Abstract
Antibody phage display libraries combined with high-throughput selections have recently demonstrated tremendous promise to create the next generation of renewable, recombinant antibodies to study proteins and their many post-translational modification states; however many challenges still remain, such as optimized antibody scaffolds. Recently, a single-chain Fab (scFab) format, in which the carboxy-terminus of the light chain is linked to the amino-terminus of the heavy chain, was described to potentially combine the high display levels of a single-chain Fv with the high stability of purified Fabs. However, this format required removal of the interchain disulfide bond to achieve modest display levels and subsequent bacterial expression resulted in high levels of aggregated scFab, hindering further use of scFabs. Here, we developed an improved scFab format that retains the interchain disulfide bond by increasing the linker length between the light and heavy chains to improve display and bacterial expression levels to 1–3 mg per liter. Furthermore, rerouting of the scFab to the co-translational signal recognition particle (SRP) pathway combined with reengineering of the signal peptide sequence results in display levels 24-fold above the original scFab format and 3-fold above parent Fab levels. This optimized scFab scaffold can be easily reformatted in a single step for expression in a bacterial or mammalian host to produce stable (81°C Tm), predominantly monomeric (>90%) antibodies at a high yield. Ultimately, this new scFab format will advance high-throughput antibody generation platforms to discover the next generation of research and therapeutic antibodies.
Keywords: antibody, signal peptide, high-throughput screening, phage display, bacterial, mammalian expression
Introduction
Antibodies are essential diagnostic and therapeutic reagents that greatly advance biomedical research. However, antibodies exist to only a fraction of human proteins and an even smaller number of antibodies exist against various post-translational modification states or splice variants of proteins1, 2. Furthermore, recent studies have highlighted the problems (e.g., poor validation, off-target binding, and a non-renewable format) with some existing antibodies3, 4. Therefore, to address these challenges, several large-scale efforts have been initiated to generate well-validated, renewable, recombinant antibodies against a large number of targets to drive new biomedical discoveries. Phage display is well suited for this application due to its rapid and in vitro nature. In particular, robust antibody phage display methods, which employ highly diverse (>109) single-chain fragment variable (scFv) or fragment antigen binding (Fab) libraries, have recently been developed5–8 and these methods have proven amenable to high-throughput automation9, 10. Advances in high-throughput phage display have created the pressing need for novel strategies such as new helper phages11–13, library diversification strategies8, 14, and reformatting methods for downstream expression15, 16 to upgrade and improve the antibody generation pipeline.
The ideal antibody format to facilitate high-throughput antibody selection and production would exhibit high display levels on phage to improve the recovery of rare clones and produce high yield, stable, and well-behaved protein upon simple reformatting for downstream bacterial or mammalian production. The high display level of scFv domains on phage greatly facilitates the discovery of many novel antibodies6, 7, 17, but two factors confound the use of the scFv as a robust scaffold. First, most scFvs possess lower stabilities than Fabs and are prone to aggregation and domain swapping during production and storage18. Additionally, reformatting of the scFvs to a more stable Fab or IgG scaffold can result in a reduction in affinity for the target antigen. Alternatively, highly stable, monomeric Fabs can be efficiently isolated from highly diverse phage display libraries8, 14. However, Fabs typically exhibit lower display levels on phage relative to the parent scFvs and reformatting Fabs into IgGs for mammalian expression can be challenging due to the presence of the bacterial intergenic region and bacterial signal peptide for the heavy chain15, 16.
To combine the advantages of both the scFv and Fab, the concept of a single-chain Fab (scFab), in which the carboxy-terminus of the constant light chain is fused to the amino-terminus of the variable heavy chain via a flexible linker, has recently been introduced (Fig. 1A)19. The resulting scFab scaffold, which contained a 36 amino acid linker, could be displayed on both phage and yeast particles, retained the binding affinity of the parent Fab or scFv, and could be easily reformatted into a single-chain IgG (scIgG) for mammalian cell expression20, 21. Despite this success, several challenges in using the scFab scaffold remain. First, the initial display of level of the scFab was quite poor relative to the parent Fab. To improve the display level of the scFab, the disulfide bond that connects the carboxy-terminus of the light and heavy chains of the Fab (indicated by spheres in Fig. 1A) was removed under the assumption that it would help increase expression. While the display level improved, the resulting bacterially expressed scFab exhibited a high level of aggregation, thus greatly complicating purification and downstream applications. Since this disulfide bond has previously been shown to contribute substantially to the stability of the Fab, removal of the disulfide likely enhances aggregation and prevents easy production of a homogeneous scFab sample22.
Figure 1. Schematic of scFab protein and vectors.
(a) Model of scFab in which a linker consisting of glycine, serine, alanine, and threonine connects the C-terminus of light chain to the N-terminus of the heavy chain. The interchain disulfide bond is highlighted as spheres. The model was generated using Modeller42. (b) Representation of Fab and scFab phage display vectors that contain a PhoA promoter driving expressing of the antibody-coat protein fusion cDNA (SP: signal peptide; VL/H: variable domain of light chain or heavy chain, respectively; CL/H: constant domain of light chain or heavy chain, respectively; cP3: truncated P3 coat protein).
