Abstract
Background
The prevalence of mental illness, particularly depression and dementia, is increased by obesity. Here we test the hypothesis that obesity-associated changes in gut microbiota are intrinsically able to impair neurocognitive behavior in mice.
Methods
Conventionally housed, non-obese, adult male C57BL/6 mice maintained on a normal chow diet were subjected to a microbiome depletion/transplantation paradigm using microbiota isolated from donors (given) on either high-fat (HFD) or control diet (CD). Following recolonization, mice were subjected to comprehensive behavioral and biochemical analyses.
Results
The mice given HFD microbiota had significant and selective disruptions in exploratory, cognitive, and stereotypical behavior compared to mice with CD microbiota in the absence of significant differences in body weight. Sequencing-based phylogenetic analysis confirmed the presence of distinct core microbiota between groups, with alterations in α- and β- diversity, modulation in taxonomic distribution, and statistically significant alterations to metabolically active taxa. HFD microbiota also disrupted markers of intestinal barrier function, increased circulating endotoxin, and increased lymphocyte expression of Iba1, TLR2, and TLR4. Finally, evaluation of brain homogenates revealed that HFD-shaped microbiota increased neuroinflammation and disrupted cerebrovascular homeostasis.
Conclusions
Collectively, these data reinforce the link between gut dysbiosis and neurologic dysfunction and suggest that dietary and/or pharmacological manipulation of gut microbiota could attenuate the neurologic complications of obesity.
Keywords: gut dysbiosis, intestinal permeability, mental health, neuroinflammation, obesity, neurobehavior
INTRODUCTION
The etiology of most neuropsychiatric disorders is likely multifactorial and based on genetic and environmental risk factors (1). One potentially important environmental driver of mental illness is obesity, which dramatically increases risk of depression, dementia, and stroke, and is associated with increased brain pathology and decreased brain function (reviewed in (2)). For example, functional studies report deficits in learning, memory, and executive function in obese compared to nonobese patients (3),(4), and likewise link obesity to enhanced depression and anxiety disorders (5),(6). However, there are contradictory reports that dispute these findings (7),(8), suggesting that the cause of obesity-associated mental illness is not obesity per se, but rather one or more of the variable manifestations of obesity.
One potential site whereby diet-induced obesity could affect physiology is the gut microbiome, as recent advances in 16S rRNA sequencing and informatics have revealed that modern diets high in fat and sugar trigger robust alterations in the core gut microbiome (9). The human gastrointestinal tract harbors as many as 100 trillion bacteria from up to 1000 distinct species, and this dynamic population of microbes participates in numerous physiologic functions including nutrition/digestion, growth, inflammation, immunity, and protection against pathogens (10),(11),(12). Accordingly, the varying combinations of bacteria within individuals has been suggested to underlie variable host susceptibility to illness (13),(14), including neuropsychiatric impairment (15),(16). For example, specific alterations in colon bacteria are associated with cognitive impairment in patients with hepatic encephalopathy (17), and clinical studies show that oral probiotics decrease anxiety and improve mental outlook (18),(19). Furthermore, animal studies have shown that behavior and synaptic plasticity are altered in germ-free mice, and that this phenotype is reversed by microbiome colonization (20). The aim of the present study was to test the hypothesis that the obesity-concomitant microbiome undermines behavior even in the absence of obesity. Non-obese, adult male C57BL/6 mice were conventionally housed and maintained on chow diet but subjected to a microbiome depletion paradigm followed by adoptive transfer of cecal plus colonic contents collected from donor mice fed either high fat (HFD) or control diet (CD). Recipient mice were subjected to a battery of neuropsychological tests, followed by sequencing of gut microbiota, and thorough biochemical evaluation of intestine, blood and brain samples.
EXPERIMENTAL METHODS
Animals and treatments
The PBRC IACUC approved all experimental protocols, which were compliant with NIH guidelines. To generate microbiota donor material, 8 week-old male C57Bl/6 mice (Jackson Laboratories) were given regular chow diet (13% fat calories, Purina LabDiet 5001) or high fat diet (60% fat calories, Research Diets D12492) for 10 weeks (see Supplementary Table S1 for diet compositions). At the time of microbiota harvest, the high-fat fed mice weighed 37, 0 ± 1.7 g and the chow fed mice weighed 24.5 ± 1.2 g. Mice were euthanatized and cecal plus colonic contents were harvested, pooled, and diluted 40-fold (weight: volume) in sterile water. After centrifugation at 800 RPM, the supernatant was aliquoted under sterile conditions for storage at −80°C. Recipient 3 month-old male C57Bl/6 mice (Jackson Laboratories) were group-housed under standard laboratory conditions with free access to water and chow diet (Purina LabDiet 5001). Mice were given a cocktail of ampicillin, gentamicin, metronidazole, neomycin (all at 0.25 mg/day), and vancomycin (0.125 mg/day) once daily for 14 consecutive days by oral gavage (21). Mice were recolonized 72 hours later via daily oral gavage of donor microbiota (100 µl) for 3 days (22), (23). To offset potential founder and/or cage effects (24) and to reinforce the donor microbiota genotype, booster inoculations were given bi-weekly throughout the study. Body weight and composition was measured regularly, and all mice were euthanatized following behavioral testing. Plasma, lymphocytes, intestines, intestinal contents, and brains were collected, with data compiled from 10 animals per group.
Behavioral Testing
All behavioral testing was conducted between 7 am and 1 pm, and was recorded/analyzed using Any-Maze software (Stoelting Co) for unbiased quantification of body location, orientation, distance, speed, and mobility/immobility. Detailed Methods on behavioral assays are provided in the Supplement. Overall anxiety and exploratory behavior was assessed using elevated plus (25) and open field assays (26). Stereotypical behavior was assessed by quantifying marble burying during a 30 minute trial in a novel cage pre-loaded with 4 cm of clean bedding and 16 evenly-spaced marbles (Figure 1D and (27)). Memory was measured using a video-based fear conditioning system (Med-Associates) that pairs a unique context (scent and cage) and unconditioned stimulus (auditory tone) with a repeated foot shock (day 1), and then quantifies freezing behavior to the context (day 2) and to the tone (day 3) as measures of memory (28). Behavioral tests were administered in the order listed above over 2 weeks, beginning 3 weeks after the end of antibiotic treatment (Figure 1A). To curtail carry-over effects, the elevated plus, open field, and marble-burying assay were conducted during the first week of testing with 48 hours recovery between each task, while fear conditioning was tested the following week (29),(30).
