU as responsive in a given mouse if its relative abundance on all LFPP timepoints was significantly different compared to its relative abundance on all HFHS timepoints (q-value<0.05, Wilcoxon rank sum test).
Modeling of dynamics
We customized the MC-TIMME model (Gerber et al., 2012) to capture dynamics of the outbred mouse time series and dietary oscillation experiments. This model uses nonparametric Bayesian methods to infer prototypical patterns of change in OTU relative abundances over time (signatures), and groups of OTUs with similar temporal behavior (signature groups). For the outbred mouse experiments, we used exponential relaxation process models to capture the dynamics of dietary shifts. For the dietary oscillation experiments we adjusted the MC-TIMME model to assess the dependence of changes in the microbiota on repeated dietary shifts. See Supplemental Experimental Procedures for justifications of our assumptions and a detailed model validation.
Analysis of gene and pathway abundances
PICRUSt version 0.9.1 (Langille et al., 2013) was used to impute the abundance of KEGG orthologous groups (KOs). We retained KOs with >100 predicted counts in ≥5 time-points in >50% of mice and a significant response to diet in >50% of mice (Wilcoxon rank-sum test, q-value<0.05). Post-filtering, we clustered the remaining KOs based on their temporal abundance patterns using the MC-TIMME algorithm. We then used the hypergeometric test to evaluate pathway-level enrichment.
Statistical analysis
Statistical analyses were performed using GraphPad Prism. Data are expressed as mean ± sem and significance was set at a two-tailed p-value<0.05, unless otherwise indicated.
Supplementary Material
Figure S1. Experimental design, related to Figures 1–7. (A) Male and female mice from 5 inbred strains were fed a LFPP or HFHS diet for ≥15 weeks prior to sample collection (Table S1a). (B) Male C56BL/6J wild-type mice and four transgenic lines were fed a LFPP diet until they reached 7 weeks of age, at which point they were switched to the HFHS diet for one week (Table S1b). (C) Male C57BL/6J and 129S1/SvImJ wild-type mice were fed defined mixtures of the LFPP and HFHS diet (Table S1c). (D) Outbred mice were weaned onto a LFPP diet (blue), prior to a 1-week of the HFHS diet (red), 1 week LFPP, and then 9 weeks of maintenance on the HFHS diet. Microbial community structure was determined over time (Table S1d). Additional mice were sampled after 21 weeks of consumption of the LFPP or HFHS diet without the sequential diet shifts (Table S1e). (E) Male C57BL/6J mice were fed an LFPP (blue) or HFHS (red) diet. The two oscillating groups were switched every three days, while the control animals were fed each diet continuously. All mice were fed the HFHS diet from days 26–33 (Table S1f).
Figure S2. A consistent microbial response to the high-fat, high-sugar diet in outbred mice, related to Figure 5a. (A) Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from an Unweighted UniFrac-based PCoA. Points are labeled based on the current diet: LFPP (blue) and HFHS (red). On average, 52 mice were sampled at each timepoint. Values are mean ± sem. (B) Diet resets inter-individual differences in microbial community structure. Mantel tests were performed for all pairwise comparisons of Bray-Curtis distance matrices representing each timepoint. Shading is proportional to the p-value using the distance matrix on the y-axis as the reference for all 18 comparisons. (C) Relative abundance of major taxonomic orders in outbred mice fed a LFPP and HFHS diet. Groups within the same bacterial phyla are indicated by different shades of the same color. Values are means; on average, 52 mice were sampled at each timepoint. All taxa with an average relative abundance >1% are shown.