To address these challenges, we sought to develop an improved scFab platform capable of efficient display on phage and robust bacterial expression of highly stable, monomeric scFabs. We discovered that changing the identity of the signal peptide as well as increasing the length of the linker elevates display levels on phage and subsequent bacterial expression, while still retaining the C-terminal disulfide. Furthermore, we found that the improved scaffold exhibits similar recovery efficiencies as the parent Fab from phage display libraries and that additional reengineering of the signal peptide can elevate scFab display levels above the levels of the parent Fab. The resulting scFabs could also be efficiently expressed in both bacterial and mammalian hosts. Finally, the expressed scFabs exhibit wild-type Fab-level thermostabilities and exist predominantly as monomers in solution. Thus, this improved scFab platform should facilitate high-throughput generation of novel renewable, recombinant antibodies.
Results
Influence of signal peptide identity and linker length on scFab display
Previous work on the scFab focused on characterizing scFabs with only a few linker lengths (34, 36, and 38 amino acids)19. Our structural modeling suggested that these linkers were likely too short to connect the carboxy-terminus of the light chain to the amino-terminus of heavy chain and thus, could hinder antibody assembly via strain. Modeling of a (Gly4Ser)2 peptide linker in PEP-FOLD suggests that a relaxed 10 residue linker is ~ 12 Å in length as opposed to 24 Å in a fully extended state23. Thus, a fully extended linker of ~28 residues would cover the linear distance (~65 Å) between the light chain carboxy-terminus and heavy chain amino-terminus. However, the linker must stretch around the outside of the Fab and a fully extended linker would likely introduce steric strain during the folding of the individual Fab domains. Our modeling thus suggested that we needed to explore longer linkers and we generated scFab constructs with linkers of 50, 60, 70, and 80 amino acids in length to ascertain whether longer linkers might enhance scFab display levels on phage as well as protein expression levels. Furthermore, we preserved the interchain, carboxy-terminal disulfide in all constructs to improve stability. We used a humanized anti-peptide (HPep) antibody as the parent Fab17 into which the various linkers were introduced. Schematics of the antibody cassettes are depicted in Figure 1B. Hereafter, we refer to each scFab construct as scX where X indicates the linker length (e.g., sc36 is a scFab with a 36 amino acid linker).
Previous fusions of scFab to the P3 coat protein were translocated via the commonly used Sec pathway19. In the Sec pathway, the unfolded protein is translocated post-translationally whereas in the signal recognition particle (SRP) pathway the protein is translocated co-translationally24. We hypothesized that a portion of the scFab might fold in the cytoplasm prior to translocation and thus limit the display level achieved via the Sec pathway. Therefore, we generated scFab constructs that contained signal peptides that direct translocation through the Sec pathway (PelB) or SRP pathway (DsbA). We employed a well-validated phage display system in which a phoA promoter drives expression of the Fab or scFab fused to a truncated form of the P3 coat protein commonly used in phage display25. This system yields monomeric display of the Fabs since the truncated P3 must compete with the full-length P3 from the helper phage for incorporation into new phage particles. Each phagemid was packaged using XL-1 Blue and M13K07 helper phage. We observed only minor differences (< 3-fold) in phage titers as measured by both OD268 and colony forming units.
To determine functional display levels of the parent Fab and scFab on phage, antigen binding was monitored by ELISA (Fig. 2). Strikingly, the original PelB sc36 exhibited display levels ~8-fold lower than the parent Fab. Changes in the growth conditions including flask type (baffled vs. nonbaffled), growth temperature (30°C vs. 37°C), and shaking speed (75 rpm vs. 200 rpm) resulted in only slight differences in display level (data not shown). As predicted, increasing the linker length for the PelB scFab resulted in a 4-fold increase in display compared to sc36 (Fig. 2A). In contrast, translocation via the SRP pathway (DsbA) resulted in elevated scFab display levels for every linker length relative to translocation via the Sec pathway (PelB). In particular, sc60, sc70, and sc80 exhibited display levels comparable to the level of the parent Fab (Fig. 2B). The results highlight a difference between scFabs and scFvs as a previous study comparing scFv antibody display using either the Sec or SRP pathway found no difference in display level24.
Figure 2. Choice of secretion pathway and linker length alters display level of scFab.

(a) Phage ELISA with dilution series of phage that use the Sec secretion pathway (PelB signal peptide) to detect binding to peptide antigen. The relative display levels were then compared via the respective EC50 values. The initial sc36 construct (red line) exhibits ~8-fold lower display level compared to the parent Fab (black line). However, upon increasing the linker length, the display level gradually improves to only ~2-fold less than the display level for the sc80 construct (blue line). (b) Phage ELISA with dilution series of phage that use the SRP secretion pathway (DsbA signal peptide) to detect binding to peptide antigen. Use of the SRP pathway results in elevated levels of scFab display with levels comparable to the parent Fab for 60, 70, and 80 amino acid linkers. All data are represented as the mean ± standard deviation (n = 3).
To demonstrate that these results also translate to scFabs based on other Fabs scaffolds, we generated a DsbA sc60 based upon a GFP-binding Fab isolated from a synthetic antibody library26. This Fab has distinct sequences in all six complementarity-determining regions (CDRs). Also, this Fab contains a variable light chain derived from the κ1 family as opposed to the κ3 family for the HPep Fab and thus may possess different biophysical properties since κ1 light chain domains have lower stabilities and protein yields when expressed as isolated domains27. Importantly, wild-type Fab levels of protein display, as measured by an anti-Fab antibody, were observed for both the HPep and anti-GFP DsbA sc60 constructs (Fig. 3A). These results highlight the robust nature of this combination of signal peptide and linker length to both CDR sequence and framework differences.
