Abstract
The yellow color of inner leaves in Chinese cabbage depends on its lutein and carotene content. To identify responsible genes for yellow pigmentation in leaves, the transcriptome profiles of white (Kenshin) and yellow leaves (Wheessen) were examined using the Br300K oligomeric chip in Chinese cabbage. In yellow leaves, genes involved in carotene synthesis (BrPSY, BrPDS, BrCRTISO, and BrLCYE), lutein, and zeaxanthin synthesis (BrCYP97A3 and BrHYDB) were upregulated, while those associated with carotene degradation (BrNCED3, BrNCED4, and BrNCED6) were downregulated. These expression patterns might support that the content of both lutein and total carotenoid was much higher in the yellow leaves than that in the white leaves. These results indicate that the yellow leaves accumulate high levels of both lutein and β-carotene due to stimulation of synthesis and that the degradation rate is inhibited. A large number of responsible genes as novel genes were specifically expressed in yellow inner leaves, suggesting the possible involvement in pigment synthesis. Finally, we identified three transcription factors (BrA20/AN1-like, BrBIM1, and BrZFP8) that are specifically expressed and confirmed their relatedness in carotenoid synthesis from Arabidopsis plants.
1. Introduction
Chinese cabbage (Brassica rapa ssp. pekinensis), which is widely used vegetable in Asia, particularly in China, Japan, and Korea, is rich in vitamin C and minerals such as potassium, calcium, magnesium, and zinc. This subspecies is typical of the Brassica A genome and has a large number of genomic resources including a mapping population and BAC libraries, due to its small genome (529 Mb) relative to other Brassica species. The inner leaves can be white, yellow, or orange. Earlier reports have revealed that the color of the inner leaves is primarily associated with their carotenoid composition. Yellow inner leaves of Chinese cabbage have higher nutritional values than white inner leaves [1]. In recent years, Asian countries have preferred yellow inner leaf cultivars due to its high nutritional values and significant economic benefits.
Carotenoids represent a class of red, orange, and yellow pigments widely distributed in nature that are mainly C40 isoprenoids and play a critical role in human nutrition and health. Because humans are unable to synthesize vitamin A de novo from endogenous isoprenoids precursors, it is primarily obtained through dietary plant carotenoids. Apart from their nutritional significance, carotenoids have been implicated in reducing the risk of cancer and cardiovascular diseases via their antioxidant activity [2, 3]. Thus, development of carotenoid enriched food crops provides the most effective and sustainable approach to maximizing the nutritional and health benefits of carotenoids to a large number of populations worldwide.
The carotenoid pigment composition has been investigated in many Brassica vegetables including broccoli, Brussels sprouts, white cabbage, red cabbage, kale, and cauliflower [4] and these investigations have indicated that lutein and β-carotene were the dominant carotenoids. Chinese cabbage also contains lutein and β-carotene as the major carotenoid pigment [5], with levels of 0.01 to 0.03 mg/100 g fresh weight generally being observed [6]. The orange-yellow pigmentation of Chinese cabbage expressed in inner leaves and petals is governed by a single recessive gene. The locus of the pigmentation has been mapped from three RFLP markers closely linked to each other [7]. Microarray technology enables genome wide analyses that can be used to identify plant gene expression under various conditions, including plant organogenesis and interactions with microorganisms [8, 9]. Microarray experiments have been employed to analyze gene expression changes in a number of crop species, including Chinese cabbage [10, 11].
The objective of this study was to identify the genes responsible for yellow color pigmentation in B. rapa inner leaves using a 300K Brassica rapa microarray with 47,548 unigenes.
2. Materials and Methods
2.1. Plant Materials
Two lines of Chinese cabbage (Brassica rapa ssp. pekinensis) were used in this study: hybrid cultivar Wheessen (Woori Seed Co. Korea) for yellow inner leaves and the inbred line Kenshin for white inner leaves (Figure S1 in Supplementary Material available online at http://dx.doi.org/10.1155/2014/204969). Seedlings were transplanted to cabbage patch at Chungnam National University, Daejeon, Korea, on September 1. After completion of the heading process (November 1), inner leaves were harvested from 10 plants. The blade portion of the leaves (excluding the midrib) was separated, frozen in liquid nitrogen, and stored at −70°C until use.
The wild-type strain and T-DNA insertional mutants (SALK_045674, SALK_008677C, SALK_098442C, and SALK_012835) of Arabidopsis thaliana (L.) Heynh var. Columbia (Col-0) were collected from ABRC (http://www.arabidopsis.org/). Seeds were sterilized with 50% bleach solution with 0.1% triton X-100 (Sigma, USA), followed by germination on 60 mm × 60 mm pots in potting soil. Plants were grown in growth chamber at 22 ± 0.5°C, with a photon flux density of 140 ± 2 μmol/m2/sec and for a 16 h light/8 h dark photoperiod. Mutants were confirmed by PCR using primers that were specific for the gene sequence and the T-DNA left border (Table S1). After 3 weeks, all leaves from 20 plants were collected and subjected to measure the carotenoid content.
2.2. Measurement of Lutein and Carotene Content from Chinese Cabbage
Leaf samples from each plant were ground in liquid nitrogen using a mortar and pestle after which 1 mL DMSO and 10 mL methanol were added to 1 g of sample and mixed well. The sample was then incubated in darkness for 30 min after which 5 mL of hexane and 10 mL of 20% NaCl were added. Next, samples were centrifuged at 2100 ×g for 10 mins, after which the supernatant was collected and amended with 0.5 g of sodium sulfate. The samples were then vortex and centrifuged at 2100 ×g for 10 min. Finally, the supernatant was filtered through a 0.45 μm PTFE and analyzed by high performance liquid chromatography (HPLC).