Figure S3. Confirmation of the impact of the HFHS diet on the gut microbiota of outbred mice, related to Figure 5b. (A) The relative abundance of the 3 most abundant bacterial orders in outbred mice fed either the LFPP or HFHS diet. Values are mean ± sem. Asterisks represent significant differences (p-value<0.05, Kruskal-Wallis test with Dunn’s correction for multiple comparisons). (B) Tukey’s box plots of the first principal coordinate from a Bray-Curtis dissimilarity-based PCoA. Outlier samples are shown for each diet. All of the outliers on the LFPP diet were from 3 cages (#16, 18, and 19), potentially representing an artifact of co-housing. Asterisks represent significant differences (p-value<0.05, Wilcoxon rank sum test). (C) Histogram depicting the relative abundance of the Akkermansia genus in mice fed a LFPP or HFHS diet. A bin width of 5% abundance was used; values represent the center of each bin. Panels A-C represent data from 71–72 mice per diet. (D) Correlation between the relative abundance of Akkermansia inferred from 16S rRNA gene sequencing and the abundance of Akkermansia measured by quantitative PCR for 24 samples drawn at random, expressed on a log10-log10 scale (R2=0.9335; p-value<0.0001; Pearson correlation). Two samples produced zero Akkermansia counts by both methods and were therefore excluded from the log-based analysis. Points are labeled based on sample properties: outbred mice (circles), inbred mice (squares), LFPP diet (black), HFHS diet (white).
Figure S4. Computational modeling of microbial dynamics, related to Figures 5c,7. (A) Outbred mouse model. An example of a prototype signature (solid red line) is depicted. The model of dynamics for each prototype signature is a function continuous in both time and values, which is used to parameterize the mean of the negative binomial distribution (NBD). The function is defined piece-wise on 4 intervals: (a) initial LFPP diet, (b) first HFHS dietary exposure, (c) second LFPP diet, and (d) second HFHS dietary exposure. The function is constant on intervals (a) and (c). On intervals (b) and (d), the function follows an exponential relaxation process; equations for the corresponding relaxation processes are shown above intervals (b) and (d). Equilibrium levels for intervals are depicted at the right of the figure. (B) Dietary oscillation model. An example of a prototype signature (red and blue line segments) is depicted. The model of dynamics for each prototype signature is used to parameterize the mean of the negative binomial distribution (NBD). The function is defined piece-wise on each HFHS and LFPP dietary interval using the linear models shown on the right of the figure. The μH and μL parameters represent baseline levels for the HFHS and LFPP dietary intervals respectively, and βH and βL represent the linear dependence (slope) of levels on the ordering of dietary intervals. For the example shown, both slopes are positive, indicating increasing levels with each subsequent interval for both diets.
Figure S5. Dietary oscillation rapidly alters host physiology and chow consumption, related to Figure 6. (A) Percentage weight change relative to baseline over time. The two counter-oscillatory groups are indicated by a solid line (group 1) or a dashed line (group 2). Timepoints are colored based on the diet consumed over the prior 24 hours; i.e. oscillator group 1 was switched onto the HFHS diet on day zero. (B) Percentage weight change over time for control mice continuously fed the LFPP (solid line) or HFHS diet (dashed line). (C) Mean % change in body weight during each 3-day feeding interval, and (D) total % change in body weight (day 26 – day zero). (E) Chow consumption over time (kcal per day). (F) Mean chow consumption on the first day following each diet shift for mice in both oscillating groups during consumption of the LFPP or HFHS diet. Values in panels A,B,E are mean ± sem (n=3–5 mice per group). Values in panels C,D,F represent mean (lines) and biological replicates (points). Asterisks represent significant differences (p-value<0.05, Wilcoxon rank-sum test).
Figure S6. Dynamic microbial responses to dietary oscillations, related to Figures 6,7a. Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from (A) an Unweighted UniFrac-based PCoA (community membership) and (B) a Bray-Curtis dissimilarity-based PCoA (community structure). Experimental groups can be distinguished by color and shape: oscillating group 1 (green circles), oscillating group 2 (orange squares), LFPP controls (blue inverted triangles), and HFHS controls (red triangles). Values are mean ± sem (n=3–5 mice/group). (C) Diet-associated OTUs with stable levels. Relative abundance of species-level OTUs that were present, responsive to diet, had consistent temporal patterns, and maintained a consistent level on each diet over time (see Methods for thresholds used). Each row represents a consensus temporal signature (aggregated model estimates across mice) for an OTU. Blue indicates relative abundances below the mean abundance for the entire signature, and red indicates relative abundances above the mean. Values represent model estimates, in units of log transformed and standardized relative abundances. The taxonomic assignments for each OTU are labeled on the right of each heatmap: Bacteroidales (black), Clostridiales (orange), Erysipelotrichales (blue), Coriobacteriales (green), and RF39 (brown). Asterisks indicate OTUs with detailed graphs shown in Figure S7f,g.