Figure 3. scFab performance is robust to variation in CDR sequence and light chain identity.

(a) Anti-Fab ELISA with parent Fab and sc60 for HPep and GFP-binding Fabs reveals similar display levels for both the sc60 and Fab. HPep Fab contains a light chain from the human Vκ3 family whereas the GFP-binding Fab contains a light chain from the human Vκ1 family. These two Fabs also differ in the sequences of all six CDRs. (b) Competition ELISAs with Fab- and sc60-phage and varying concentrations of soluble antigen reveals no substantial difference in binding affinity between the sc60 and Fab. All data are represented as the mean ± standard deviation (n = 3).
Affinity characterization and functional selection of sc60 phage
We sought to demonstrate that this DsbA sc60 phage can be efficiently isolated from a phage display library. We first performed competitive ELISAs to quantify the binding constant for sc60 phage relative to the parent Fab phage. Importantly, we observed no change in competitive binding curves between the two phage samples, thus indicating that the insertion of the 60 amino acid linker does not alter the binding affinity of the parent Fab against two distinct antigens, a peptide and a protein (Fig. 3B).
To create a stringent test to demonstrate that the sc60 performs as well as the parent Fab during normal phage display selections, we generated a series of mock antibody libraries by doping HPep Fab, HPep sc60, or HPep Fab and sc60 phage into negative control GFP scFab phage at dilutions of 1:107 and 1:1010, representing the typical diversities of antibody phage display libraries generated by restriction cloning or Kunkel mutagenesis, respectively. After two rounds of selection against the peptide antigen, HPep sc60 and Fab phage were efficiently recovered from both mock libraries (Table 1). Impressively, HPep sc60 phage were also isolated when initially mixed with an equal concentration of competing HPep Fab phage further highlighting that the phage packaging, display levels, and affinities are similar for both the sc60 and parent Fab phage. Furthermore, we have performed affinity maturation on multiple phospho-specific antibodies using phage display libraries constructed using the sc60 scaffold and obtained antibodies with 10- to 100-fold improvements in affinity (J. Koerber and J. Wells, manuscript in preparation).
Table 1. Results of mock selection with DsbA sc60 format.
Forty-eight single clones were screened by ELISA after rounds 1 and 2 of selection. PCR was used to classify each ELISA-positive clone as scFab or Fab.
| Dilution | Round 1 ELISA+ Clones |
Round 2 ELISA+ Clones |
|
|---|---|---|---|
| HPep scFab | 1:107 | 9/48 | 46/48 |
| HPep scFab | 1:1010 | 2/48 | 46/48 |
| HPep Fab | 1:107 | 14/48 | 44/48 |
| HPep Fab | 1:1010 | 7/48 | 47/48 |
| HPep scFab+ Fab | 1:107 | 42/48 (24 scFab) | 48/48 (30 scFab) |
| HPep scFab+ Fab | 1:1010 | 18/48 (12 scFab) | 48/48 (32 scFab) |
Selection of new signal peptides that enhance scFab display
Since we observed that switching signal peptides from PelB to DsbA improved display of the scFab scaffold, we hypothesized that further increases in display might be possible via additional reengineering of the signal peptide sequence. Therefore, we generated two antibody phage display libraries using the sc60 scaffold in which either the wild-type PelB or DsbA signal peptide was subjected to soft randomization. After three rounds of selection against decreasing peptide concentrations, we screened individual phage clones via ELISA against both the peptide antigen and an anti-Fab antibody to identify clones with elevated scFab display levels. In total, we isolated five new signal peptides (three clones derived from PelB and two clones derived from DsbA) that mediate higher scFab display levels than either the PelB or DsbA parent signal peptides (Fig. 4A). The new signal peptides differed by three to six amino acids from the parent signal peptides and interestingly, we found new variants of the PelB signal peptide that mediated higher display than the wild-type DsbA signal peptide (Fig. 4B). We employed a quantitative ELISA method to measure the amount of scFab/Fab per phage particle, as previously described28. Parent Fab-phage and wild-type DsbA sc60 contained ~0.15 Fab per phage whereas the best variant (DsbA11) contained 0.5 scFab per phage. In total, the best signal peptide mediates a 3-fold higher scFab display compared to both the parent Fab and sc60.
Figure 4. Evolved signal peptides mediate higher scFab display than parent signal peptides.

(a) A single-point phage ELISA with 3×1010 phage per mL (as measured by OD268) bound to peptide antigen demonstrates that three PelB-derived signal peptides and two DsbA-derived signal peptides mediate enhanced sc60 display levels compared to the parent PelB and DsbA signal peptides. All data are represented as the mean ± standard deviation (n = 3). (b) Protein sequence alignment of five new signal peptides and parent PelB and DsbA peptides. The DsbA clones contain one additional amino acid from the mutagenesis reaction. Each clone contains both synonymous (S) and non-synonymous (NS) mutations: PelB9 (5 S, 5 NS); PelB12 (1 S, 5 NS); PelB15 (4 S, 5 NS); DsbA7 (5 S, 4 NS); and DsbA11 (3 S, 7 NS) (Supplemental Figure 1).