The lutein and carotene content were analyzed using an HPLC (Waters Corp., Milford, Mass., USA) equipped with a 2707 autosampler, a 1525 binary pump, and a C18 reversed-phase symmetry analytical column (2.6 μm × 100 mm × 4.6 mm) with a guard column containing the same packing material as the analytical column (Phenomenex Kinetex). The following binary mobile phases were used during analysis: solvent A, 75% methanol in HPLC grade water; solvent B, ethyl acetate. The gradient for the HPLC analysis was linearly applied as follows: 0% B at zero min, 75% B at 10 min, and 100% B at 14 min. Flow rate was set to 1.0 mL/min at constant room temperature, the Waters UV/visible detector was set at 450 nm and carotenoid standards [lutein xanthophyll (alfalfa sigma) and β-carotene (wako)] were used to generate characteristic UV visible spectra and calibration curves.
Individual carotenoids in the samples were tentatively identified based on comparison of their individual UV-visible spectrum and retention time. Lutein and β-carotene content were analyzed for each sample on the day of analysis. The percentage of lutein and β-carotene was calculated per 1 g of raw sample.
2.3. Measurement of Total Carotenoid Content from Arabidopsis Plants
The content of total carotenoids from Arabidopsis plants was assayed by a simple and efficient method by Porra et al. [12]. Leaves were ground in liquid nitrogen with mortar and pestle. One mL of DMF (dimethylformamide) was added to ca. 100 mg of powder in the dark. The suspension was microfuged by 12,000 rpm at 4°C for 10 min and supernatant was diluted by 10 times with DMF. The absorbance of the resulting solution was measured at 461 and 664 nm. The carotenoid content (μg/mL) was calculated by an equation: [A461 − (0.046 × A664)] × 4. Finally, its value was transformed into μg/g tissue.
2.4. Construction of the Br300K Oligomeric Chip and Microarray Analysis
A 300k microarray chip (Br50K; version 2.0) for B. rapa designed from 47,548 unigenes (Table S2) was manufactured by NimbleGen, Inc. (http://www.nimblegen.com/) as recently described [13]. To assess the reproducibility of the microarray analysis, we repeated the experiment two times using independently prepared total RNA. The data were then normalized and processed with cubic spline normalization using quantile to adjust signal variations between chips and robust multichip analysis (RMA) using a median polish algorithm implemented in NimbleScan [14, 15]. RNA preparation, GeneChip hybridization, and data analyses were also conducted following previously described methods [16].
2.5. RT-PCR Analysis
Total RNA (5 μg) from each sample was added with random hexamer primers in a superscript first-strand cDNA synthesis system according to the manufacturer's instructions (Invitrogen, USA). Complementary DNA was diluted 10-fold and 1 μL of the diluted cDNA was used in a 20 μL PCR mixture. RT-PCR primers are listed in Table S1 and the primers for BrACT1, which was used as a control, were 5′-AGATCGTCCCCGGCTTCAAA-3′ (forward) and 5′-CAAGGCGTAAACGACGCAGG-3′ (reversed). Standard PCR was performed by subjecting the samples to 5 min initial denaturation at 94°C, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s. PCR products were electrophoresed on a 1% agarose gel.
3. Results
3.1. Carotenoid Analysis
We compared the carotenoid composition including lutein and β-carotene in the inner leaves of Wheessen (yellow inner leaves) with those of Kenshin (white inner leaves) using HPLC analysis. The content of lutein and β-carotene is listed in Table 1. A lutein-representing peak was present in both Wheessen and Kenshin, although Wheessen showed a peak area higher than that found in Kenshin (data not shown). The content of lutein and β-carotene in Wheessen inner leaves was higher than that of Kenshin inner leaves by 250- and 17-fold, respectively. These data were consistent with those observed for standard lutein and β-carotene. The results supported that carotenoid becomes the main pigment in inner leaves of Chinese cabbage and is responsible for their yellow color.
Table 1.
Content of lutein and β-carotene in two contrasting Chinese cabbage cultivars.
| B. rapa cultivar | Lutein (μg/g dry weight) |
β-carotene (μg/g dry weight) |
|---|---|---|
| Kenshin (white inner leaves) | 0.19 ± 0.08 | 0.69 ± 0.08 |
| Wheessen (yellow inner leaves) | 47.49 ± 1.59 | 12.00 ± 0.61 |
3.2. Microarray Analysis
To evaluate the pigmentation of yellow inner leaves in Chinese cabbage, we conducted microarray analyses using the version 2 Br300K chip (Table S2). Among 47,548 genes on the Br300K chip, 11,543 genes showed probe intensity (PI) values of less than 500 from two tested leaf samples. To reduce the false positives, we ignored these 11,543 genes in subsequent analyses, while the remaining 36,005 genes were subjected to significance analysis of microarray (SAM) [17]. Thus, genes with adj.P.Value or false (FDR) discovery rate below 0.05 were collected and further selected for those genes with expression greater than 2-fold or less than −2-folds. A total of 10,199 were differentially expressed: 4,807 that were upregulated and 5,392 that were downregulated in yellow inner leaves (Table S3). Highly expressed genes in yellow leaves, like the top 20 upregulated genes in yellow inner leaves (Tables S3 and S4), appeared to be new genes whose functions have not been identified in plants up to now. Some of these novel genes might be directly or indirectly involved in the determination of yellow color in Chinese cabbage.
Interestingly, Br300K microarray included 8,542 unigenes classified as no hit found upon initial analysis with Arabidopsis thaliana annotation. Among these, 180 unigenes were specifically expressed in yellow leaves (Table S5). When these sequences were subjected to BlastN analysis, some of the unigenes showed low similarity to Brassica sp. and other plant unigenes without known domain up to now (data not shown). Identification of function of these genes will be indispensable for further researches if the efficient and fast method of B. rapa transformation is available.
