Abstract
Following central nervous system (CNS) injury, activated astrocytes form glial scars, which inhibit axonal regeneration, leading to long-term functional deficits. Engineered nanoscale scaffolds guide cell growth and enhance regeneration within models of spinal cord injury. However, the effects of micro-/nanosize scaffolds on astrocyte function are not well characterized. In this study, a high throughput (HTP) microscale platform was developed to study astrocyte cell behavior on micropatterned surfaces containing 1 μm spacing grooves with a depth of 250 or 500 nm. Significant changes in cell and nuclear elongation and alignment on patterned surfaces were observed, compared to on flat surfaces. The cytoskeleton components (particularly actin filaments and focal adhesions) and nucleus-centrosome axis were aligned along the grooved direction as well. More interestingly, astrocytes on micropatterned surfaces showed enhanced mitochondrial activity with lysosomes localized at the lamellipodia of the cells, accompanied by enhanced adenosine triphosphate (ATP) release and calcium activities. These data indicate that the lysosome-mediated ATP exocytosis and calcium signaling may play an important role in astrocytic responses to substrate topology. These new findings have furthered our understanding of the biomechanical regulation of astrocyte cell–substrate interactions, and may benefit the optimization of scaffold design for CNS healing.
Cells sense and respond to biomaterial surface topography via phenotypic modification1,2,3. These reactions, in turn, profoundly influence cell–substrate interaction, leading to changes in cell morphology, directional migration, and even differentiation4,5,6,7,8,9,10. In particular, arrays of parallel nanogrooves and nanoridges are used frequently as topographical cues to elucidate effects of geometrical constraints on cell functions in the nervous system, such as directional neurite growth of hippocampal neurons on micropatterned grooves11 and elevated migration of Schwann cells in engineered nerve guidance conduits12. However, topological regulation of astrocyte function has not been well characterized.
The physiological and structural relevance of astrocytes in the central nervous system (CNS) has attracted interests in their functional significance in the brain. In particular, after CNS injury, activated astrocytes form glial scars that block axon extension and migration and cause nerve regeneration failure13. Development of new materials to improve tissue healing by optimizing physical characteristics of synthetic scaffolds and/or to mobilize astrocytes and neurons by using nanotechnology-based strategies is the focus of several recent studies14,15,16,17,18,19. The goal of these studies is to direct cellular growth and to enhance axonal extension into CNS lesions where typically such migration and extension is limited by astrogliosis20,21,22. One such strategy may be to combine a topographical approach (such as electrospun aligned fibers) with drug release (fibers releasing therapies) to prevent gliosis while still promoting dorsal root ganglia neurite extension23.
Recent studies provide insight into the ability of different topographical attributes (geometry, roughness, size, shape) and material to alter astrocytic function and to promote neurite extension18,24,25. Many of the neuron-astrocyte reciprocal signaling events and Ca2+ excitability functions have been recently discovered in lieu of advances in microfabrication techniques26. However, most of these strategies have utilized costly and time consuming techniques to produce large volume of micro- and nano-fabricated scaffolds1. In addition, research is also hindered by the limited precision of small features, which are constructed by low throughput means, and rarely thoroughly assess cellular behavior on such substrates27,28.
Here we report a high throughput (HTP) technique to place micro-/nano-scale features onto the surfaces of PDMS wells and evaluate multiple cell activities. The established platform enables studying many cellular functions simultaneously and efficiently. We observed highly oriented and aligned actin filaments, altered focal adhesion complexes (FACs), phagosome like specific localization of acidic subcellular lysosomal and mitochondrial organelles in astrocytes on patterned substrates. Most interestingly, enhanced adenosine triphosphate (ATP) release and intracellular calcium signaling activity were noticed during initial cell attachment and spreading on micropatterned surfaces. These findings suggest that the lysosome-mediated ATP exocytosis may dictate astrocyte interaction with micro-nanopatterned substrates.