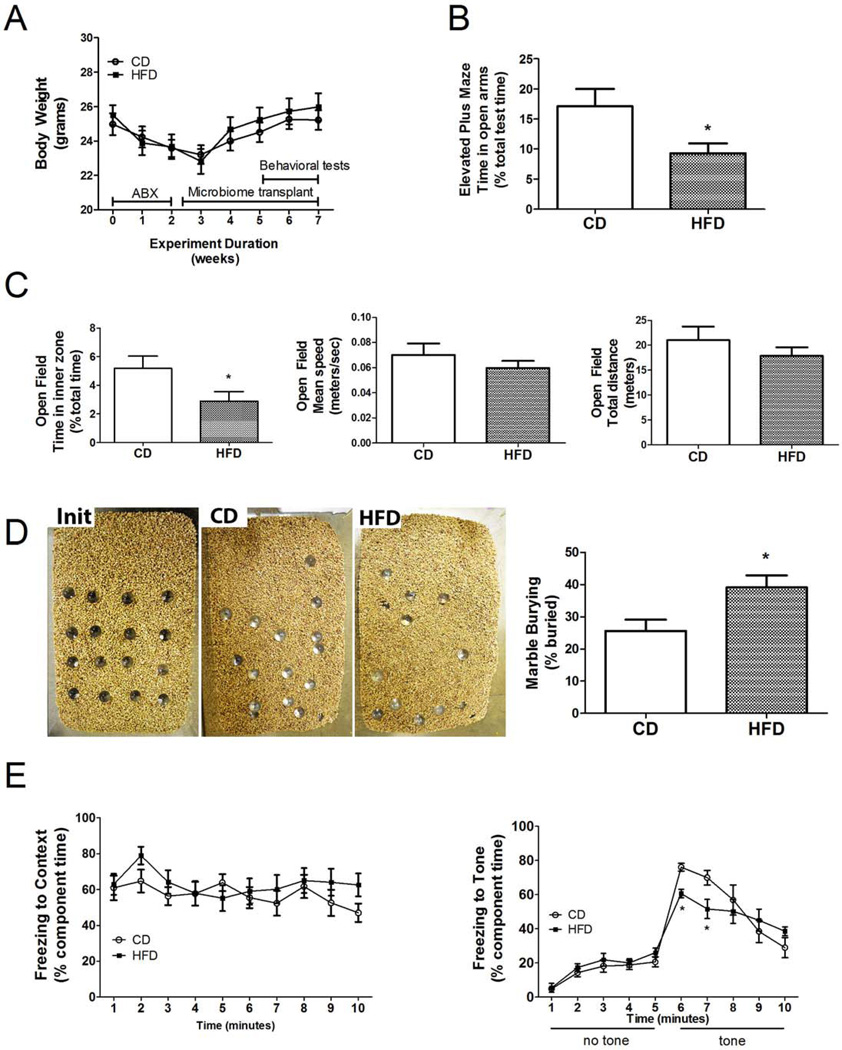
Figure 1.
High fat diet associated microbiota increases anxiety and stereotypical behaviors, but decreases memory in mice. (A) Body weight during depletion (ABX), recolonization (Microbiome transplant), and behavioral protocols shows no difference between mice transplanted with microbiota from high fat diet fed donors (HFD) or control diet fed donors (CD). (B) Time spent exploring the open arms of the Elevated Plus Maze was significantly reduced in HFD as compared to CD mice. (C) Time spent in the inner zone of the Open Field (left panel), but not mean speed (center panel) or total distance traveled (right panel) was significantly decreased in HFD mice as compared to CD mice. (D) Marble burying behavior was significantly increased in HFD vs. CD mice, as shown by representative images of marble placement before (Init) or after the 30 minute trail with mice transplanted with CD- or HFD– associated microbiota, and quantitative analysis (right panel). (E) Following fear conditioning, freezing behavior to context on training day 2 was not different between groups (left panel), but conditioned freezing to the tone on day 3 was significantly reduced in HFD as compared to CD mice (right panel). All data are presented as mean ± SEM of 10 mice per group, and *p < 0.05 based on t-tests or ANOVA.
16S Metagenomic Sequencing
Fecal samples were collected under aseptic conditions from all mice during the final week of behavioral testing, while cecal samples were collected aseptically at euthanasia. Sequencing and bioinformatics were performed by the LSU Microbial Genomics Resource Center. DNA was isolated using QIAamp DNA Stool kits (Qiagen) modified to include a bead-beating step. After DNA isolation, 16S rDNA hypervariable regions V3-V4 were PCR amplified using primers with the V3F CCTACGGGAGGCAGCAG and V4R GGACTACHVGGGTWTCTAAT gene-specific sequences, Illumina adaptors, and molecular barcodes as described (31) to produce 430bp amplicons. Samples were sequenced on an Illumina MiSeq using V3 sequencing kit (300bp paired end reads). The forward read files were processed through the UPARSE pipeline (32), truncating reads to a uniform length of 250 bp, then removing reads with quality scores less than 16. Additional filtering removed reads that appeared only once throughout all samples (“singletons”) and remaining unique reads were clustered into Operational Taxonomic Units (OTU) at 97% similarity. Chimeric OTUs were removed as identified by UCHIME run against a gold standard reference database of non-chimeric sequences. Finally, the original filtered reads (before dereplication) were mapped to the OTUs using USEARCH at 97% identity. QIIME 1.8 was used to pick and align a representative set. The Ribosomal Database Project (RDP) classifier was used to assign a taxonomic classification to each read in the representative set and a phylogentic tree was constructed from the representative sequences. Among samples, the minimum read count after filtering was 21182, with a median read count of 57537. Relative abundance of each OTU was examined at Phylum, Class, Order, Family, Genus and Species levels. Alpha (within a community) and beta (between communities) diversity metrics as well as taxonomic community assessments were produced using QIIME 1.8 scripts.