3.3. Functional Categorization of Genes
Gene functions were analyzed based on gene ontology (GO) to simplify the annotation process using controlled vocabularies and hierarchies including three major categories, cellular component (CC), biological process (BP), and molecular function (MF) [18]. All EST sequences that showed greater than 2-fold increases were classified into 2,312, 2,334, and 2,546 ESTs of CC, BP, and MF, respectively, while there were 2,499, 2,546, and 2,662 downregulated genes of CC, BP, and MF, respectively. The majority of the differently expressed genes were shown to be involved in metabolic and cellular process, and some were components of the plasma membrane and plastids (chloroplast). The unique sequences were grouped under the first category of CC (Figure 1), which contained cell wall, chloroplast, cytosol, ER, extracellular region, Golgi apparatus, mitochondria, nucleus, other cellular components, other cytoplasmic components, other intracellular components, other membranes, plasma membranes, plastid, ribosome, and unknown cellular components. Previous studies have reported that carotenoids are also synthesized and accumulated in lipid bodies inside the chromoplasts and plastids which accumulate pigments in flowers, fruits, and storage roots. Carotenoids are more stable in chromoplasts than in plastids because they are protected from light [19]. The second category included BP (Figure 2), with subcategories such as cell organization and biogenesis, development processes, DNA or RNA metabolism, electron transport or energy pathways, other biological processes, cellular processes, metabolic processes, protein metabolism, response to abiotic or biotic stimulus, stress, signal transduction, transcription, transport, and unknown biological processes. Although enzymes appear to be of particular importance during biological process responses, some carotenogenic enzymes such as geranylgeranyl diphosphate (GGDP) synthase (GGPS) and phytoene synthase (PSY) from the isoprenoid pathway are part of a soluble large protein complex that catalyzes the formation of phytoene in plastid stroma [20]. A large protein complex PDS catalyzes the synthesis of α- and β-carotene from phytoene in the plant plastid membrane [21]. This phytoene enzyme plays a key role in the carotenoid biosynthetic pathway. The large numbers of unique sequences were grouped under the third category of MF (Figure 3), with DNA or RNA binding, hydrolase activity, kinase activity, nucleic acid binding, nucleotide binding, other binding, other enzyme binding, other molecular functions, protein binding, receptor binding or activity, structural molecule activity, transcription factor activity, transferase activity, transporter activity, and unknown molecular functions. The expression of many genes targeted to plastids is regulated through a signal between the nucleus and the plastids. These signals, whether environmental or developmental, are supposed to include reactive oxygen species (ROS), carotenoids, and hormones. Thus, plastids seem to be important to carotenoid biosynthesis [22].
Figure 1.
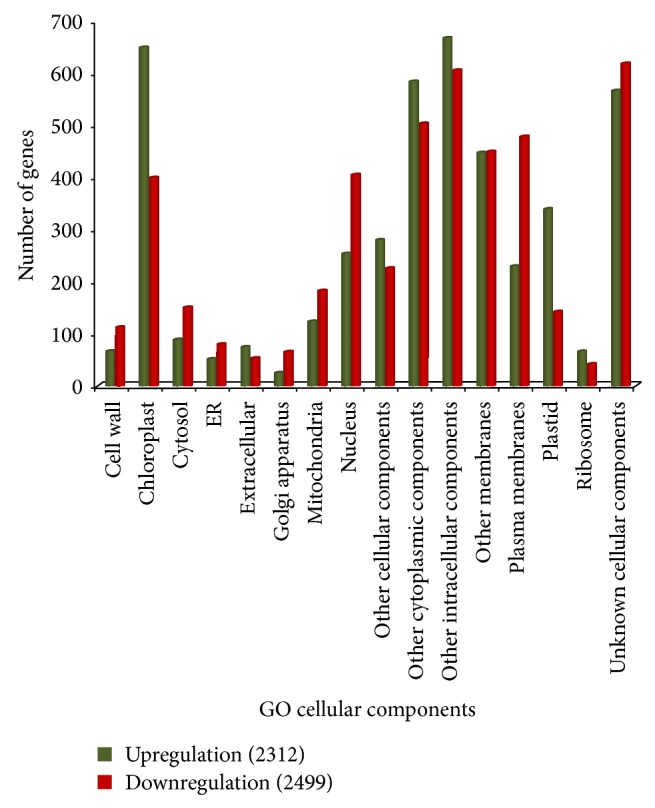
Gene ontology characterization by loci for cellular components.
Figure 2.
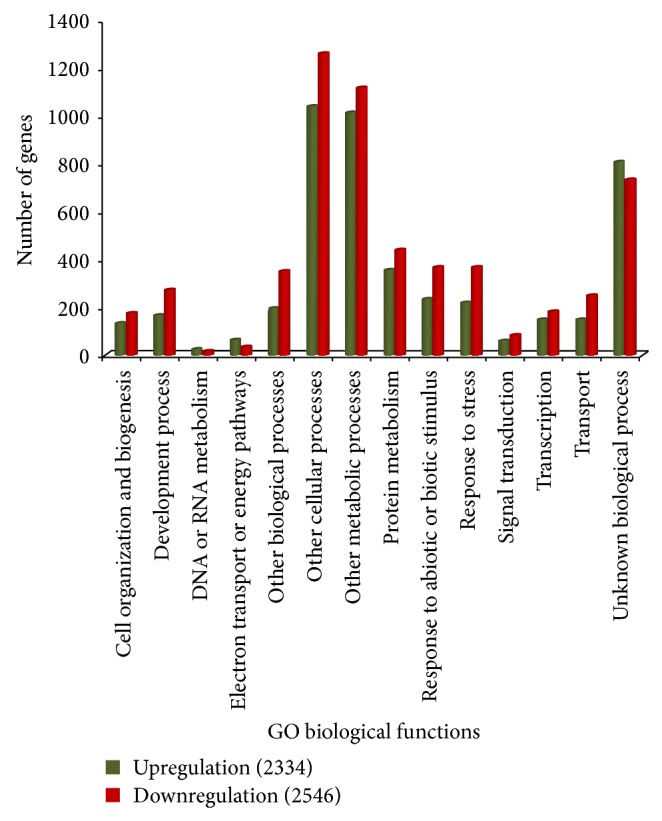
Gene ontology characterization by loci for biological functions.
Figure 3.
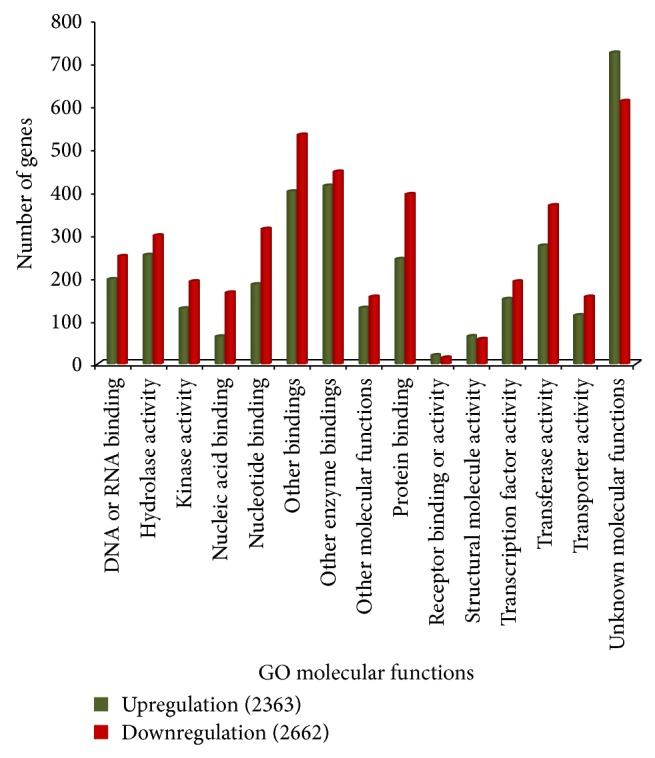
Gene ontology characterization by loci for molecular functions.
3.4. Expression of Lutein and β-Carotene Synthesis-Related Genes
To understand the relatedness of known genes that regulate the carotenoid level, expression of several categories of B. rapa (Br) orthologous genes was analyzed (Table 2). These genes include α- and β-carotene synthesis-related genes (phytoene synthase (PSY), PDS, carotene isomerase (CRTISO), and lycopene epsilon cylase (LUT2)), lutein and zeaxanthin synthesis-related genes (carotene epsilon-monooxygenase (RUT1), lutein deficient 5 (LUT5), carotene β-hydroxylase 1 (HYDB1), and HYDB2), and carotenoid degradation genes (9-cis-epoxycarotenoid dioxygenase 3 (NCED3), NCED4, and NCED6). Three known color-related genes (pale CRESS1 (PAC1), cauliflower orange (OR), and immutans (IM)) were used for references. As shown in Table 2, the expression of genes associated with carotenoid synthesis was higher in yellow inner leaves compared to that in white inner leaves, whereas the expression of degradation related genes was lower. The level of Or gene that controls β-carotene content [23] was similar in both inner leaves, while PAC1 level that controls early chloroplast development [24] was high in white leaves, suggesting these genes may not be related to the determination of yellow color. However, the level of IM which determines source-sink interactions which appeared to be little higher in yellow leaves rather than white inner leaves, suggesting somewhat relatedness of chloroplast development with carotenoid biosynthesis. Microarray analysis was conducted in duplicate using biological repeated samples and correlation coefficient between replicated microarray analyses was found to be greater more than 0.96 (Figure S2). The RT-PCR expression ratio was compared with our microarray results and correlation coefficient values were found to be greater than 0.8 (Figure S3). Unfortunately, some RT-PCR results were not consistent with PI values, suggesting requirement of real-time PCR experiment.
Table 2.
Carotene and lutein synthesis-related genes.
| Classification | Gene name | Br Chip ID | At locus | PI value | RT-PCR | ||
|---|---|---|---|---|---|---|---|
| White | Yellow | W | Y | ||||
| Alpha-, beta-carotene synthesis | BrPSY | Brapa_ESTC045764 | At5g17230 | 2,591 | 8,978 |