Results
Cell proliferation, orientation and elongation response to groove depth-variant substrates
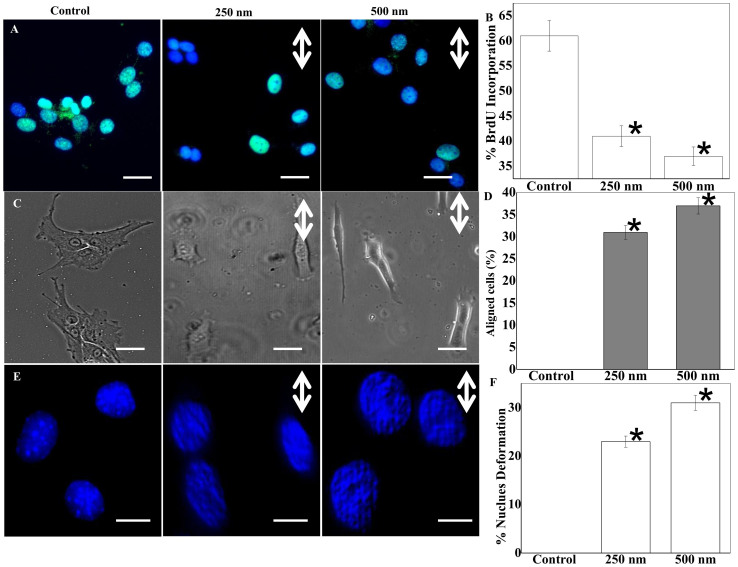
Using optical grating glasses with microscale patterns that can be faithfully copied into PDMS, a HTP experimental platform was made using a sticky-miliwell slide (Fig. 1). We analyzed astrocyte proliferation with the BrdU assay and noticed a ~35% drop in cell proliferation on patterned surfaces compared to flat or routine cell culture substrates (Fig. 2A–B). Cell morphology was assessed quantitatively as shown in Fig. 2C–D. Astrocytes polarized largely along the direction of micro- and nano-structured ridges/grooves. The extent of cell alignment and elongation appeared sensitive to the groove depth at a nanometer scale29,30. For example, cells on the deeper 500 nm patterns aligned more along the direction of ridges/grooves, compared to those on the 250 nm deep patterns, but the aspect ratio is similar on two patterned surfaces (Fig. 2C–D & S1A). On the flat surfaces, the cells became less oriented and more rounded. Furthermore, irrespective of pattern depth, at leading edges astrocytes formed flattened lamellipodia preferentially in the direction of pattern depth (Fig. 2C).
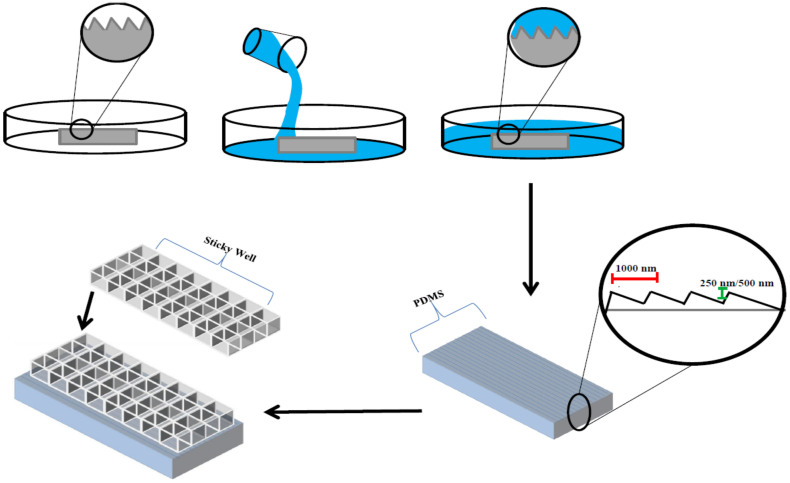
Figure 1. Schematic highlighting the high throughput procedure for fabricating the micro-nanopatterned device for investigating astrocyte mechanosensitivity in this study.
The upper panel illustrates key steps taken to copy micro- and nanoscale features from optical gratings in PDMS. The lower panels show creation of multiwelled, patterned substrate through the bonding of a sticky well onto PDMS with copied features.
Figure 2. Quantitative analysis of astrocyte proliferation, alignment and nuclei deformation.
(A) BrdU (bromine derivative Uracil) assay indicates higher BrdU uptake (green) of residue in cells on control surfaces (scale bar: 20 μm). (B) Bar plot results from BrdU assay demonstrate significantly higher proliferation on control surfaces. (C) Phase contrast images clearly demonstrate astrocytes seeded on microgrooves show much greater elongation and alignment (scale bar: 20 μm). (D) Quantification of the percentage of aligned cells. (E) Maximum intensity projection of Z-stack images of DAPI stained nucleus show altered nuclear morphology on the patterned surfaces. (F) Quantification reveals that nuclei deformation is greatest in astrocytes seeded on the microgroove (Scale bars: 5 μm; Double-sided arrows show groove directions). * stands for significant difference from the control.