Plasma and Tissue Analyses
Whole blood was collected by cardiac puncture of terminally anesthetized mice into EDTA-treated tubes, and plasma and lymphocytes were isolated and analyzed immediately or stored at −80°C. Endotoxin levels in plasma were measured using a kinetic limulus amebocyte lysate test (Lonza Group, Limited). Levels of bioactive lipids and hormones/adipokines were measured as previously reported (33). Lymphocyte, colon, jejunum, and brain (medial prefrontal cortex) samples were homogenized and processed for Western blot with chemiluminescence as previously reported (33). For accurate quantification across blots, samples from both treatment groups were included in each individual blot, and data were first calculated as a ratio of expression over tubulin expression, and then expression in mice with HFD microbiota was calculated/presented as percent expression in control (CD) mice.
Statistical analyses
All behavioral and biochemical data were analyzed using Prism software (GraphPad Software, Inc.), and are displayed as mean ± standard error of measurement. Body weight and fear conditioning behavior was analyzed with 2-way repeated measures ANOVA to determine main effects of treatment and duration, followed by planned Bonferroni post-hoc comparisons to determine differences between groups. All other behavioral and biochemical data were analyzed by unpaired t-tests. Statistical significance for all analyses was accepted at p < 0.05, and *, **, and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively.
For sequencing data, alpha diversity rarefaction curves were produced by plotting several diversity metrics against the number of sequences considered from a sample. Subsequent analysis of diversity was performed at a depth of 20000 sequences per sample. Statistical significance was compared using a non-parametric permutation test for a pairwise comparison of categories. The p-values were Bonferroni-corrected for multiple testing. Beta diversity, principle coordinates analysis (PCoA) plots were produced, using both weighted (considers abundance of each species) and unweighted (considers presence/absence of species) UniFrac metrics. Plots were visualized using the Emperor 3D Viewer. Statistical significance was assessed with a non-parametric permutation test to compare a chosen category. ANOVA was used to test for differences in relative abundance of specific OTUs for each group. An unweighted g-test was used to evaluate the statistical significance of the presence/absence of OTUs across categories. DESeq2 software was also used to test for differential representation of OTUs, and also to identify in an unbiased manner all individual OTUs in which the group difference reached statistical significance using mean normalized sequence count level higher than 30 counts and FDR<0.05 criteria (Wald statistics with Benjamini-Hochberg correction) in either the CD or HFD group.
RESULTS
HFD-derived gut microbiota impair behavioral performance in mice
All animals tolerated the antibiotic regimen with no overt effects other than a mild, approximate 10% loss of body weight (Fig 1A). qPCR-based analyses of 16S RNA levels in fecal samples collected from mice mid-way through the antibiotic treatment revealed an approximate 90–95% reduction in fecal bacterial burden compared to matched but untreated mice (fecal DNA concentration 82,502.1 + 18,255 µg/g in control samples, 3,417.4 + 1,212 µg/g in samples following antibiotic exposure). Mice were subjected to thorough behavioral phenotyping starting 3 weeks after recolonization with either microbiota from high-fat or chowfed mice (Fig 1A). Exploratory and anxiety-based behavior assessed using the Elevated Plus maze revealed that mice with HFD-associated microbiota spent significantly less time (t(18)=2.32, p<0.05) in the open arms of the maze (Fig. 1B). The Open Field assay likewise revealed that mice with HFD-associated microbiota spent significantly less time (t(18)=2.13, p<0.05) in the inner zone of the open field (Fig. 1C). Overall locomotor activity assessed in the Open Field showed no differences in mean speed or total distance traveled between CD and HFD groups (Fig. 1C), suggesting that decreased exploratory behavior in mice with HFD microbiota reflect increased anxiety not decreased motor function. Mice were tested for marble burying, a measure of compulsive, anxiety-like behavior (34). HFD-shaped microbiota were associated with a significant increase (t(18)=2.64, p<0.05) in marble burying (Fig. 1D). Finally, the fear conditioning assay was used to measure memory. Significant differences in freezing behavior were observed on the third day of the fear conditioning test, when cued learning (freezing in a novel context in response to the tone) was assessed. Post-hoc analyses revealed that freezing in response to tone was significantly decreased in mice with HFD microbiota as compared to mice with CD microbiota (Fig. 1E). In addition, the averaged slopes of behavioral waveforms depicting freezing of individual mice in response to tone were significantly different between the groups (−11.8 ± 1.48 in CD mice; −5.5 ± 0.6 in HFD mice; t(18)=4.05, p<0.001) suggesting attenuated within-session extinction of fear behavior in mice with HFD microbiota.