|
|
| BrPDS | Brapa_ESTC002927 | At4g14210 | 198 | 644 |

|
||
| BrCRTISO | Brapa_ESTC016346 | At1g06820 | 199 | 565 |

|
||
| BrRUT2 | Brapa_ESTC013530 | At5g57030 | 172 | 522 |

|
||
|
| |||||||
| Lutein and zeaxanthin synthesis | BrRUT1 | Brapa_ESTC043629 | At3g53130 | 956 | 1,544 |

|
|
| BrRUT5 | Brapa_ESTC000882 | At1g31800 | 173 | 444 |

|
||
| BrHYDB1 | Brapa_ESTC013493 | At4g25700 | 1,995 | 2,768 |

|
||
| BrHYDB2 | Brapa_ESTC006182 | At5g57030 | 1,044 | 4,856 |

|
||
|
| |||||||
| Degradation | BrNCED3 | Brapa_ESTC007729 | At3g14440 | 1,848 | 1,622 |

|
|
| BrNCED4 | Brapa_ESTC026102 | At4g19170 | 8,361 | 4,453 |

|
||
| BrNCED6 | Brapa_ESTC022333 | At3g24220 | 113 | 106 |

|
||
|
| |||||||
| Color | BrPAC1 | Brapa_ESTC007240 | At2g48120 | 4,656 | 1,719 |

|
|
| BrOR | Brapa_ESTC013943 | At5g61670 | 901 | 702 |
|
||
| BrIM | Brapa_ESTC010363 | At4g22260 | 1,285 | 1,523 |