Astrocyte nuclei deformation in response to microgroove patterned substrates
We noted that majority of astrocyte nuclei were deformed on patterns, and the percentage was significantly different from that on flat surfaces (Fig. 2E–F). On 500 nM grooves, more than 30% of cells demonstrated modulation in nuclear shape and geometry, as demonstrated by the strip-like patterns on the nucleus. We also observed that astrocytes cultured on microgrooves with nanometric depth on PDMS have a higher percentage of the nucleus-centrosome (NC) axis oriented in the direction of pattern depth (Fig. S1B–D). In contrast, NC axis exhibited random orientation on the other surfaces.
Grooves induce alignment of actin stress fibers, focal adhesions, and intermediate filaments
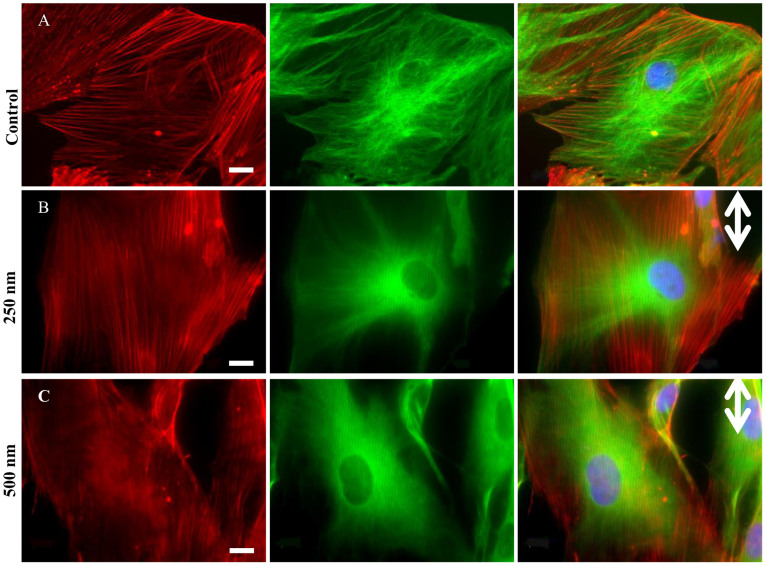
Fig. 3A–C demonstrates cytoskeleton (actin-tubulin) modulation in patterned astrocytes on control, 250 nm, and 500 nm grooves, respectively. Actin filaments are aligned and extended along the microgrooves. As shown with red lines in Fig. S2A–C, cable-like actin filaments exhibit greater alignment along microgrooves, in the directions of nanometric depth (indicated by double sided arrows). In addition, more than 65% of astrocytes show visible actin cables on patterns, compared to 30% on a flat substrate (Fig. S3A), indicating significant cytoskeletal modulation induced by microgrooves.
Figure 3. Cytoskeletal alignment analysis.
(A–C) single channel and Overlay of immunostained cytoskeletal elements: actin (TRITC), tubulin (FITC), and nuclei (DAPI) at 63× magnification. Scale bars: 10 μm. Double-sided arrows represent groove directions.
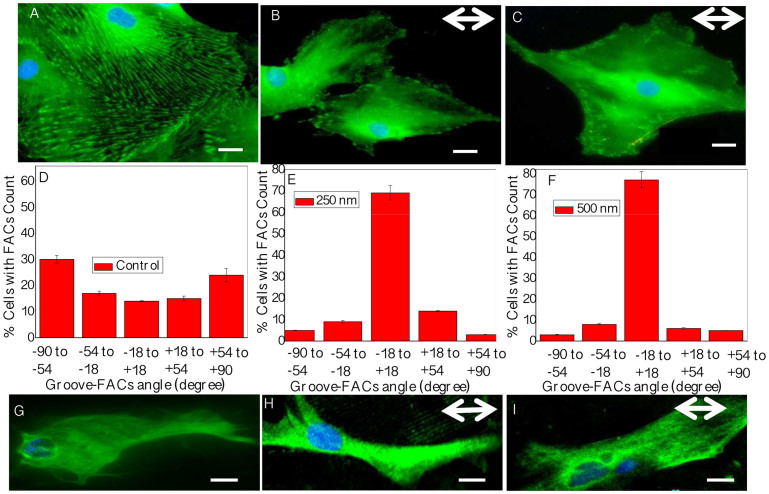
We further investigated the interaction between microtopography and vinculin structured focal adhesions. We observed significant variations in FACs quantity, quality, and orientation (Fig. 4A–F). Cells on unpatterned surfaces exhibited classical focal contacts (spindle shaped, large area, randomly distributed all over the cell cytoskeleton). In contrast, the FACs of astrocytes on patterned surfaces were accumulated in the close vicinity of the cell edges, aligned in the groove direction, merged with actin cables, and extended along individual microridges (Fig. S3B–D).