Microbiota transplantation results in distinct phylogenetic profiles
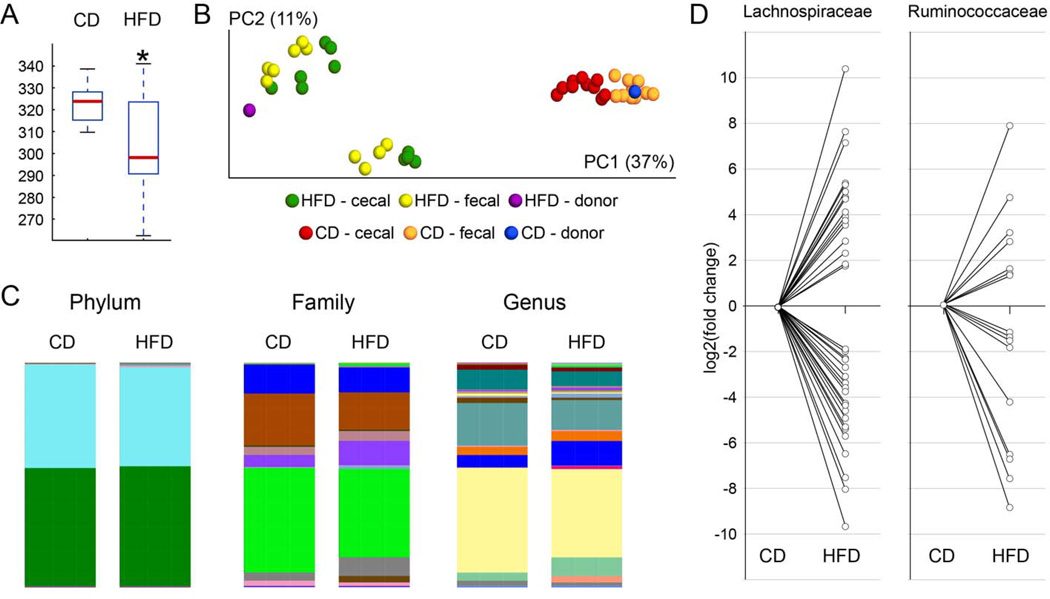
Fecal and cecal microbiota compositions in recipient mice were analyzed by V3 16S rDNA phylogenetics as described in Methods. Analysis of cecal and fecal samples from all mice demonstrated that the adoptive transfer protocol was successful in producing distinct core microbiomes in the 2 groups of mice (Supplementary Tables S2 and S3). Additionally, HFD microbiota demonstrated significantly reduced alpha diversity relative to CD samples, as well as greater evenness (cecal samples: Chao1 p = 0.0362, Observed Species p = 0.0302; fecal samples: Chao1 p = 0.0033, Observed Species p = 0.004; Fig. 2A and Supplementary Fig. S1). Rarefaction curves of fecal and cecal diversity (Supplementary Figs. S1 and S2) also support this interpretation as well. Evaluation of beta diversity metrics based on unweighted UniFrac distances showed that the community structures observed in the HFD samples are significantly different (p = 0.0001) from the communities detected in the CD samples (Fig. 2B). Visualization by Principal Coordinate Analysis demonstrates that CD and HFD samples form distinct clusters, and that for each condition, cecal and fecal samples form a cluster around the initial inoculum (Fig. 2B). The taxonomical distribution within groups at Phylum, Family and Genus levels for the cecal (Fig. 2C and Supplementary Fig. S3) and fecal samples (Supplementary Fig. S4) revealed the divergent composition of communities in CD compared to HFD samples.
Figure 2.
Effects of transplantation protocol on recipient gut microbiome diversity and population. Cecal and fecal microbiome populations from both donor and recipient mice were analyzed using 16S rRNA sequencing, and (A) Box Plots were generated to depict differences in Chao1 α-diversity show that mice with HFD microbiota exhibited a statistically significant (p = 0.0362) reduction in α-diversity compared to mice with CD mirobiota. Red line: median; black lines: range of values. (B) Scaled principal coordinate analysis to visualize the unweighted UniFrac distances of both cecal and fecal samples from individual recipient mice. Red and orange circles depict cecal and fecal samples from CD-treated mice; green and yellow circles represent cecal and fecal samples from HFD-treated mice. The pooled samples used as donor microbiota are show in blue for the CD donor pool, and in magenta for the HFD donor pool. α- diversity was found to be statistically significantly different between the CD- and HFD-treated groups (p = 0.0001). (C) Microbiota membership is reflected in bar diagrams depicting the taxonomic distribution within cecal samples within the CD and HFD groups at the Phylum, Family and Genus levels. Microbiota from the HFD-treated group show higher representation of Clostridiales (Family: purple; Genus: blue). Higher resolution images together with the detailed color codes are shown in Supplementary Figures S3 and S4. (D) Microbiome differences between CD- and HFD-treated mice at the level of Lachnospiraceae and Ruminococcaceae, two families within the order of Clostridiales. Statistically significant fold-changes between the HFD-treated vs. the CD-treated group were determined in DESeq2. Significant fold-changes for individual OTUs belonging either to Lachnospiraceae or to Ruminococcaceae (family level as maximum taxonomical depth for these OTUs) were log2-transformed and plotted relative to the CD group. This analysis demonstrated shifts in representation within each family.
The UPARSE pipeline (32) was used to identify operational taxonomic units (OTUs), and ANOVA revealed significant differences in the relative abundance of OTUs between HFD and CD microbiota (see Supplementary Tables S4 and S5 for Bonferroni corrected and false discovery rate (FDR) controlled values). To corroborate ANOVA data with statistical methods better suited for sequence count data, DESeq2 software was used to test for differential representation of OTUs in cecal samples. Individual OTUs for which the group difference reached statistical significance using mean normalized sequence count level higher than 30 counts and FDR<0.05 criteria (Wald statistics with Benjamini-Hochberg correction) in either the CD or HFD group were identified (Supplementary Table S6). Of the 104 OTUs passing these DESeq2 filters, 53 OTUs were higher in the HFD samples compared to CD, whereas 51 OTUs were higher in CD. Of the 104 significantly different OTUs, 91 belong to the phylum Firmicutes, with 90 of these coming from class Clostridiales. These unbiased analyses show that the overall distinction between HFD and CD is based on shifts in the representation of individual OTUs within Clostridiales rather than a binary shift from, for example, Bacteroidetes in the CD samples to Firmicutes in the HFD samples. Looking at specific orders within Clostridiales (Fig. 2D and Supplementary Fig. S5), some orders are present at significantly higher levels in the HFD group (17 members of Lachnospiraceae; 9 members of Ruminococcaceae), whereas others are lower (21 members of Lachnospiraceae; 9 members of Ruminococcaceae). Similar findings were obtained in fecal samples (data not shown). Finally, the list of differently represented OTUs was queried for presumed ‘beneficial’ bacteria, such as Akkermansia muciniphila. A. muciniphila was 5.4-fold lower in HFD samples compared to CD (FDR=0.06), indicating that this species of bacteria may be associated with a healthier microbiome, as suggested previously (35). Likewise, the presumably detrimental Bilophila sp. (belonging to Desulfovibrionaceae) was strongly enriched (~ 300-fold; FDR=2.5×10−25) in HFD microbiota, comprising 0.78% of the microbial community in this group. Conversely, Bilophila sp. was barely detectable in CD samples at 0.0024%.