High-level expression in both bacteria and mammalian hosts
Having validated the DsbA sc60 as a robust format for antibody phage display, we next sought to demonstrate that this scaffold could be efficiently expressed in a bacterial host. Therefore, in a single cloning step using Sfi I, we moved the HPep scFabs into a bacterial expression vector (pJK6). This vector contains a pTac promoter that drives expression of a scFab with a PelB signal peptide. Based upon our phage results with the DsbA signal peptide, we also tested a pJK6 variant that used a DsbA signal peptide to drive secretion, but we observed no change in expression levels (data not shown). The parent Fab along with all five scFabs constructs were expressed in a protease-deficient C43 Pro+ strain29. Strikingly, we observed ~30-fold lower functional antibody yields for the sc36 scaffold compared to the parent Fab, whereas scFab scaffolds with 60-, 70-, and 80-mer linkers exhibited 5-fold decrease in yield compared to the parent Fab (Fig. 5A). Additionally, the comparable levels of maximum ELISA signal present within each soluble lysate suggests the absence of substantial levels of aggregated antibody, a fact later confirmed by gel filtration. Soluble antibodies were purified from lysate using Protein A chromatography and assayed for antigen binding by ELISA. Soluble yields of each construct were determined by measuring binding to the peptide antigen. All purified antibodies also bound to the peptide antigen at similar levels with comparable maximum signals, confirming both the wild-type affinities and monomeric state (Fig. 5B). Analysis of each purified antibody by SDS-page and Coomassie staining confirmed the expected molecular weights and high purity of these scaffolds (Fig. 5C).
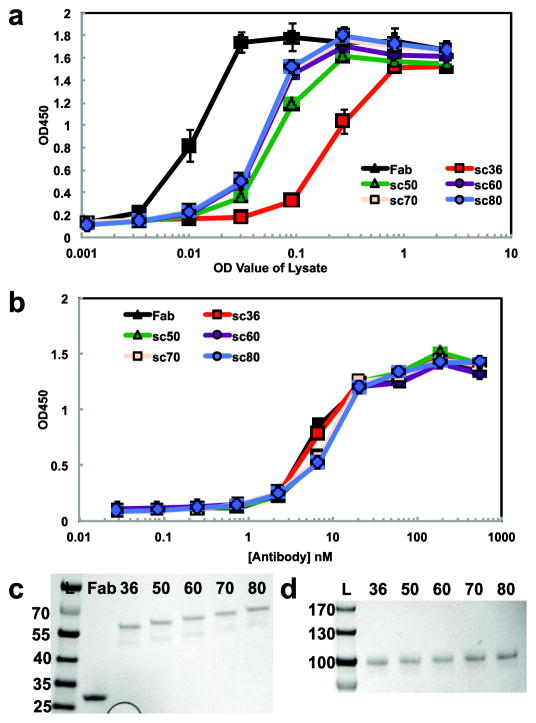
Figure 5. Robust bacterial and mammalian expression of scFab proteins.
(a) The soluble fraction of bacterial lysates containing expressed Fab/scFab was analyzed by ELISA. The original sc36 construct (red line) expressed poorly (~30-fold less) relative to the parent Fab (black line) as previously observed19. However, increasing the linker length beyond 50 residues led to a plateau of expression yields only ~5-fold less than the Fab. (b) Analysis of Protein A-purified scFabs reveals identical binding to the parent Fab as determined by titration ELISAs. (c) SDS-page analysis of Fab and sc36, sc50, sc60, sc70, and sc80 proteins confirms that the two chains of the Fab remain together when linked and migrate at the expected molecular weights. All antibodies were purified via Protein A chromatography prior to analysis. (d) SDS-page analysis of scIgG constructs from sc36, sc50, sc60, sc70, and sc80 reveals a single polypeptide at the expected molecular weight upon mammalian expression. All scIgGs were produced by transient transfection of HEK293T and purified by Protein A chromatography. All data are represented as the mean ± standard deviation (n = 3).
Finally, we sought to demonstrate that these scFab constructs could be readily expressed in mammalian cells as singe chain IgGs (scIgGs) and thus using Sfi I, we moved the HPep scFabs into a mammalian expression vector (pJK7). This vector contains a hEFI-HTLV promoter that drives expression of a cDNA cassette consisting of an IL2 signal peptide, the scFab, and a rabbit Fc. Upon transient transfection of HEK293T cells and subsequent Protein A purification, we obtained scIgGs at yields of 5 μg per mL media (Fig. 5D). Interestingly, the yields did not vary substantially among the various linker lengths, suggesting the mammalian secretory pathway likely provides a better environment for assembly of the scIgGs.
Stability characterization of scFabs
Removal of the stabilizing interchain disulfide bond between the light chain and heavy chain in previous scFab scaffolds resulted in reduced stability and a heterogeneous mixture of multimeric states19. Therefore, we sought to characterize the stability and multimerization state of our scFabs, which retain the disulfide bond, relative to the parent Fab. Protein A-purified Fab and scFabs were separated by size exclusion chromatography. As expected, the parent Fab eluted as a single monomeric species (Fig. 6A). For every scFab, the major peak corresponded to a monomer with low amounts of multimers observed for sc60 and a higher amount for sc70 and sc80 (Fig. 6B and Supplemental Figure 2). These species are particularly prominent in the sc70 and sc80 preparations, potentially due to the ability of light chains to pair with heavy chains from a second scFab molecule to create a dimeric scFabs.
Figure 6. Stability characterization of bacterially-expressed scFabs.