|
||
| BrACT1 |

|
||||||
3.5. Identification of Yellow-Specific Responsive Genes
In addition to carotenoid biosynthetic genes, some of genes that are predominantly expressed in yellow inner leaves will be either directly or indirectly related to the yellow color determination in Chinese cabbage. To identify leaf-color specific genes, we selected genes as follows. Yellow-specific genes were defined as those that had PI values of over 1000 in yellow leaves but less than 500 in white leaves (Table S6), while white specific genes were those that had PI values of over 1000 in white leaves but less than 500 in yellow leaves (Table S7). The total numbers of yellow and white specific genes were 761 and 647, respectively, implying that the expression of a large number of genes is essential for determination of pigmentation and other traits in each line. These inner color-specific genes also include many unidentified genes up to now. We confirmed expression of some of color-specific genes by RT-PCR (Table 3). As shown in Table 3, many genes were unidentified genes or B. rapa specific genes. In addition, some genes, like chitinase and taumatin protein gene, might not be related to leaf color determination. These results indicate that many of color-specifically expressed genes could be due to cultivar traits, while some of them might be related to color determination.
Table 3.
Genes expressed specifically in either yellow or white inner leaves.
| Expression pattern | Gene name | Br SEQ ID | At locus | PI value | RT-PCR | ||
|---|---|---|---|---|---|---|---|
| White | Yellow | W | Y | ||||
| Yellow specific | Unknown protein | Brapa_ESTC005061 | At4g30660 | 37 | 11,124 |

|
|
| Chitinase | Brapa_ESTC005522 | At3g47540 | 34 | 13,990 |

|
||
| CA18 | Brapa_ESTC013802 | At5g14740 | 38 | 8,063 |

|
||
| Dynein light chain | Brapa_ESTC010078 | At4g27360 | 124 | 20,072 |

|
||
| Unknown protein | Brapa_ESTC010121 | At4g10300 | 24 | 3,243 |

|
||
| Reductase | Brapa_ESTC013704 | At5g52100 | 462 | 18,255 |

|
||
| Calcium binding | Brapa_ESTC035004 | At1g15860 | 497 | 11,041 |

|
||
| APX4 | Brapa_ESTC013724 | At4g09010 | 437 | 12,439 |

|
||
| SHM2 | Brapa_ESTC017490 | At5g26780 | 466 | 4,033 |

|
||
| SBPase | Brapa_ESTC047526 | At3g55800 | 440 | 7,571 |

|
||
| Thylakoid protein | Brapa_ESTC011620 | At5g52970 | 456 | 6,459 |

|
||
| Unknown protein | Brapa_ESTC006516 | At3g19800 | 458 | 4,961 |

|
||
| Brapa_ESTC048334 | no_hits | 13 | 21,498 |

|
|||
| Brapa_ESTC042517 | no_hits | 16 | 25,294 |

|
|||
| Brapa_ESTC028538 | no_hits | 32 | 16,831 |

|
|||
| Brapa_ESTC006072 | no_hits | 13 | 4,401 |

|
|||
| Brapa_ESTC000684 | no_hits | 91 | 28,921 |

|
|||
| Brapa_ESTC041578 | no_hits | 86 | 17,882 |

|
|||
| Brapa_ESTC002681 | no_hits | 175 | 29,165 |

|
|||
| Brapa_ESTC002391 | no_hits | 197 | 27,114 |

|
|||
| Brapa_ESTC048903 | no_hits | 403 | 15,662 |

|
|||
| Brapa_ESTC002008 | no_hits | 444 | 19,448 |

|
|||
|
| |||||||
| White specific | Thaumatin protein | Brapa_ESTC024359 | At4g36010 | 7,666 | 125 |