HFD-derived gut microbiota increase intestinal permeability, systemic inflammation, and brain inflammation
Postmortem studies were conducted to identify potential pathways whereby altered gut microbiota impaired behavior. Analysis of blood glucose and bioactive hormones/lipids in plasma as well as body weight and body composition revealed no significant group differences, demonstrating that obesity and metabolic syndrome/dysfunction was not induced by HFD microbiota (Supplementary Table S7).
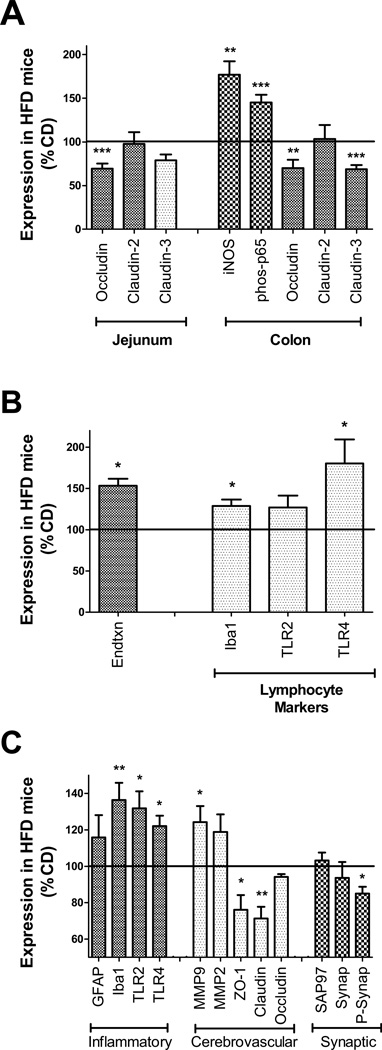
To determine if transplantation with HFD-shaped microbiota altered intestinal barrier function, the expression of markers of intestinal inflammation and permeability, and also circulating endotoxin and inflammatory markers, was assessed (see Supplementary Fig. S6 for representative Western blots). Compared to CD, mice with HFD microbiota had significantly decreased occludin (t(18)= 4.95, p<0.001) expression in the jejunum (Fig. 3A). Additionally, expression of iNOS (t(18)= 3.70, p<0.01) and phosphorylation of the p65 subunit of NFκB (t(18)=4.13, p<0.001) was increased, while occludin (t(18)=3.32, p<0.01) and claudin-3 (t(18)=4.13, p<0.001) were decreased, in colons of HFD mice (Fig. 3A), indicating increased intestinal inflammation and permeability in mice with HFD microbiota. In addition, data show significantly increased plasma endotoxin (t(18)=2.64, p<0.05) in mice with HFD microbiota (Fig. 3B, while evaluation of isolated lymphocytes revealed increased expression of the macrophage marker Iba1 (t(16)=2.59, p<0.05) and TLR4 (t(16)=2.73, p<0.05) in mice with HFD microbiota (Fig. 3B).
Figure 3.
Transplantation with microbiota shaped by high fat diet disrupts intestinal barrier proteins and increases systemic and brain inflammation. (A) Relative expression of tight junction proteins occludin, claudin-2, and claudin-3 in jejunum (left). Expression of inducible nitric oxide synthase (iNOS), phosphorylated p65 (phos-p65), and tight junction proteins occludin, claudin-2, and claudin-3 in colon (right) in mice with HFD microbiota relative to CD mice. (B) Levels of plasma endotoxin, and lymphocyte expression of macrophage markers (Iba1) and TLR4, in mice with HFD microbiota as compared to CD mice. (C) Markers of inflammation, cerebrovascular integrity, and synaptic density in tissue homogenates prepared from the medial prefrontal cortex. Graphs depict increased microgliosis (Iba1) and TLR2 and TLR4 expression, increased matrix metalloproteinase 9 (MMP9) expression, and decreased expression of endothelial tight junction proteins (ZO-1 and Claudin-5) and phosphorylated synapsin-1 (P-Synap) in HFD mice. All data depict mean ± SEM expression in mice with HFD microbiota presented as % CD mice (100% line on graph), and *, **, and *** depict p < 0.05, p < 0.01, and p < 0.001, respectively, based on t-tests. See Supplementary Fig. S6 for representative images of all Western blot Data.
To determine the effects of HFD-shaped microbiota on brain, protein markers of brain injury and inflammation were quantified (see Supplementary Fig. S6 for representative Western blots). Analyses were thematically split into evaluations of inflammation/gliosis, cerebrovascular integrity, and synaptic density, and were conducted in the medial prefrontal cortex, a brain structure involved in both anxiety and cognitive behaviors in mice (36). Compared to mice with CD-shaped microbiota, expression of the microglial marker Iba1 (t(18)=3.48, p<0.01), TLR2 (t(18)=2.72, p<0.05), and toll-like receptor 4 (TLR4; t(18)=2.83, p<0.05) were increased in HFD mice (Fig. 3B). Additionally, mice with HFD-associated microbiota had decreased levels of the tight junction proteins ZO-1(t(18)=2.32, p<0.05) and claudin-5 (t(18)=4.11, p<0.001), and increased expression of matrix metalloproteinase 9 (MMP9; t(18)=2.29, p<0.05; Fig. 3B). While overall expression of the synaptic marker proteins SAP97 and synapsin 1 were similar in both groups, levels of phosphorylated synapsin 1 were significantly reduced (t(18)=2.26, p<0.05) in mice with HFD-shaped microbiota (Fig. 3B). Finally, levels of brain derived neurotrophic factor (BDNF) were assessed in the medial prefrontal cortex, but there were no significant differences in soluble BDNF in mice with HFD microbiota as compared to mice with CD microbiota (Supplementary Table S7).