(a) Gel filtration analysis of purified Fab yields a single monomeric species that elutes at the expected molecular weight. (b) Gel filtration analysis of sc60 reveals a mostly monomeric species with a small amount (<10 %) of higher molecular weight species. (c) Thermostabilities of the Fab and scFabs were determined by DSF. All antibodies exhibited identical thermostabilites with a Tm of 81°C, highlighting the Fab-level stability of the scFabs.
To characterize scFab stability, we performed a differential scanning fluorimetry (DSF) assay to measure the thermal stabilities of the same Fab and scFab samples that were analyzed by size exclusion. Importantly, all scFabs exhibited similar thermostabilities (Tm ~ 81°C) as the parent Fab (Fig. 6C). Interestingly, the sc36 exhibited a secondary peak with a Tm of 68°C, suggesting the presence of the less stable species within the population. Therefore, the combination of the longer linker and retained intermolecular disulfide yields predominantly monomeric, stable scFabs, thus facilitating easier production and downstream characterization (e.g., affinity measurement, storage, or functional assays).
Discussion
This improved scFab format that employs a DsbA signal peptide, a 60 amino acid linker, and retains the intermolecular disulfide bond results in a more robust platform for antibody phage display. In a direct competition experiment involving mock libraries, this scFab format efficiently competes with the parent Fab and thus, represents an improvement over the existing scFab scaffold (Table 1)19. Furthermore, we have shown that the signal peptide can be further engineered to improve scFab display levels by another factor of 2- to 3-fold, exceeding the parent Fab display levels (Fig. 4). Soluble scFab that possesses Fab-like stability can also be easily produced in both bacterial and mammalian hosts with only a small decrease in expression yield (~5-fold). We envision that further optimization of the signal peptide, promoter, and bacterial strain will help improve expression yields.
Several strategies have been employed to optimize Fab expression in bacteria as both the expression rate and amount of the light chain are critical factors for proper Fab assembly30. For example, optimization of the translational initiation region and signal peptide can alleviate backup of the secretion pathway and help minimize cellular stress during antibody production. As antibodies typically contain both multiple disulfides and cis/trans-prolines, expression of chaperones that aid in disulfide bond formation or aid in proline isomerization has been shown to improve bacterial expression of both scFvs and IgGs30, 31. We conducted a preliminary test of sc60 expression in a C43 Pro+ strain that harbors a chaperone plasmid32 that co-expresses thiol-disulfide oxidoreductases (DsbA and DsbC) and peptidyl-prolyl cis/trans-isomerases (FkpA and SurA) and observed a 4-fold increase in expression level (data not shown).
As the linker length increases beyond 50 amino acids in length, we began to observe the presence of a greater degree of multimeric contaminants in the Protein A-purified scFabs. Two strategies may alleviate this problem. First, analysis of the display levels, expression yields, and gel filtration profiles suggests the optimal linker length lies somewhere between 50–60 amino acids in the length. Second, we have observed that incorporating a heat treatment step after bacterial lysis during Fab purification greatly improves Fab purity by removing less stable multimeric and degradation products (M. Paduch and A. Kossiakoff, personal communication). The high scFab thermostabilities will allow us to incorporate this step into the scFab purification to further improve purity.
Signal peptide optimization represents a promising, but challenging strategy to improve both scFab phage display and recombinant expression levels. To date, studies have only attempted to systematically characterize the effect of different, naturally occurring signal peptides on scFv display levels24, likely due to the ease of focusing on a signal peptide in each construct. On the other hand, Fabs require one signal peptide for each chain for efficient phage display, making identification of improved signal peptides challenging. We have exploited the single polypeptide nature of our scFab construct to demonstrate that rerouting the scFab into the SRP secretion pathway results in >4-fold increase in display level relative to the Sec secretion pathway (Fig. 2). A similar change in the secretion pathway has also led to improved display of proteins that exhibit high thermostabilities and fold rapidly in the bacterial cytoplasm, such as DARPins and thioredoxin33, 34. Potentially, the introduction of the linker increases the folding rate of the scFab by tethering the light and heavy chains in a close proximity and thus, rerouting the scFab through the SRP co-translational pathway results in more efficient display.
In addition to discovering the utility of the DsbA signal peptide for scFab display, we engineered enhanced variants of both the DsbA and PelB signal peptides. Interestingly, the three variants of the PelB signal peptide mediate 3- to 4-fold increases in scFab display level contain different combinations of mutations, thus making it difficult to identify common traits of the improved variants (Fig. 4). However, all of the variants contain mutations within the hydrophobic core of PelB. Previous mutations of this region have been shown to reroute the original PelB from the Sec pathway to the SRP pathway35 and thus, these mutations may result in rerouting of the scFab to the SRP pathway. In the case of the two DsbA variants, the only common mutation is replacement of helix-breaking Gly10 with either Val or Ala. The recent structure of a signal peptide bound to SRP revealed that the signal peptide adopts an alpha-helical conformation to promote multiple interactions with SRP36. The DsbA mutations we observe likely stabilize the alpha-helical structure of the hydrophobic core and may enhance binding to SRP, resulting in higher display.
We envision applying this sc60 format to generate next-generation synthetic antibody libraries to target proteins and post-translational modifications8, 17. In particular, the single polypeptide sc60 format may facilitate the incorporation of new helper phages11–13 to achieve robust polyvalent scFab display and enhance the recovery of rare clones against challenging targets such as peptides, conformational states of proteins, or cell surface proteins17, 29, 37. Additionally, as this scaffold retains the stability properties of the parent Fab, it can be seamlessly integrated into existing pipelines for improved high-throughput antibody generation5–8. For example, the scFab scaffold greatly simplifies antibody reformatting to scIgG or fusion to other tags and thus will aid in efficient downstream antibody validation. Reformatting lead scFabs for an in vitro transcription-translation format capable of high-level IgG production38, 39 could yield purified scIgG in only one day compared to more than one week for mammalian cell production. Ultimately, this scFab format will help improve the generation of novel antibodies from recombinant antibody libraries to advance biomedical research and therapeutics.