|
|
| Brapa_ESTC002914 | no_hits | 5,441 | 62 |

|
|||
| Brapa_ESTC048544 | no_hits | 2,635 | 27 |

|
|||
| Brapa_ESTC028228 | no_hits | 1,891 | 21 |

|
|||
| BrACT1 |

|
||||||
3.6. Identification of Transcription Factor Genes Induced Specifically in Yellow Leaves
Transcription factors control the expression of a genome and play vital roles in the plant life cycle. Therefore, we examined the transcription factors associated with yellow pigmentation [25]. Among 2,336 transcription factor genes, 536 clones were found to be expressed in low levels with a PI value < 500 for both yellow and white leaves, while 312 clones were constitutively expressed in both leaves (Table S8). Twenty-one transcription factors responsive to yellow color in B. rapa were identified by comparison with Arabidopsis information (Table 4). Although these TFs have not been reported for their function with respect to carotenoid biosynthesis yet, there is a possibility of involvement of these genes in color determination in B. rapa, directly or indirectly. Particularly, three genes showing high levels of expression in yellow inner leaves will be good candidates for yellow color regulation. These include Brapa_ESTC013161 (A20/AN1-like zinc finger protein), Brapa_ESTC025847 (BIM1 (BES1-interacting MYC-like protein 1)), and Brapa_ESTC006452 (ZFP8 (Zinc finger protein8)). These genes may be responsible for the carotenoid synthesis or related to pigment metabolism during carotenoid production.
Table 4.
List of transcription factors expressed specifically in yellow inner leaves.
| B. rapa SEQ_ID | TAIR7_cds ID | A_thaliana_homologue_Description | PI value | Fold Change (Y/W) | |
|---|---|---|---|---|---|
| White leaves | Yellow leaves | ||||
| Brapa_ESTC013161 | AT1G51200 | A20/AN1-like zinc finger protein | 161 | 2674 | 16.6 |
| Brapa_ESTC025847 | AT5G08130 | BIM1 (BES1-interacting Myc-like protein 1) | 280 | 3517 | 12.5 |
| Brapa_ESTC006452 | AT2G41940 | ZFP8 (ZINC FINGER PROTEIN 8) | 269 | 3323 | 12.4 |
| Brapa_ESTC024811 | AT5G64810 | WRKY51 (WRKY DNA-binding protein 51) | 155 | 1002 | 6.5 |
| Brapa_ESTC017689 | AT1G51600 | ZML2 (ZIM-LIKE 2) | 472 | 3009 | 6.4 |
| Brapa_ESTC017762 | AT2G18328 | DNA binding | 401 | 2336 | 5.8 |
| Brapa_ESTC004343 | AT4G16420 | ADA2B (PROPORZ1) | 286 | 1593 | 5.6 |
| Brapa_ESTC051029 | AT1G49950 | ATTRB1/TRB1 (TELOMERE REPEAT BINDING FACTOR 1) | 396 | 1918 | 4.8 |
| Brapa_ESTC047297 | AT1G49950 | ATTRB1/TRB1 (TELOMERE REPEAT BINDING FACTOR 1) | 400 | 1902 | 4.8 |
| Brapa_ESTC032962 | AT4G31060 | AP2 domain-containing transcription factor, putative | 382 | 1582 | 4.1 |
| Brapa_ESTC006124 | AT5G64810 | WRKY51 (WRKY DNA-binding protein 51) | 312 | 1166 | 3.7 |
| Brapa_ESTC001675 | AT4G17810 | Nucleic acid binding/transcription factor/zinc ion binding | 442 | 1601 | 3.6 |
| Brapa_ESTC011816 | AT3G51960 | bZIP family transcription factor | 309 | 1079 | 3.5 |
| Brapa_ESTC037364 | AT4G12020 | WRKY19 (WRKY DNA-binding protein 19) | 412 | 1394 | 3.4 |
| Brapa_ESTC014402 | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54) | 322 | 1021 | 3.2 |
| Brapa_ESTC035091 | AT3G57600 | AP2 domain-containing transcription factor, putative | 455 | 1411 | 3.1 |
| Brapa_ESTC011257 | AT4G34680 | GATA transcription factor 3, putative (GATA-3) | 371 | 1095 | 3.0 |
| Brapa_ESTC025927 | AT3G17609 | HYH (HY5-HOMOLOG) | 392 | 1152 | 2.9 |
| Brapa_ESTC035196 | AT4G30410 | Transcription factor | 480 | 1248 | 2.6 |
| Brapa_ESTC025999 | AT3G50890 | ATHB28 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 28) | 464 | 1189 | 2.6 |
| Brapa_ESTC030153 | AT1G79180 | AtMYB63 (myb domain protein 63) | 470 | 1023 | 2.2 |
Four T-DNA knock-out Arabidopsis mutants corresponding to three B. rapa genes (Brapa_ESTC013161, Brapa_ESTC025847, and Brapa_ESTC006452) were analyzed for carotenoid content by comparison with Arabidopsis wild type (Table S9). The content of total carotenoid was greatly reduced in T-DNA knock-out mutants, implying their possible function in the carotenoid biosynthesis. Particularly knock-out of A20/AN1-like zinc finger protein gene greatly affected the carotenoid content.
4. Discussion
Transcriptome studies using microarray analyses are increasingly being used to identify key genes involved in plant growth, development, and responses to environmental changes. Nevertheless, microarray technology has not been widely applied to the study of Brassica species owing to the absence of a high density Brassica microarray with sufficient coverage of the entire genome. Instead, Arabidopsis microarray has been widely employed for analysis of Brassica species because both species belong to the same family (Brassicaceae) and have an evolutionary close relationship. The Brassica genome has generally diverged from Arabidopsis [26, 27] and consists of approximately 46,000 genes [28]. Although the Arabidopsis microarray chip may provide some information regarding gene regulation of Brassica plants, it is not sufficient to provide information regarding Brassica biology [29]. Therefore, to accurately conduct transcriptome studies, a Brassica specific microarray Br300K was used in the present study. This microarray will also be useful in investigations of other Brassica species, including B. napus and B. oleracea.
Carotene and lutein contents are known key factors associated with yellow pigmentation in Chinese cabbage. However, no reports of specific genes involved in yellow color determination in Chinese cabbage have been published to date, except the involvement of BrCRTISO in orange color determination [30]. Researches to identify genes regulating yellow inner leaves and to confirm inheritance of its characteristics in Chinese cabbage are essential for the improvement of Chinese cabbage quality. We identified many genes that were specifically expressed in yellow inner leaves using microarray experiment and confirmed some of them by RT-PCR. Furthermore, a high correlation was observed between microarray data of the two samples. In addition, a high correlation between RT-PCR analysis and microarray data indicated that our data from the Br300K microarray were reliable and reproducible.