DISCUSSION
The present findings represent the first definitive evidence that high fat diet-induced changes to the gut microbiome are sufficient to disrupt brain physiology and function in the absence of obesity. Specifically, data show that transplantation of microbiota shaped by high fat diet, but not control low fat diet, caused significant and selective disruptions in exploratory, cognitive, and stereotypical behavior in conventionally housed, non-obese, diet-naive mice. Overall, these data are in agreement with the extensive body of literature describing the sensitivity of the brain to diet-induced obesity ((37),(28)) and the growing number of studies linking gut microbiota to CNS health and behavior (38),(20),(39),(20). For example, there is a reported high co-morbidity between psychiatric syndromes including depression and anxiety with gastrointestinal disorders; while conversely, recent studies link probiotics to positive changes in mood and behavior (reviewed in (40)). Furthermore, changes in microbiota appear to mediate weight gain commonly associated with antipsychotic-administration (41),(42), which has been likewise linked to improvements in core schizophrenia symptoms, depression, and overall mental functioning (43). It should be pointed out, however, that reports have shown that high fat diet consumption can allay anxiety and depressive-like behaviors in mice subjected to chronic social stress (44). Thus, these data underscore the strong but complex influence of dietinduced changes to the gut microbiome on stress-induced behaviors, and emphasize the clinical utility of the gut-brain-axis as a target for future therapeutic intervention
The significant behavioral phenotype of the mice described in this report, combined with the established association between psychiatric conditions and gastrointestinal symptoms, support the concept of a microbiome-gut-brain axis (45),(46), but the mechanisms whereby gut microbes affect behavior are not understood. Gut microbial metabolism is known to produce catecholamines, histamine, and/or other neuroactive mediators that can directly stimulate the local enteric nervous system and/or primary afferent fibers of vagal or dorsal root origin (38; 47). Indeed, reports have shown that the probiotic bacterium Lactobacillus rhamnosus can directly increase single- and multiunit firing rate of the mesenteric nerve bundle and can decrease stress-induced corticosterone and anxiety/depression in mice (48),(49). Moreover, the positive behavioral effects of L. rhamnosus are abolished by vagotomy (48). In addition to direct interactions with neural processes, immune activation and inflammation participate in nearly all neurologic/psychiatric disorders (50),(51), and gut dysbiosis might alter brain function via this pathway. Indeed, the increases in gram-negative Proteobacteria within the gut, endotoxin in the blood, and inflammatory markers in the brain collectively suggest that intestinal permeability and inflammation links HFD microbiota to behavioral dysfunction. In further support of this scenario, transplantation of microbiota from obese donors to germ-free recipients has been shown to disrupt intestinal tight junction protein and increase translocation of bacteria into the bloodstream (52). In relation to neurologic impairment, alterations to gut microbiota and disrupted intestinal barrier function are seen in mouse models of autism-spectrum disorder (53). Collectively, these data suggest that unhealthy diet-induced alterations to gut microbiota could boost the prevalence and/or severity of numerous neurologic conditions that involve inflammation, including autoimmune disease, autism, and Alzheimer’s disease.
While previous reports have shown that gut microbiome transplantations into germ-free mice can replicate many aspects of the obese phenotype (54),(55), this is the first demonstration that high fat-shaped gut microbiota can intrinsically and adversely affect neurologic function/physiology in conventionally housed mice, even in the absence of altered diet, adiposity, or metabolic syndrome. A variety of tools and techniques have been developed to study the gut microbiome, microbiota, including introduction into germ-free recipients, antibiotic use, administration of prebiotics and probiotics, and specific GI infection. Indeed, the use of gnotobiological methods on experimental animals has been indispensable in establishing the significance of gut microbiota to mammalian physiology (56). However, there are several characteristics of germ free mice, outside of changes in cecal size and bowel motility, which undermine their utility and physiologic relevance. For example, germ-free mice are well-known to be smaller than conventional mice, with decreased cardiac output and notably underdeveloped immune systems (57), (58). As any of these confounds could affect the development and function of the brain, we opted instead to use a strategy whereby donor microbiota were adoptively transferred to conventionally housed mice following antibiotic-based microbial depletion. While both the depletion and recolonization protocols are based on established methods (21),(22), (23),(24), there are limitations of the antibiotic-based model that could have affected the outcome of our study. It is likely that the antibiotic regimen did not entirely deplete the recipient microbiome, which could differentially affect recolonization by specific bacteria. It is also possible that repeated gavage and/or sustained systemic antibiotic exposure could contribute to some of the behavioral alterations obseved. However, as both groups were treated equally, the impact of such potential artifacts is minimized.