Figure S7. Representative diet-responsive OTUs with stable or oscillation number dependent abundances, related to Figures 7a, S6c. Detailed plots are shown for representative examples (see asterisks in Figures 7a, S6c) that display oscillation number dependent (panels A–E) or stable (panels F,G) abundances for OTUs in individual mice. Dashed red and blue lines represent median inferred signatures on the HFHS and LFPP diets respectively (widths of line segments are proportional to length of each dietary regimen for oscillator group 1); shading represents 95% credible intervals for estimates. Vertical axes represent model estimates of counts per 10K sequencing reads.
Table S1. 16S rRNA gene sequencing metadata, related to Figures 1–7.
Table S3. Taxonomic associations with host genotype, related to Figure 2.
Table S4. Species-level OTUs with changes in abundance identified using MC-TIMME, related to Figures 5c,7a.
Acknowledgments
We would like to thank Gary Churchill (Jackson Laboratory), Lawrence David, Rachel Dutton, and Andrew Murray (Harvard FAS Center for Systems Biology) for insightful comments; Christian Daly, Michelle Clamp, and Claire Reardon for sequencing support; and Jennifer Brulc and Ravi Menon (General Mills) for helpful discussions. This work was supported by the National Institutes of Health (P50 GM068763; P50 GM076468; F32 DK101154), the Brigham and Women’s Department of Pathology and Center for Clinical and Translational Metagenomics, and the General Mills Bell Institute of Health and Nutrition, Minneapolis, MN.
Footnotes
ACCESSION NUMBERS
Sequencing reads are in MG-RAST (Meyer et al., 2008) under the accession number 11172.
Supplemental Information includes 7 figures, 4 tables, Supplemental Experimental Procedures, and Supplemental Results and can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atkinson RL, Dietz WH, Foreyt JP, Goodwin NJ, Hill JO, Hirsch J, Pi-Sunyer FX, Weinsier RL, Wing R, Hoofnagle JH, et al. Weight cycling. National Task Force on the Prevention and Treatment of Obesity. JAMA. 1994;272:1196–1202. [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard OJ, Summers LH. Hysteresis and the European Unemployment Problem. Nat Bur of Econ Res. 1987;(1950) [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Gatti DM, Munger SC, Svenson KL. The diversity outbred mouse population. Mamm Genome. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber GK, Onderdonk AB, Bry L. Inferring dynamic signatures of microbes in complex host ecosystems. PLoS Comput Biol. 2012;8:e1002624. doi: 10.1371/journal.pcbi.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K, O’Connell JF, Jones NG. Hunting income patterns among the Hadza: big game, common goods, foraging goals and the evolution of the human diet. Philos Trans R Soc Lond B Biol Sci. 1991;334:243–250. doi: 10.1098/rstb.1991.0113. [DOI] [PubMed] [Google Scholar]
- Hildebrand F, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiles DC, Atherton DL. Theory of Ferromagnetic Hysteresis. J Magn Magn Mater. 1986;61:48–60. [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci U S A. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, Zhang T, Rohlf FJ, Zhu W, Gu C, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7:e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JW, Wang X, Kachman SD, Auwerx J, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer M, Carpenter S, Foley JA, Folke C, Walker B. Catastrophic shifts in ecosystems. Nature. 2001;413:591–596. doi: 10.1038/35098000. [DOI] [PubMed] [Google Scholar]