Figure 4. Analyses of focal adhesions and glial fibrillary acidic protein (GFAP) expression.
(A–C) Focal adhesion pattern is guided by topography as indicated by vinculin stain (green). (D–F) Histograms show measured FACs aligned in the direction of microgrooves. (G–I) GFAP stained astrocytes show greater intermediate filament expression on patterned surfaces. Scale bars: 20 μm in (A–C), and 10 μm in (G–I). Double-sided arrows represent groove directions.
We also stained for the astrocyte specific intermediate filament (IF) marker, GFAP (Fig. 4G–I). In control, intermediate filament distribution was limited to perinuclear regions, but this type III intermediate filament appeared more fibrous, and aligned along the microgrooves.
Topography modulates mitochondrial activity and lysosome localization of astrocytes cultured on microgrooves
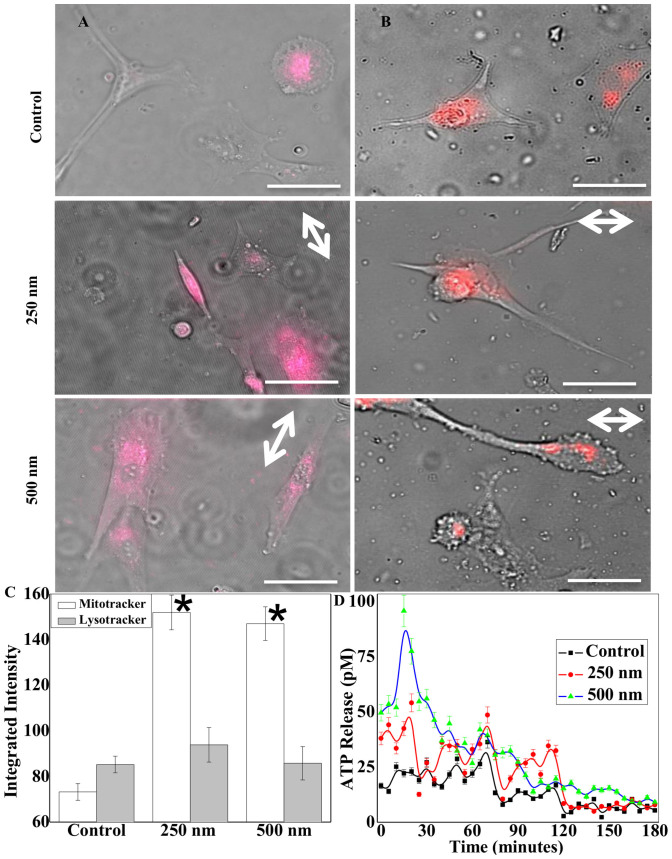
We observed higher Mitochondrial activities in astrocytes seeded on micropatterned surfaces. MitoTracker dye distributed across the entire cell body when the cells were on the patterned grooves, in contrast to the central localization on flat surfaces (Fig. 5A left panel). On the patterned surfaces, both 250 nm and 500 nm in depth, the mitochondria were present not only near the nucleus but also at the protruding edges of the cell. On unpatterned surfaces, mitochondria were localized to the vicinity of the nucleus. Unlike mitochondria, acidic subcellular organelles, as indicated by the lysosome staining, were found to also locate at the tip of growing or expanding lamellipodial structures in the cells on grooves (Fig. 5B right panel), while the lysosomes were limited to interior regions when the cells were on flat surfaces. Interestingly, mean fluorescence and degree of LysoTracker dye uptake, were quantitatively similar (Fig. 5C). Conversely, the integrated intensity for MitoTracker was significantly higher for the cells on nanogrooves than those on flat surfaces.
Figure 5. Mitochondrial and lysosomal activity.
(A–B) Live cell mitochondrial (left) and lysosomal (right) tracking of cells seeded on flat and patterned PDMS surfaces after 90 minutes of cell seeding (scale bars: 50 μm). (C) Quantification of integrated average intensity of MitoTracker and LysoTracker positive cells demonstrating significantly greater mitochondrial activity in astrocytes seeded over micropatterned surfaces. (D) In rationale with MitoTracker activity on patterned surfaces, bar graph demonstrates enhanced ATP release events from astrocytes at the onset of cell spreading and alignment. Double-sided arrows show groove directions. * stands for significant difference from the control.