Wheessen and Kenshin were used for HPLC analysis to compare carotenoid composition. In a previous study, lutein was found to be predominantly accumulated in yellow inner leaves [5, 31]. Carotenoid composition, especially lutein in the inner leaves of yellow cultivar, was higher than that in the inner leaves of orange cultivar upon HPLC analysis [30]. Similar results were obtained from yellow inner leaves compared to white inner leaves (Table 1). Collectively, these data confirmed that carotenoid profiles of the yellow inner leaf cultivars of Chinese cabbage were obviously higher than and different from those of common white leaf cultivars [1]. However, there is no report on the involvement of related genes and its expression. We found out that expression of lutein and β-carotene biosynthesis-related genes (BrPSY, BrPDS, BrRUT, BrHYDB, and BrCRTISO) is high in yellow inner leaves, whereas expression of its degradation-related genes is low (Table 2). In the carotenoid biosynthetic pathway, phytoene synthase (PSY) catalyzes the first committed reaction of the head to head condensation of two geranylgeranyl diphosphate (GGPP) molecules [20]. BrHYDB acts as a catalyst for the production of zeaxanthin hydrolyzed from β-carotene [32–34]. BrPSY and BrHYDB are believed to be responsible for β-carotene and zeaxanthin synthesis [20]. The primary role of BrCRTISO is as an enzyme that converts prolycopene into lycopene in B. rapa plants. In this study, comparative analysis with Arabidopsis counterparts revealed that BrCRTISO is likely responsible for carotenoid biosynthesis. These results are similar to those reported for Arabidopsis, which encodes a functional carotenoid isomerase and causes prolycopene accumulation when mutated [35]. Lee et al. [30] reported that BrCRTISO1 was not normally expressed in orange cultivars and this result indicates that a lack of BrCRTISO1 transcript was the cause of sequence variation in the orange cultivars when compared to the counterpart gene in the yellow leaves cultivars. This could be one of the causes responsible for the yellow inner leaves phenotype.
We identified 1 unknown gene and 9 putative genes responsible for yellow pigmentation (Table 3). These genes showed high expression in yellow leaves when compared with white leaves. Moreover, some stress-related genes-like such as chitinase, dynein light chain, and reductase genes [36] also showed high expression in yellow leaves (Table 3). Although these genes are not involved in carotenoid biosynthesis, this might be one of the causes of pigmentation. In addition, a large number of no_hit_found genes (novel genes in B. rapa) (180 genes) were specifically expressed in yellow leaves (Table S5). It will be essential for identifying function of these genes in relation to leaf color determination. Based on these findings, functional studies should be conducted to clarify the nature of these unknown genes in Chinese cabbage.
We identified 21 putative transcription factor genes associated with yellow color in B. rapa. Basic helix-loop-helix proteins and MYB proteins also function together to control flower pigmentation in snapdragon [37] and petunia [38]. MYB transcription factors act as the primary component of color differences between plant varieties such as potato, tomato, pepper [16], and grape [39] and in some species of Antirrhinum [40]. Zinc binding proteins are triggered via specific mechanisms related to transcription factors involved in important biological processes [41]. These facts may support that these TFs will participate in the regulation of carotenoid content in B. rapa.
Regarding the three transcription factors that were highly expressed in yellow inner leaves (Table 4, Table S9), none of them has been reported as gene as determining color formation. Arabidopsis orthologous (A20/AN1-like zinc finger protein) of Brapa_ESTC013161 shows induced expression by stress with redox-dependent regulation [42], and Arabidopsis BIM1 controls embryonic patterning through brassinosteroid signaling [43]. There is no report on the function of Brapa_ESTC006452 orthologous (ZFP8) yet. All these results suggest that transcription factors showing yellow-specific expression might be related to control the carotenoid biosynthesis in B. rapa.
In conclusion, we identified three genes as functionally novel genes that are involved in yellow color pigmentation and also a large number of unknown genes exhibit functionally diverse transcription factor activity, using newly developed 300K Brassica rapa microarrays. These unknown genes may be responsible for yellow pigmentation in Chinese cabbage and the results showed that biological functions of particular transcription factors may be activated in the pigmentation pathways. Despite identification of some stress related genes not related to carotenoid biosynthesis-like chitinase, reductase proteins might be one of the causes for the yellow color trait. Overall, the results presented herein will provide a tool for future transcriptome studies and investigation of genes involved in carotenoid biosynthesis of Chinese cabbage as well as valuable insight into the response to yellow pigmentation production.
Supplementary Material
In this study, we used Br300K B. rapa microarray and identified three genes as functionally novel genes that involved in yellow color pigmentation. Supplementary figures (Figure S1 to S3) explains the information about plant materials and correlation coefficient of two experimental results. The three identified transcription factor genes are specifically expressed and confirmed their relatedness in yellow pigmentation. Table S1 to S9 supports our results and provides detailed annotation information of some differentially expressed genes.
Acknowledgments
This research was supported by Golden Seed Project (Center for Horticultural Seed Development, nos. 213003-04-2-CG100 and 213003-04-2-SB230), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Chen P. F. The Study of Identify and Accumulation Mechanisms and Regulation Control of Carotenoid on Chinese Cabbage. Yangzhou University; 2008. [Google Scholar]
- 2.Giovannucci E. Tomatoes, tomato-based products, lycopene, and cancer: review of the epidemiologic literature. Journal of the National Cancer Institute. 1999;91(4):317–331. doi: 10.1093/jnci/91.4.317. [DOI] [PubMed] [Google Scholar]
- 3.Hadley C. W., Miller E. C., Schwartz S. J., Clinton S. K. Tomatoes, lycopene, and prostate cancer: progress and promise. Experimental Biology and Medicine. 2002;227(10):869–880. doi: 10.1177/153537020222701006. [DOI] [PubMed] [Google Scholar]
- 4.Podsedek A. Natural antioxidants and antioxidant capacity of Brassica vegetables: a review. LWT—Food Science and Technology. 2007;40(1):1–11. doi: 10.1016/j.lwt.2005.07.023. [DOI] [Google Scholar]
- 5.Wills R. B. H., Rangga A. Determination of carotenoids in Chinese vegetables. Food Chemistry. 1996;56(4):451–455. doi: 10.1016/0308-8146(95)00226-X. [DOI] [Google Scholar]
- 6.Singh J., Upadhyay A. K., Prasad K., Bahadur A., Rai M. Variability of carotenes, vitamin C, E and phenolics in Brassica vegetables . Journal of Food Composition and Analysis. 2007;20(2):106–112. doi: 10.1016/j.jfca.2006.08.002. [DOI] [Google Scholar]
- 7.Matsumoto E., Yasui C., Ohi M., Tsukada M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis) Euphytica. 1998;104(2):79–86. doi: 10.1023/a:1018370418201. [DOI] [Google Scholar]
- 8.Qin H., Feng T., Harding S. A., Tsai C.-J., Zhang S. An efficient method to identify differentially expressed genes in microarray experiments. Bioinformatics. 2008;24(14):1583–1589. doi: 10.1093/bioinformatics/btn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Kou X., Fugal K., McLaughlin J. Comparison of HPLC methods for determination of anthocyanins and anthocyanidins in bilberry extracts. Journal of Agricultural and Food Chemistry. 2004;52(4):688–691. doi: 10.1021/jf034596w. [DOI] [PubMed] [Google Scholar]
- 10.Kim C., Kikuchi S., Satoh K., et al. Genetic analysis of seed specific gene expression for pigmentation in colored rice. Biochip Journal. 2009;3(2):125–129. [Google Scholar]