- Scholz F, Badgley BD, Sadowsky MJ, Kaplan DH. Immune mediated shaping of microflora community composition depends on barrier site. PLoS One. 2014;9:e84019. doi: 10.1371/journal.pone.0084019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W, Moore J, Chen K, Lassaletta AD, Yi CS, Tyson JJ, Sible JC. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci U S A. 2003;100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen PL, Schalkwijk J, van der Meer JW, Xavier RJ, Netea MG, Gevers D. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J Innate Immun. 2014;6:253–262. doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the Collaborative Cross. Genetics. 2012;190:291–294. doi: 10.1534/genetics.111.138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012;6:1848–1857. doi: 10.1038/ismej.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Experimental design, related to Figures 1–7. (A) Male and female mice from 5 inbred strains were fed a LFPP or HFHS diet for ≥15 weeks prior to sample collection (Table S1a). (B) Male C56BL/6J wild-type mice and four transgenic lines were fed a LFPP diet until they reached 7 weeks of age, at which point they were switched to the HFHS diet for one week (Table S1b). (C) Male C57BL/6J and 129S1/SvImJ wild-type mice were fed defined mixtures of the LFPP and HFHS diet (Table S1c). (D) Outbred mice were weaned onto a LFPP diet (blue), prior to a 1-week of the HFHS diet (red), 1 week LFPP, and then 9 weeks of maintenance on the HFHS diet. Microbial community structure was determined over time (Table S1d). Additional mice were sampled after 21 weeks of consumption of the LFPP or HFHS diet without the sequential diet shifts (Table S1e). (E) Male C57BL/6J mice were fed an LFPP (blue) or HFHS (red) diet. The two oscillating groups were switched every three days, while the control animals were fed each diet continuously. All mice were fed the HFHS diet from days 26–33 (Table S1f).
Figure S2. A consistent microbial response to the high-fat, high-sugar diet in outbred mice, related to Figure 5a. (A) Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from an Unweighted UniFrac-based PCoA. Points are labeled based on the current diet: LFPP (blue) and HFHS (red). On average, 52 mice were sampled at each timepoint. Values are mean ± sem. (B) Diet resets inter-individual differences in microbial community structure. Mantel tests were performed for all pairwise comparisons of Bray-Curtis distance matrices representing each timepoint. Shading is proportional to the p-value using the distance matrix on the y-axis as the reference for all 18 comparisons. (C) Relative abundance of major taxonomic orders in outbred mice fed a LFPP and HFHS diet. Groups within the same bacterial phyla are indicated by different shades of the same color. Values are means; on average, 52 mice were sampled at each timepoint. All taxa with an average relative abundance >1% are shown.
Figure S3. Confirmation of the impact of the HFHS diet on the gut microbiota of outbred mice, related to Figure 5b. (A) The relative abundance of the 3 most abundant bacterial orders in outbred mice fed either the LFPP or HFHS diet. Values are mean ± sem. Asterisks represent significant differences (p-value<0.05, Kruskal-Wallis test with Dunn’s correction for multiple comparisons). (B) Tukey’s box plots of the first principal coordinate from a Bray-Curtis dissimilarity-based PCoA. Outlier samples are shown for each diet. All of the outliers on the LFPP diet were from 3 cages (#16, 18, and 19), potentially representing an artifact of co-housing. Asterisks represent significant differences (p-value<0.05, Wilcoxon rank sum test). (C) Histogram depicting the relative abundance of the Akkermansia genus in mice fed a LFPP or HFHS diet. A bin width of 5% abundance was used; values represent the center of each bin. Panels A-C represent data from 71–72 mice per diet. (D) Correlation between the relative abundance of Akkermansia inferred from 16S rRNA gene sequencing and the abundance of Akkermansia measured by quantitative PCR for 24 samples drawn at random, expressed on a log10-log10 scale (R2=0.9335; p-value<0.0001; Pearson correlation). Two samples produced zero Akkermansia counts by both methods and were therefore excluded from the log-based analysis. Points are labeled based on sample properties: outbred mice (circles), inbred mice (squares), LFPP diet (black), HFHS diet (white).