Cellular ATP release and calcium activity are enhanced on microgrooves
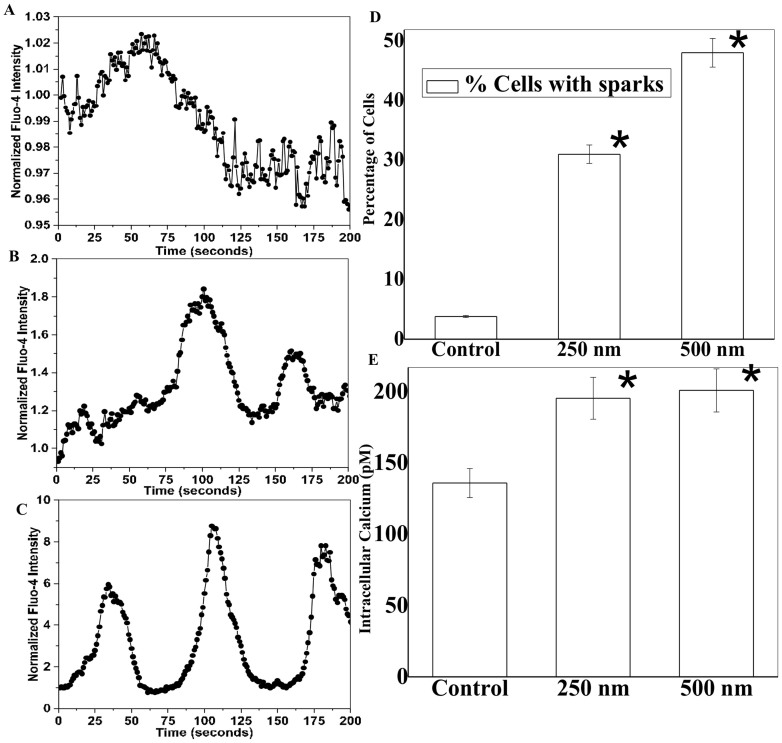
We further quantified ATP release and calcium signaling in control and patterned cells at different time points. We found that more ATP release was observed on patterned samples, particularly during initial cell seeding phase (within 1–2 hours) of cell-microgroove interactions as shown in Fig. 5D. After 2 hours, the ATP level decreased, but it remained significantly higher than the control. This difference coincided with elevated MitoTracker staining and calcium activity, which suggests that ATP release via lysosomal exocytosis triggers elevated intracellular calcium activity. Thus, we investigated calcium events in astrocytes with temporal imaging (Fig. 6). We observed 40–50% higher intracellular calcium concentration with spontaneous peaks on microgroove patterns (Fig. 6E). This result was corroborated by the fact that overall astrocytic activity is enhanced by underneath topography, as there are higher MitoTracker positive cells on the patterns. Intriguingly, micro-nanopatterns are sufficient to induce significant calcium activity only during initial events (<3 hours) of substrate-cell interactions, while these events were absent on unpatterned control surfaces (Movie S1–S3). We performed overnight cells seeding on patterns and measured calcium activity after 12–16 hours; however, we could not see the characteristic calcium peaks after the initial 2–3 hours seeding of the astrocytes (data not shown). We analyzed all spontaneous calcium peaks and found significant differences in spatiotemporal properties between groups, including amplitude, intensity level, time-to-peak, and relaxation time between peak, and time interval between two neighboring peaks (Fig. S4A). On patterned surfaces, amplitude and peak intensity of calcium peaks were higher and time-to-peak and relaxation time were shorter (Fig. S4B).
Figure 6. Calcium signaling on patterned surfaces.
(A–C) Typical normalized [Ca2+]i intensity oscillation curves recorded in control cells (A), and on 250 nm (B) and 500 nm (C) deep micropatterned surfaces. (D–E) Patterned surfaces induce significantly more calcium peaks and intracellular calcium release in astrocytes. * stands for significant difference from the control.
Discussion
In this study, we established an HTP platform for patterning astrocytes and studying cell-substrate interactions. We found that astrocytes align along the microgrooves with elongated and deformed nuclei. These cells can be discriminated in their response to flat and microgrooves with nanometric features via distinct calcium signaling. Based on these findings, we propose that in vitro implant or stent device design needs to incorporate both micro- and nanoscale features at the biomaterial–cell interface to exploit the regenerative capability of the cells for CNS tissue regeneration31. Phagosome like activity were seen in the growing ends of lamellipodia using acidic compartment specific LysoTracker probe, exploring a novel mechanistic insight in contact guidance of glial specific cells. The presence of more mitochondrial activity of astrocytes on patterned surfaces suggests that cell-surface interaction is a calcium-dependent, energy-intensive process.