- 11.Yang K. A., Lim C. J., Hong J. K., et al. Identification of Chinese cabbage genes up-regulated by prolonged cold by using microarray analysis. Plant Science. 2005;168(4):959–966. doi: 10.1016/j.plantsci.2004.11.011. [DOI] [Google Scholar]
- 12.Porra R. J., Thompson W. A., Kriedemann P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica. 1989;975:384–394. doi: 10.1016/s0005-2728(89)80347-0. [DOI] [Google Scholar]
- 13.Dong X., Feng H., Xu M., et al. Comprehensive analysis of genic male sterility-related genes in Brassica rapa using a newly developed Br300K oligomeric chip. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0072178.e72178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31(4, article e15) doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Workman C., Jensen L. J., Jarmer H., et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biology. 2002;3(9) doi: 10.1186/gb-2002-3-9-research0048.research0048.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jong W. S., Eannetta N. T., de Jong D. M., Bodis M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theoretical and Applied Genetics. 2004;108(3):423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- 17.Tusher V. G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Gene Ontology Consortium. The gene ontology in 2010: extensions and refinements. Nucleic Acids Research. 2009;38(supplement 1):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vishnevetsky M., Ovadis M., Vainstein A. Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends in Plant Science. 1999;4(6):232–235. doi: 10.1016/s1360-1385(99)01414-4. [DOI] [PubMed] [Google Scholar]
- 20.Lu S., Li L. Carotenoid metabolism: biosynthesis, regulation, and beyond. Journal of Integrative Plant Biology. 2008;50(7):778–785. doi: 10.1111/j.1744-7909.2008.00708.x. [DOI] [PubMed] [Google Scholar]
- 21.Lopez A. B., Yang Y., Thannhauser T. W., Li L. Phytoene desaturase is present in a large protein complex in the plastid membrane. Physiologia Plantarum. 2008;133(2):190–198. doi: 10.1111/j.1399-3054.2008.01058.x. [DOI] [PubMed] [Google Scholar]
- 22.Egea I., Barsan C., Bian W., et al. Chromoplast differentiation: current status and perspectives. Plant and Cell Physiology. 2010;51(10):1601–1611. doi: 10.1093/pcp/pcq136. [DOI] [PubMed] [Google Scholar]
- 23.Lu S., van Eck J., Zhou X., et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. The Plant Cell. 2006;18(12):3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiter R. S., Coomber S. A., Bourett T. M., Bartley G. E., Scolnik P. A. Control of leaf and chloroplast development by the Arabidopsis gene pale cress . Plant Cell. 1994;6(9):1253–1264. doi: 10.1105/tpc.6.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim C. K., Park S. H., Kikuchi S., et al. Genetic analysis of gene expression for pigmentation in Chinese cabbage (Brassica rapa) Biochip Journal. 2010;4(2):123–128. doi: 10.1007/s13206-010-4206-9. [DOI] [Google Scholar]
- 26.O'Neill C. M., Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. The Plant Journal. 2000;23(2):233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- 27.Rana D., van den Boogaart T., O'Neill C. M., et al. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant Journal. 2004;40(5):725–733. doi: 10.1111/j.1365-313x.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang T.-J., Kim J. S., Kwon S.-J., et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa . Plant Cell. 2006;18(6):1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S. C., Lim M. H., Kim J. A., et al. Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K Oligo microarray. Molecules and Cells. 2008;26(6):595–605. [PubMed] [Google Scholar]
- 30.Lee S., Lee S.-C., Byun D. H., et al. Association of molecular markers derived from the BrCRISTO1 gene with prolycopene-enriched orange-colored leaves in Brassica rapa . Theoretical and Applied Genetics. 2014;127(1):179–191. doi: 10.1007/s00122-013-2209-3. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe M., Musumi K., Ayugase J. Carotenoid pigment composition, polyphenol content, and antioxidant activities of extracts from orange-colored Chinese cabbage. LWT-Food Science and Technology. 2011;44(9):1971–1975. doi: 10.1016/j.lwt.2011.04.010. [DOI] [Google Scholar]
- 32.Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):402–407. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galpaz N., Ronen G., Khalfa Z., Zamir D., Hirschberg J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18(8):1947–1960. doi: 10.1105/tpc.105.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J., DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3474–3479. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H., Kreunen S. S., Cuttriss A. J., DellaPenna D., Pogson B. J. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14(2):321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed N. U., Park J.-I., Jung H.-J., et al. Molecular characterization of stress resistance-related chitinase genes of Brassica rapa . Plant Physiology and Biochemistry. 2012;58:106–115. doi: 10.1016/j.plaphy.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Goodrich J., Carpenter R., Coen E. S. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68(5):955–964. doi: 10.1016/0092-8674(92)90038-E. [DOI] [PubMed] [Google Scholar]
- 38.Quattrocchio F., Wing J. F., van der Woude K., Mol J. N. M., Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant Journal. 1998;13(4):475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi S., Ishimaru M., Hiraoka K., Honda C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 2002;215(6):924–933. doi: 10.1007/s00425-002-0830-5. [DOI] [PubMed] [Google Scholar]
- 40.Schwinn K., Venail J., Shang Y., et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the Genus antirrhinum. Plant Cell. 2006;18(4):831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takatsuji H. Zinc-finger transcription factors in plants. Cellular and Molecular Life Sciences. 1998;54(6):582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ströher E., Wang X. J., Roloff N., Klein P., Husemann A., Dietz K. J. Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in arabidopsis thaliana. Molecular Plant. 2009;2(2):357–367. doi: 10.1093/mp/ssn084. [DOI] [PubMed] [Google Scholar]
- 43.Chandler J. W., Cole M., Flier A., Werr W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNRÖSCHEN and DORNRÖSCHEN-LIKE. Plant Molecular Biology. 2009;69(1-2):57–68. doi: 10.1007/s11103-008-9405-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In this study, we used Br300K B. rapa microarray and identified three genes as functionally novel genes that involved in yellow color pigmentation. Supplementary figures (Figure S1 to S3) explains the information about plant materials and correlation coefficient of two experimental results. The three identified transcription factor genes are specifically expressed and confirmed their relatedness in yellow pigmentation. Table S1 to S9 supports our results and provides detailed annotation information of some differentially expressed genes.