Materials and Methods
Construction of Vectors
All scFab and Fab constructs were generated in pJK1, a previously described phage display vector17. Briefly, pJK1 contains a phoA promoter that drives expression of a PelB signal peptide, a cDNA cassette flanked by unique Sfi I sites, and a truncated form of the P3 coat protein (Fig. 1B). To characterize the influence of the class of signal peptide on scFab display, a modified version of pJK1 was generated to contain a DsbA signal peptide (pJK8). To generate the scFab construct, flexible linkers of varying length (36, 50, 60, 70, and 80 amino acids) were used to connect the C-terminus of the light chain to the N-terminus of the heavy chain of the parent HPep Fab gene, a humanized Fab that binds a peptide antigen from EGFRvIII17. Each linker consisted of a mixture of A, T, G, and S residues (Supplemental Table 1). Analogous scFab constructs were also generated for a GFP-binding Fab isolated from a synthetic phage display library26. While both the HPep Fab and the GFP-binding Fab contain variable heavy chains from the VH3 family, they contained distinct CDR sequences in all 6 CDRs variable light chains from the Vκ3 and Vκ1 families, respectively. All restriction enzymes and DNA polymerases were purchased from NEB (Ipswich, MA). Oligonucleotides were purchased from IDT and all constructs were verified by DNA sequencing (Quintara Biosciences).
Phage and Fab ELISAs
All phage stocks were prepared in XL-1 Blue cells and M13K07 helper phage. Phage samples were purified by PEG precipitation and phage concentrations were determined by OD268. Phage clones and soluble proteins were functionally characterized by ELISA. Briefly, 384-well Maxisorp plates were coated with 10 ug/mL NeutrAvidin overnight at 4°C and then blocked with 0.5% BSA for 2 hr at 20°C. Three-fold dilutions of the phage supernatant were incubated with 20 nM biotinylated peptide for 30 min and captured on the NeutrAvidin-coated plates. Bound phage were then detected using a horseradish peroxidase (HRP)-conjugated anti-M13 antibody (1:5000) (GE Healthcare). Alternatively, levels of displayed Fab/scFab were determined by ELISA against immobilized anti-Fab antibody. Briefly, 384-well Maxisorp plates were coated with 1 ug/mL anti-human Fab (Sigma-Aldrich) overnight at 4°C and then blocked with 0.5% BSA for 2 hr at 20°C. Three-fold dilutions of phage stocks were then incubated on the plate for 1 hr at 20°C. The bound phage were then detected with the HRP-conjugated anti-M13 antibody. To measure the amount of Fab displayed per phage particle, we performed the same ELISA except using 1ug/mL anti-M13 antibody (GE Healthcare), as previously described28. The OD450 values in the linear range were used to construct a standard curve to quantify the number of Fab molecules at a given phage concentration in anti-Fab or peptide ELISAs.
To characterize Fab/scFab protein binding, dilutions of soluble lysate or purified Fab/scFab were incubated with 20 nM biotinylated peptide for 30 min and captured on the NeutrAvidin-coated plates. Bound Fabs were then detected using HRP-conjugated Protein A (1:5000) (Pierce).
Mock Phage Selections
Six mock phage libraries were generated by dilution of phage that displayed either the HPep Fab or HPep scFab into phage that displayed a negative control scFab that binds GFP. Libraries A and B contained the HPep scFab phage diluted at 1:107 and 1:1010 to represent medium and high diversity libraries. Libraries C and D contain the HPep Fab phage diluted at 1:107 and 1:1010. Libraries E and F contain equal amounts of the HPep Fab and HPep scFab phage diluted at 1:107 and 1:1010. Each library was then subjected to two rounds of selection against the biotinylated peptide (biotin-GEKKGNYVVTDH). Briefly, 5 × 1012 cfu phage were incubated with 100 nM peptide for 30 min at 20°C. Bound phage were subsequently captured with streptavidin-coated magnetic beads (Promega). After 5 washes in PBS with 0.1% Tween, bound phage were eluted with 0.1 M glycine pH2.2. Eluted phage were then amplified overnight. The second round of selection was performed using the same conditions as round one.
To determine the identity of individual clones after each round of selection, forty-eight individual clones were picked and grown in 96-well deep well blocks to produce individual phage stocks. The resulting phage were characterized by ELISA as described. The identity (scFab vs. Fab) of each ELISA-positive phage clone was then verified using PCR with a forward primer that bound in the intergenic region (Fab) or linker (scFab) and a common reverse primer that bound in the variable heavy chain region.