Alterations in microbiome composition following manipulation have been evaluated in the past in an attempt to identify beneficial core microbiota. For example, studies on high-fat dietinduced gut microbiota have reported a shift in the relative abundance of two major phyla, a reduction in Bacterioidetes and an increase in Firmicutes (54),(55). Furthermore, abundance of these two phyla was shifted in the opposite direction after weight loss or gastric bypass surgery (59),(60) and thus has been proposed that the balance between these 2 phyla might reflect the balance of “unhealthy” and “healthy” microbiota. However, our data suggest that this binary distinction does not sufficiently reflect the complexity of diet-induced changes to the gut microbiome, as has been suggested in previous investigations of diet-induced obesity and gut microbiota (61). Specifically, our data indicate that shifts within the Firmicute phylum drive the overall distinction of HFD from CD, rather than phylum-wide shifts from Bacteroidetes to Firmicutes (see Figure 2 and Supplementary Figure S5). While it is currently not possible to identify the specific alterations or population shifts that drive the observed behavioral/biochemical alterations, the relative abundance of purportedly beneficial and harmful species in each group was probed. Akkermansia muciniphila, a presumed beneficial species, was 5.4-fold lower in HFD samples compared to CD, indicating that this species of bacteria may be associated with a healthier microbiome, as suggested previously (35). Likewise, the presumed detrimental Bilophila sp. (belonging to Desulfovibrionaceae) was strongly enriched (~ 300-fold; FDR=2.5×10−25) in HFD microbiota, comprising 0.78% of the microbial community in this group. Conversely, Bilophila sp. was barely detectable in CD samples at 0.0024%. As member(s) of the phylum Protobacteria, Bilophila sp. may be partially responsible for the increase in endotoxin observed in the serum from HFD-treated mice compared to CD-treated mice (see Fig. 3). Indeed, higher Bilophila wadsworthia have been repeatedly found in human patients suffering from intestinal diseases (62), (63). The collective identification of specific bacterial species/populations driving adverse physiologic responses to diet will facilitate the future design of personalized mirobiomes that optimize physiologic function in the context of modern diets/ lifestyles. Overall, these data strongly suggest that therapeutic manipulation of the microbiome, which should be highly responsive compared to existing clinical targets, could dramatically mitigate the prevalence and/or severity of neuropsychiatric disorders.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr. Barry Robert for expert veterinary assistance related to antibiotic administration. This work was supported by the National Institutes of Health (DK047348 to HRB), and also used PBRC (Animal Phenotyping) and LSU (Microbial Genomics Resource Center) that are funded in part by the National Institutes of Health (P20-RR021945, P30-DK072476, and P60-AA009803).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Shih RA, Belmonte PL, Zandi PP. A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int. Rev. Psychiatry. 2004;16:260–283. doi: 10.1080/09540260400014401. [DOI] [PubMed] [Google Scholar]
- 2.Bruce-Keller AJ, Keller JN, Morrison CD. Obesity and vulnerability of the CNS. Biochim. Biophys. Acta. 2009;1792:395–400. doi: 10.1016/j.bbadis.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elias MF, Elias PK, Sullivan LM, Wolf PA, D'Agostino RB. Lower cognitive function in the presence of obesity and hypertension: the Framingham Heart Study. Int. J. Obes. Relat. Metab. Disord. 2003;27:260–268. doi: 10.1038/sj.ijo.802225. [DOI] [PubMed] [Google Scholar]
- 4.Waldstein SR, Katzel LI. Interactive relations of central versus total obesity and blood pressure to cognitive function. Int. J. Obes. (Lond.) 2006;30:201–207. doi: 10.1038/sj.ijo.0803114. [DOI] [PubMed] [Google Scholar]
- 5.Needham BL, Epel ES, Adler NE, Kiefe C. Trajectories of change in obesity and symptoms of depression: the CARDIA study. Am. J. Public Health. 2010;100:1040–1046. doi: 10.2105/AJPH.2009.172809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J, Xiao L. Obesity and depression in US women: results from the 2005–2006 National Health and Nutritional Examination Survey. Obesity (Silver Spring) 2010;18:347–353. doi: 10.1038/oby.2009.213. [DOI] [PubMed] [Google Scholar]
- 7.Atlantis E, Baker M. Obesity effects on depression: systematic review of epidemiological studies. Int. J. Obes. (Lond) 2008;6:881–891. doi: 10.1038/ijo.2008.54. [DOI] [PubMed] [Google Scholar]
- 8.Rivenes AC, Harvey SB, Mykletun A. The relationship between abdominal fat, obesity, and common mental disorders: results from the HUNT study .J Psychosom. Res. 2009;66:269–275. doi: 10.1016/j.jpsychores.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7(10):e47713. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robles Alonso V, Guarner F. Linking the gut microbiota to human health. Br. J. Nutr. 2013;109(Suppl 2):S21–S26. doi: 10.1017/S0007114512005235. [DOI] [PubMed] [Google Scholar]
- 11.Collins SM, Kassam Z, Bercik P. The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol. 2013;16:240–245. doi: 10.1016/j.mib.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aziz Q, Doré J, Emmanuel A, Guarner F, Quigley EM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol. Motil. 2013;25:4–15. doi: 10.1111/nmo.12046. [DOI] [PubMed] [Google Scholar]
- 14.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas-Escobar M, Elliott E, Neu J. Effect of intestinal microbial ecology on the developing brain. JAMA Pediatr. 2013;167:374–379. doi: 10.1001/jamapediatrics.2013.497. [DOI] [PubMed] [Google Scholar]
- 16.Dinan TG, Quigley EM. Probiotics in the treatment of depression: science or science fiction? Psychiatry. 2011;45:1023–1025. doi: 10.3109/00048674.2011.613766. [DOI] [PubMed] [Google Scholar]
- 17.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bested AC, Logan AC, Selhub EM. Intestinal microbiota, probiotics and mental health: from Metchnikoff to modern advances. Part III - convergence toward clinical trials. Gut Pathog. 2013;5:4. doi: 10.1186/1757-4749-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, D’Souza R, Hong ST. The role of gut microbiota in the gut-brain axis: current challenges and perspectives. 2013;4:403–414. doi: 10.1007/s13238-013-3017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heimesaat MM, Plickert R, Fischer A, Göbel UB, Bereswill S. Can microbiota transplantation abrogate murine colonization resistance against Campylobacter jejuni? Eur. J. Microbiol. Immunol. (Bp) 2013;3:36–43. doi: 10.1556/EuJMI.3.2013.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, Taur Y, Jenq RR, van den Brink MR, Xavier JB, Pamer EG. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCafferty J, Mühlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxietyrelated behavior in rodents. Nat. Protoc. 2007;2:322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choleris E, Thomas AW, Kavaliers M, Prato FS. A detailed ethological analysis of the mouse open field test: effects of diazepam, chlordiazepoxide and an extremely low frequency pulsed magnetic field. Neurosci. Biobehav. Rev. 2001;25:235–260. doi: 10.1016/s0149-7634(01)00011-2. [DOI] [PubMed] [Google Scholar]