Compared to HTP methods, traditional methods to investigate cell-surface interactions in neurobiology, taken a single-step approach where one assay at a time, has been used to study a particular characteristic24. As proof of concept, we have showed that using 8 well sticky chambers, each well would allow for the collection of 3 data points, allowing for 24 data points in total. Throughput could be raised to 72 data points per experiment if a 24-well sticky chambered slide were utilized. In past, PDMS based device for studying cell-cell (axon-glia) interactions at a higher throughput has been demonstrated32. However, few HTP reports show mechanistic aspects of cell-substrate interactions in vitro. The proposed system reported in this study enabled us to analyze four variables at the same time, two wells for each experimental condition including cell morphology/alignment, live calcium signaling, mitochondrial activity, and lysosomal localization.
Decreased proliferation observed could be due to significant change in cell aspect ratio and nuclear shape. Large variation in cell shape index is reported to cause drop in cellular proliferation due to increased chromatin condensation resulting from a deformed nucleus under large anisotropic forces33. Cells of different types are reported to align along micro- and nanoscale grooves, but the localization of mechanical force-related proteins is poorly understood, particularly in CNS related cell types28,34. In this study, we observed highly aligned astrocyte cytoskeleton in groove directions. More interestingly, the nucleus has a repeating stripe pattern in the direction of pattern depth. Similar findings have been reported in other microfabricated systems11,28. Indeed, micro-topographic geometry and spacing can strongly influence nuclear shape and size35.
In this study, we observed quantitatively higher ATP in astrocyte culture when seeded on patterned surfaces. Increases in ATP concentration are often tied to the increase in lysosomal activity27,36. Astrocytes are known to communicate via calcium signaling37, and exhibit excitability with changes in [Ca2+]i38,39. In addition, differential calcium signaling in response to micropit restrictions of single astrocytes has been reported in recent literature40. We, therefore, speculate that there might be differential calcium responses with astrocytes as they interact with various topographical surfaces. Indeed, we observed increased [Ca2+]i intensity oscillation and related parameters on patterned surfaces. This enhanced calcium activity may associate with lysosomal and mitochondrial functions. Lysosomes have been shown to store high concentrations of ATP in astrocytes, and more ATP release into the extracellular space is accompanied by the production of calcium peaks27. It was speculated that the release of ATP causes the discharge of the calcium from endoplasmic reticulum store into the intracellular space41. The ATP stored within the lysosomes is initially produced by mitochondria through action of different metabolic pathways. Therefore, one would see an increase in mitochondrial activity of the cells on micropatterned surfaces to compensate the ATP exocytosis. This is also confirmed by the fact that the elevation of extracellular ATP level temporally coincides with high calcium activity at the initial phase of astrocyte seeding on micropatterns. Thus, ATP release through lysosome activity may promote the astrocyte calcium activities.
The in vitro high-throughput platform is a very useful tool for deciphering complex cell responses to biomaterial, and therefore may be used to guide the design of optimal scaffolds for CNS tissue repair.
Methods
Fabrication of topographically defined micro- and nanopatterned arrays for HTP analysis
Microgroove features were transferred from commercial reflective ruled diffraction gratings (Edmund optics, Barrington, NJ) to polydimethylsiloxane (PDMS; Sylgard 184, Dow Corning, Corning, NY), by mixing prepolymer and curing agents at a 10:1 ratio and cured for 24 hours at 70°C (Fig. 1). The optical gratings used have a 25 × 25 mm surface area with 1200 grating pattern/mm and a depth of 250 or 500 nm. The pattern transfer was evident by the distinct colorful reflection on the surface that is clearly visible without any magnification. The grooves transferred onto the PDMS can also be seen in 63× phase contrast images (Fig. 2). Subsequently, sticky-miliwell slides (ibidi) were positioned and bonded with PDMS sheets for the HTP cell-surface interaction study. We used 8 well sticky chambers for each sample type containing microgroove and flat surfaces.