Construction and selection of diverse signal peptide library
The sc60 construct in pJK1 was used to prepare a stop template, which contains six consecutive stop codons in place of the PelB signal peptide. The C-terminal region (AQPAMA) of the PelB signal peptide was retained in order to keep the Sfi I site for cloning. This stop template was used as a template for Kunkel mutagenesis with oligonucleotides designed to randomize the codons encoding for either the PelB or DsbA signal peptides. To obtain a highly functional library of mutant signal peptides, we opted for a “soft” diversification strategy in which the frequency of the wild-type nucleotide was set to 79% and the frequency of every other nucleotide was set to 7%. The two oligonucleotides were PelB 5′-gtagaggttgaggtgattttatg(WWW)(ZWX)(XZW)(ZZY)(XXZ)(WXY)(YXW)(YXX)(YXZ)(YYW) (ZZY)(XZW)(ZZW)(XZX)(YXg)gcccagccggc-3′ and DsbA 5′-gtagaggttgaggtgattttatg(WWW)(WWY)(WZZ)(ZYY)(XZY)(YXY)(XZY)(YXZ)(YYZ)(ZZW)(YZZ)(ZZW)(YYg)gcccagccggc-3′. W is a mixture of 79% A, 7%T, 7%G, and 7%C, X is a mixture of 7% A, 79%T, 7%G, and 7%C, Y is a mixture of 7% A, 7%T, 79%G, and 7%C, and Z is a mixture of 7% A, 7%T, 7%G, and 79%C. The resulting mutagenesis reactions were electroporated and phage were produced as previously described40. The final diversities of the PelB and DsbA libraries were 1.82 × 109 and 1.92 × 109, respectively.
To isolate signal peptides that facilitated higher levels of scFab display, each library was subjected to three rounds of selection as described above, except with gradually decreasing concentrations of the peptide antigen (round 1 – 5 nM, round 2 – 0.5 nM, and round 3 – 0.1 nM). Individual phage clones were analyzed for antigen binding by ELISA as described above.
Bacterial expression and purification of scFab
Each scFab construct (sc36, sc50, sc60, sc70, and sc80) was cloned using Sfi I into pJK4, a previously described bacterial expression vector17. Briefly, pJK4 contains a pTac promoter that drives expression of a cDNA consisting of the PelB signal peptide followed by the scFab gene. ScFabs and the parent Fab were expressed in the protease-deficient C43 Pro+ strain29. One and a half liter cultures were inoculated with an overnight starter culture and grown to an OD600 of 1. Protein expression was induced with 1 mM IPTG and the cultures were grown for ~20 hr at 30°C. Cells were harvested and lysed in PBS containing 1 mM PMSF using a microfluidizer. Fabs/scFabs were then purified by Protein A chromatography. Briefly, soluble lysates were loaded onto a pre-equilibrated 5 mL HiTrap Protein A column (GE Healthcare) and washed with 5 column volumes of PBS. Bound Fabs/scFabs were then eluted with 0.1 M acetic acid, immediately neutralized with 1 M Tris pH 8 and buffered exchanged into PBS for short-term storage at 4°C. Alternatively, Fabs/scFabs were flash frozen in PBS with 10% glycerol for long-term storage at −80°C. For analytical gel filtration, Protein A-purified Fabs/scFabs were loaded onto a HiTrap Superdex 200 10/300 GL equilibrated in PBS.
Mammalian expression of scIgG
Each scFab construct (sc36, sc50, sc60, sc70, and sc80) was cloned using Sfi I into pJK6, a previously described mammalian expression vector17. Briefly, pJK6 contains a hEFI-HTLV promoter that drives expression of a cDNA cassette containing the IL-2 signal peptide, the scFab gene, and the rabbit IgG Fc domain (CH2 and CH3 domains). Approximately 5 × 106 HEK293T cells were seeded in one 10-cm dish and transfected with 20 ug of each pJK6 construct using a standard calcium phosphate protocol. Eight hours later, media was removed and replaced with fresh media that contained SuperLow IgG FBS (Hyclone). Supernatant was then collected at two and five days post-transfection. Supernatant was neutralized with 10x PBS and secreted scIgG was purified from the supernatant via Protein A chromatography.
Thermostability analysis
The melting temperature (Tm) of each Fab/scFab was determined by differential scanning fluorimetry (DSF)41. Briefly, 25 uL reactions containing 5 uM Fab, 4x Sypro Orange (Invitrogen) and PBS were prepared and 10 uL was transferred to a 384-well PCR plate. In a LightCycler 480 (Roche), the reaction was heated from 25°C to 95°C with a ramp rate of ~0.5°C per 30 s. The first derivative of the resulting fluorescence curves was used to determine the Tm for each Fab.
Supplementary Material
Highlights.
High-throughput antibody generation requires improved scaffolds.
Improved phage display for a scFab with long linkers and optimized signal peptide.
Rapid reformatting for high yield, stable scFab from bacterial and mammalian hosts.
New, optimized scFab to advance antibody discovery.
Acknowledgments
We thank members of the Wells lab for helpful discussions regarding this manuscript. We thank Brian Lee, Tet Matsuguchi, and Karolina Wypisniak for providing the anti-GFP antibody and assisting with the DSF assay. James Koerber is a Fellow of the Life Sciences Research Foundation. This work was supported by a grant from the National Institutes of Heath (R01 CA154802 to JAW) and (U54 HG006436 to JAW).