- 27.Angoa-Pérez M, Kane M, Briggs DI, Francescutti DM, Kuhn DM. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. J. Vis. Exp. 2013;82:50978. doi: 10.3791/50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeman LR, Zhang L, Nair A, Dasuri K, Francis J, Fernandez-Kim SO, Bruce-Keller AJ, Keller JN. Obesity increases cerebrocortical reactive oxygen species and impairs brain function. Free Radic Biol Med. 2013;56:226–233. doi: 10.1016/j.freeradbiomed.2012.08.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paylor R, Spencer CM, Yuva-Paylor LA, Pieke-Dahl S. The use of behavioral test batteries, II: effect of test interval. Physiol. Behav. 2006;87:95–102. doi: 10.1016/j.physbeh.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Vöikar V, Vasar E, Rauvala H. Behavioral alterations induced by repeated testing in C57BL/6J and 129S2/Sv mice: implications for phenotyping screens. Genes Brain Behav. 2004;1:27–38. doi: 10.1046/j.1601-183x.2003.0044.x. [DOI] [PubMed] [Google Scholar]
- 31.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 33.Pepping JK, Freeman LR, Gupta S, Keller JN, Bruce-Keller AJ. NOX2 deficiency attenuates markers of adiposopathy and brain injury induced by high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2013;304:E392–E404. doi: 10.1152/ajpendo.00398.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Njung'e K, Handley SL. Evaluation of marble-burying behavior as a model of anxiety. Pharmacol Biochem Behav. 1991;38:63–67. doi: 10.1016/0091-3057(91)90590-x. [DOI] [PubMed] [Google Scholar]
- 35.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, Hong S, Lee D, Lee MH, Choi JS, Koh MJ, Sun W, Kim H, Lee HW. Altered expression of synaptotagmin 13 mRNA in adult mouse brain after contextual fear conditioning. Biochem. Biophys. Res. Commun. 2012;425:880–885. doi: 10.1016/j.bbrc.2012.07.166. [DOI] [PubMed] [Google Scholar]
- 37.Stranahan AM, Norman ED, Lee K, Cutler RG, Telljohann RS, Egan JM, Mattson MP. Diet-induced insulin resistance impairs hippocampal synaptic plasticity and cognition in middle-aged rats. Hippocampus. 2008;18:1085–1088. doi: 10.1002/hipo.20470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011:255–264. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 39.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu E, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 40.Dinan TG, Cryan JF. Melancholic microbes: a link between gut microbiota and depression? Neurogastroenterol. Motil. 2013;25:713–719. doi: 10.1111/nmo.12198. [DOI] [PubMed] [Google Scholar]
- 41.Davey KJ, O'Mahony SM, Schellekens H, O'Sullivan O, Bienenstock J, Cotter PD, Dinan TG, Cryan JF. Gender-dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology (Berl) 2012;221:155–169. doi: 10.1007/s00213-011-2555-2. [DOI] [PubMed] [Google Scholar]
- 42.Davey KJ, Cotter PD, O'Sullivan O, Crispie F, Dinan TG, Cryan JF, O'Mahony SM. Antipsychotics and the gut microbiome: olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Transl. Psychiatry. 2013;3:e309. doi: 10.1038/tp.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ascher-Svanum H, Stensland M, Zhao Z, Kinon BJ. Acute weight gain, gender, and therapeutic response to antipsychotics in the treatment of patients with schizophrenia. BMC Psychiatry. 2005;5:3. doi: 10.1186/1471-244X-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finger BC, Dinan TG, Cryan JF. High-fat diet selectively protects against the effects of chronic social stress in the mouse. Neuroscience. 2011;192:351–360. doi: 10.1016/j.neuroscience.2011.06.072. [DOI] [PubMed] [Google Scholar]
- 45.Montiel-Castro AJ, González-Cervantes RM, Bravo-Ruiseco G, Pacheco-López G. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 2013;7:70. doi: 10.3389/fnint.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochoa-Repáraz J, Mielcarz DW, Begum-Haque S, Kasper LH. Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann. Neurol. 2011;69:240–247. doi: 10.1002/ana.22344. [DOI] [PubMed] [Google Scholar]
- 47.Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013;9:e1003726. doi: 10.1371/journal.ppat.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Burgos A, Wang B, Mao YK, Mistry B, McVey Neufeld KA, Bienenstock J, Kunze W. Psychoactive bacteria Lactobacillus rhamnosus (JB-1) elicits rapid frequency facilitation in vagal afferents. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G211–G220. doi: 10.1152/ajpgi.00128.2012. [DOI] [PubMed] [Google Scholar]
- 50.Raison CL, Miller AH. Is depression an inflammatory disorder? Curr. Psychiatry Rep. 2011;13:467–475. doi: 10.1007/s11920-011-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyman M, Lloyd DG, Ji X, Vizcaychipi MP. Neuroinflammation: The role and consequences. Neurosci. Res. 2014;79C:1–12. doi: 10.1016/j.neures.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Duca FA, Sakar Y, Lepage P, Devime F, Langelier B, Doré J, Covasa M. Replication of obesity and associated signaling pathways through transfer of microbiota from obese prone rat. Diabetes Care. 2014 doi: 10.2337/db13-1526. [Epub ahead of print]: [DOI] [PubMed] [Google Scholar]
- 53.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 55.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell. Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi P, Li L. The germfree murine animal: an important animal model for research on the relationship between gut microbiota and the host. Vet. Microbiol. 2012;157:1–7. doi: 10.1016/j.vetmic.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 57.Luckey TD. Effects of microbes on germfree animals. Adv. Appl. Microbiol. 1965;7:169–223. doi: 10.1016/s0065-2164(08)70387-3. [DOI] [PubMed] [Google Scholar]
- 58.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sweeney TE, Morton JM. The human gut microbiome: a review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013;148:563–569. doi: 10.1001/jamasurg.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy EF, Cotter PD, Hogan A, O'Sullivan O, Joyce A, Fouhy F, Clarke SF, Marques TM, O'Toole PW, Stanton C, Quigley EM, Daly C, Ross PR, O'Doherty RM, Shanahan F. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut Pathog. 2013;62:220–226. doi: 10.1136/gutjnl-2011-300705. [DOI] [PubMed] [Google Scholar]
- 62.Jia W, Whitehead RN, Griffiths L, Dawson C, Bai H, Waring RH, Ramsden DB, Hunter JO, Cauchi M, Bessant C, Fowler DP, Walton C, Turner C, Cole JA. Diversity and distribution of sulphate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS Immunol. Med. Microbiol. 2012;65:55–68. doi: 10.1111/j.1574-695X.2012.00935.x. [DOI] [PubMed] [Google Scholar]
- 63.Baron EJ. Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe. 1997;3:83–86. doi: 10.1006/anae.1997.0075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.