Astrocyte culture
Primary astrocyte cultures were prepared from the cerebral cortex of newborn Sprague-Dawley rats as previously described42,43. All animal procedures in this study strictly followed NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Rensselaer Polytechnic Institute. Postnatal 1-day old rat pups were euthanized by rapid decapitation. The cerebral cortices were separated from the meninges, hippocampi, and basal ganglia. The cortical tissue from four animals was dissociated in OptiMEM (Life Technologies, Carlsbad, CA) and transferred into a solution containing a 1:1 mix of recombinant protease TrypLE (Life Technologies) and OptiMEM. Cells were separated from the tissue using three 10-minute incubations with TrypLE/OptiMEM supplemented with 1 mg/ml DNAse I (Sigma-Aldrich, St. Louis, MO). The second and third extractions were combined with Dulbecco's Minimal Essential Medium (DMEM, Life Technologies) containing 10% Heat Inactivated Horse Serum (HIHS; Life Technologies) and 50 U/mL penicillin plus 50 μg/mL streptomycin (P/S, Life Technologies). Cells were pelleted using centrifugation, re-suspended in media, and plated on poly-D-lysine coated T75 culture flasks at a density of 200,000 cells/flask. The astrocytes were cultured until reaching confluency and used for subsequent procedures as described below. Purity of astrocyte cultures (>95% pure) was periodically verified using immunocytochemistry staining protocols with an antibody against astrocytic marker glial fibrillary acidic protein (GFAP; Dako, Glostrup, Denmark)42. Cells were seeded onto unpatterned and microgroove arrays that were pre-coated with 10 μg/mL fibronectin for 1 hour, and cultured for 72 hours (unless noted otherwise) prior to different characterization techniques described subsequently.
BrdU assay to quantify astrocyte proliferation
Astrocytes were incubated for 4 hours with 5-bromo-2-deoxyuridine, BrdU (Amersham Biosciences, Piscataway, NY), fixed, and stained with mouse anti-BrdU antibody (Abcam, Cambridge, England). Alexa Fluor 546-conjugated anti-mouse antibody (Vector Laboratories, Burlingame, CA) was used as a secondary antibody. The samples were mounted in Vectashield Media with 4′,6-diamidino-2-phenylindole (DAPI). Five samples of each patterned and unpatterned PDMS surface were imaged with an epifluorescence microscope. The images were analyzed with ImageJ (NIH, Bethesda, MD) for the co-localization and the quantification of BrdU positive cells.
Immunocytochemistry
The linear patterns with seeded astrocytes were fixed with 4% formaldehyde in cytoskeletal buffer (10 mM MES, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, and 0.32 M sucrose) for 30 minutes. For cytoskeleton (actin/tubulin) double staining, the cells were incubated with phalloidin-TRITC (1:400; Life Technologies) and anti-Tubulin-FITC (1:200; Sigma) for 1 hour. For the centrosome and focal complex staining on control and patterned surfaces, the cells were incubated in anti-pericentrin and anti-vinculin (Abcam), respectively, for 1 hour. Samples were then mounted in Vectashield mounting media with DAPI (Vector Laboratories). Pericentrin, also denoted as PCNT, was stained using anti-PCNT antibody (Abcam). PCNT is a protein which is expressed in the centrosome and binds to calmodulin44. For astrocytic intermediate filament specific marker, anti-Glial Fibrillary Acidic Protein (GFAP) were used (1:700; Abcam) for 1 hour.
Elongation and alignment quantification
Five samples each of patterned and control (flat PDMS) surfaces were seeded with astrocytes and fixed at day 3 to quantify alignment. Ten separate regions of each sample were imaged. Cell elongation and alignment were quantified from phase contrast images using ImageJ. Cell elongation was represented by cell aspect ratio (CAR). The longest vertical (a) and horizontal (b) distances between the leading and trailing edge of the cell, passing through the nucleus, define CAR, which was calculated as a/b ratio. An elongated cells was defined as those having a/b ratio greater than 1.56. Nuclear deformation was evaluated by the percentage of the cells whose nuclei (stained with DAPI) have stripped structures present in the fluorescence image. To evaluate cell alignment, cells were considered aligned if the angle between the long cellular axis and the grooves was less than 15°. For each group, around 150 cells were quantified. For control group, since there were no reference groove directions, we assign zero value for cell alignment. The relative orientation of the vinculin–positive focal adhesion complexes (FACs) was manually determined by measuring the alignment angle with respect to the direction of the microgrooves. Percentage of aligned FACs was plotted between −90° to +90°, with an interval of 36°. Finally, the alignment of actin filaments was also measured with a custom-written MatLab program, based on the image intensity gradient7. Briefly, the intensity gradient was first determined on subregions (16 by 16 pixels), and the direction perpendicular to this gradient is considered as the orientation of local actin filaments. Short red lines are then super-imposed onto the fluorescence image to indicate local actin filament orientation.