Abbreviations
- Fab
fragment antigen binding
- scFv
single-chain fragment variable
- scFab
single-chain Fab
- scIgG
single-chain IgG
- CDR
complementarity-determining region
- SRP
signal recognition particle
- HPep
human anti-peptide
- DSF
differential scanning fluorimetry
- HRP
horseradish peroxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 2.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 3.Bordeaux J, et al. Antibody validation. BioTechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaastad H, et al. Multiplexed immuno-precipitation with 1725 commercially available antibodies to cellular proteins. Proteomics. 2011;11:4578–4582. doi: 10.1002/pmic.201000744. [DOI] [PubMed] [Google Scholar]
- 5.Colwill K, Graslund S Renewable Protein Binder Working G. A roadmap to generate renewable protein binders to the human proteome. Nature methods. 2011;8:551–558. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- 6.Schofield DJ, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hust M, et al. A human scFv antibody generation pipeline for proteome research. Journal of biotechnology. 2011;152:159–170. doi: 10.1016/j.jbiotec.2010.09.945. [DOI] [PubMed] [Google Scholar]
- 8.Fellouse FA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Paduch M, et al. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 2013;60:3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turunen L, Takkinen K, Soderlund H, Pulli T. Automated panning and screening procedure on microplates for antibody generation from phage display libraries. Journal of biomolecular screening. 2009;14:282–293. doi: 10.1177/1087057108330113. [DOI] [PubMed] [Google Scholar]
- 11.Rondot S, Koch J, Breitling F, Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nature biotechnology. 2001;19:75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 12.Kramer RA, et al. A novel helper phage that improves phage display selection efficiency by preventing the amplification of phages without recombinant protein. Nucleic acids research. 2003;31:e59. doi: 10.1093/nar/gng058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek H, Suk KH, Kim YH, Cha S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic acids research. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prassler J, et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol. 2011;413:261–278. doi: 10.1016/j.jmb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Jones ML, et al. A method for rapid, ligation-independent reformatting of recombinant monoclonal antibodies. Journal of immunological methods. 2010;354:85–90. doi: 10.1016/j.jim.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Tesar D, Hotzel I. A dual host vector for Fab phage display and expression of native IgG in mammalian cells. Protein engineering, design & selection: PEDS. 2013;26:655–662. doi: 10.1093/protein/gzt050. [DOI] [PubMed] [Google Scholar]
- 17.Koerber JT, Thomsen ND, Hannigan BT, Degrado WF, Wells JA. Nature-inspired design of motif-specific antibody scaffolds. Nature biotechnology. 2013;31:916–921. doi: 10.1038/nbt.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 19.Hust M, et al. Single chain Fab (scFab) fragment. BMC biotechnology. 2007;7:14. doi: 10.1186/1472-6750-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, Bowley DR, Burton DR. Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J Mol Biol. 2009;389:365–375. doi: 10.1016/j.jmb.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirrmann T, et al. Oligomeric forms of single chain immunoglobulin (scIgG) mAbs. 2010;2:73–76. doi: 10.4161/mabs.2.1.10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347:773–789. doi: 10.1016/j.jmb.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 23.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic acids research. 2009;37:W498–503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thie H, Schirrmann T, Paschke M, Dubel S, Hust M. SRP and Sec pathway leader peptides for antibody phage display and antibody fragment production in E. coli. New biotechnology. 2008;25:49–54. doi: 10.1016/j.nbt.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu SS, et al. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Persson H, et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J Mol Biol. 2013;425:803–811. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 28.Junutula JR, et al. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. Journal of immunological methods. 2008;332:41–52. doi: 10.1016/j.jim.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen ND, Koerber JT, Wells JA. Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation. Proc Natl Acad Sci U S A. 2013;110:8477–8482. doi: 10.1073/pnas.1306759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly DE, Yansura DG. Production of Monoclonal Antibodies in E. coli. Biotechnol Pharm Asp. 2010;11:295–308. [Google Scholar]
- 31.Schaefer JV, Pluckthun A. Improving Expression of scFv Fragments by Co-expression of Periplasmic Chaperones. Antibody Engineering. (2) 2010;2:345–361. [Google Scholar]
- 32.Schlapschy M, Grimm S, Skerra A. A system for concomitant overexpression of four periplasmic folding catalysts to improve secretory protein production in Escherichia coli. Protein engineering, design & selection: PEDS. 2006;19:385–390. doi: 10.1093/protein/gzl018. [DOI] [PubMed] [Google Scholar]
- 33.Steiner D, Forrer P, Stumpp MT, Pluckthun A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nature biotechnology. 2006;24:823–831. doi: 10.1038/nbt1218. [DOI] [PubMed] [Google Scholar]
- 34.Schierle CF, et al. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. Journal of bacteriology. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams H, Scotti PA, De Cock H, Luirink J, Tommassen J. The presence of a helix breaker in the hydrophobic core of signal sequences of secretory proteins prevents recognition by the signal-recognition particle in Escherichia coli. European journal of biochemistry/FEBS. 2002;269:5564–5571. doi: 10.1046/j.1432-1033.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- 36.Janda CY, et al. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 38.Stafford RL, et al. In vitro Fab display: a cell-free system for IgG discovery. Protein engineering, design & selection: PEDS. 2014;27:97–109. doi: 10.1093/protein/gzu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin G, et al. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs. 2012;4:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bostrom J, Lee CV, Haber L, Fuh G. Improving antibody binding affinity and specificity for therapeutic development. Methods in molecular biology. 2009;525:353–376. xiii. doi: 10.1007/978-1-59745-554-1_19. [DOI] [PubMed] [Google Scholar]
- 41.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 42.Eswar N, et al. Comparative protein structure modeling using MODELLER. In: Coligan John E, et al., editors. Current protocols in protein science/editorial board. Unit 2. Chapter 2. 2007. p. 9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.