Figure S4. Computational modeling of microbial dynamics, related to Figures 5c,7. (A) Outbred mouse model. An example of a prototype signature (solid red line) is depicted. The model of dynamics for each prototype signature is a function continuous in both time and values, which is used to parameterize the mean of the negative binomial distribution (NBD). The function is defined piece-wise on 4 intervals: (a) initial LFPP diet, (b) first HFHS dietary exposure, (c) second LFPP diet, and (d) second HFHS dietary exposure. The function is constant on intervals (a) and (c). On intervals (b) and (d), the function follows an exponential relaxation process; equations for the corresponding relaxation processes are shown above intervals (b) and (d). Equilibrium levels for intervals are depicted at the right of the figure. (B) Dietary oscillation model. An example of a prototype signature (red and blue line segments) is depicted. The model of dynamics for each prototype signature is used to parameterize the mean of the negative binomial distribution (NBD). The function is defined piece-wise on each HFHS and LFPP dietary interval using the linear models shown on the right of the figure. The μH and μL parameters represent baseline levels for the HFHS and LFPP dietary intervals respectively, and βH and βL represent the linear dependence (slope) of levels on the ordering of dietary intervals. For the example shown, both slopes are positive, indicating increasing levels with each subsequent interval for both diets.
Figure S5. Dietary oscillation rapidly alters host physiology and chow consumption, related to Figure 6. (A) Percentage weight change relative to baseline over time. The two counter-oscillatory groups are indicated by a solid line (group 1) or a dashed line (group 2). Timepoints are colored based on the diet consumed over the prior 24 hours; i.e. oscillator group 1 was switched onto the HFHS diet on day zero. (B) Percentage weight change over time for control mice continuously fed the LFPP (solid line) or HFHS diet (dashed line). (C) Mean % change in body weight during each 3-day feeding interval, and (D) total % change in body weight (day 26 – day zero). (E) Chow consumption over time (kcal per day). (F) Mean chow consumption on the first day following each diet shift for mice in both oscillating groups during consumption of the LFPP or HFHS diet. Values in panels A,B,E are mean ± sem (n=3–5 mice per group). Values in panels C,D,F represent mean (lines) and biological replicates (points). Asterisks represent significant differences (p-value<0.05, Wilcoxon rank-sum test).
Figure S6. Dynamic microbial responses to dietary oscillations, related to Figures 6,7a. Analysis of the microbial response to the HFHS diet over time, using the first principal coordinate from (A) an Unweighted UniFrac-based PCoA (community membership) and (B) a Bray-Curtis dissimilarity-based PCoA (community structure). Experimental groups can be distinguished by color and shape: oscillating group 1 (green circles), oscillating group 2 (orange squares), LFPP controls (blue inverted triangles), and HFHS controls (red triangles). Values are mean ± sem (n=3–5 mice/group). (C) Diet-associated OTUs with stable levels. Relative abundance of species-level OTUs that were present, responsive to diet, had consistent temporal patterns, and maintained a consistent level on each diet over time (see Methods for thresholds used). Each row represents a consensus temporal signature (aggregated model estimates across mice) for an OTU. Blue indicates relative abundances below the mean abundance for the entire signature, and red indicates relative abundances above the mean. Values represent model estimates, in units of log transformed and standardized relative abundances. The taxonomic assignments for each OTU are labeled on the right of each heatmap: Bacteroidales (black), Clostridiales (orange), Erysipelotrichales (blue), Coriobacteriales (green), and RF39 (brown). Asterisks indicate OTUs with detailed graphs shown in Figure S7f,g.
Figure S7. Representative diet-responsive OTUs with stable or oscillation number dependent abundances, related to Figures 7a, S6c. Detailed plots are shown for representative examples (see asterisks in Figures 7a, S6c) that display oscillation number dependent (panels A–E) or stable (panels F,G) abundances for OTUs in individual mice. Dashed red and blue lines represent median inferred signatures on the HFHS and LFPP diets respectively (widths of line segments are proportional to length of each dietary regimen for oscillator group 1); shading represents 95% credible intervals for estimates. Vertical axes represent model estimates of counts per 10K sequencing reads.
Table S1. 16S rRNA gene sequencing metadata, related to Figures 1–7.
Table S3. Taxonomic associations with host genotype, related to Figure 2.
Table S4. Species-level OTUs with changes in abundance identified using MC-TIMME, related to Figures 5c,7a.