Time-lapse microscopy for subcellular organelle tracking on micropatterns
For mitochondria and lysosomal staining, MitoTracker® Deep Red FM and LysoTracker® Red DND-99 (Life Technologies) were used at 75 nM and 200 nM concentration, respectively, as per manufacturer's instructions. Astrocytes were incubated with MitoTracker or LysoTracker before seeded on the PDMS patterned substratum and flat PDMS surfaces. To observe short-term phenotypic changes, the environmental chamber containing a custom-made Pelicon dish was mounted onto the stage of a motorized inverted microscope equipped with an Axiocam 512B II CCD camera (Axiovert 200 M, Zeiss, Germany). Phase-contrast and fluorescence images of the astrocytes were recorded for 10 hours at 10 minutes intervals.
Live cell calcium signaling
To study the intracellular calcium signaling, astrocytes were seeded on the patterned and control PDMS surfaces at 5,000 cells per well (200 μL) for 2–4 hours. Afterwards, 10 μM fluo-4 AM was added into cell suspension media to dye cell calcium36. Cells were kept in the dark at room temperature for 20 min until the complete de-esterification of AM esters and ready for Ca2+ imaging with a confocal microscope. Cell culture media with Fluo-4 AM was replaced with fresh media containing anion-transport inhibitor probenecid (2 mM) to the cells to reduce the leakage of de-esterified indicator. Fluorescence images of the astrocyte were taken every 3 seconds to record the intracellular calcium signaling by a cooled CCD camera (Zeiss).
The intracellular calcium concentration ([Ca2+]i) amplitude was also quantified utilizing method described previously45,46. The calcium concentration was calculated by [Ca2+]i = Kd*(F − Fb)/(Fmax − F), where F is the fluorescence intensity inside of the cell, and Fmax and Fb is the fluorescence intensity at saturated and in absence of Ca2+. Kd is the ion dissociation constant, and is assumed to be 345 nM47. The peak fluorescence Fmax was obtained by treating astrocytes with the calcium ionophore.
Adenosine triphosphate (ATP) quantification
Measurement of ATP release was carried out by using an ATP assay kit based on the luciferase–luciferin test (Sigma-Aldrich). The astrocytes were seeded on micropatterned and flat PDMS surfaces and 50 μL supernatant was collected from each sample at every 5 minutes until three hours of cell seeding and spreading. Samples were stored at 4°C until analyzed. A 50-μL sample was added to 50 μL of ATP assay mix containing luciferase–luciferin buffer. Luminescence was measured with a Synergy H1 moncohromater plate reader (Biotek, Winooski, VT). Three samples from three independent experiments were evaluated. ATP release was calculated in picomolar (pM) concentration using ATP standards supplied by the manufacturer48.
Statistics
All data are presented as mean ± standard deviation (SD). One-way ANOVA followed by post-hoc Tukey's test was used to evaluate the statistical significance. Significance level was set at α = 0.05.
Author Contributions
A.V.S., M.R., A.C. and L.Q.W. designed research, performed experiments, and analyzed data. Z.M.Z. and C.A.M. isolated astrocytes. F.P. analyzed calcium signals. X.L.L. and R.G. analyzed some data. A.V.S. and L.Q.W. wrote the paper.
Supplementary Material
Supplementary info
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Acknowledgments
The authors would like to thank National Science Foundation (RJG and LQW), American Heart Association (LQW), and March of Dimes (LQW) for funding support. Leo Q. Wan is a Pew Scholar in Biomedical Sciences, supported by the Pew Charitable Trusts.
References
- Singh A. V., Patil R., Thombre D. K. & Gade W. N. Micro-nanopatterning as tool to study the role of physicochemical properties on cell–surface interactions. J Biomed Mater Res A 101, 3019–3032 (2013). [DOI] [PubMed] [Google Scholar]
- Worley K., Certo A. & Wan L. Q. Geometry–Force Control of Stem Cell Fate. BioNanoScience 3, 43–51 (2013). [Google Scholar]
- Freytes D. O., Wan L. Q. & Vunjak-Novakovic G. Geometry and force control of cell function. J Cell Biochem 108, 1047–1058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim E. K. F. et al. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 26, 5405–5413 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Sunyer R. & Trepat X. Mechanical guidance of cell migration: lessons from chemotaxis. Curr Opin Cell Biol 25, 543–549 (2013). [DOI] [PubMed] [Google Scholar]
- Singh A. V. et al. Carbon Nanotube-Induced Loss of Multicellular Chirality on Micropatterned Substrate Is Mediated by Oxidative Stress. ACS Nano 8, 2196–2205 (2014). [DOI] [PubMed] [Google Scholar]
- Wan L. Q., Ronaldson K., Guirguis M. & Vunjak-Novakovic G. Micropatterning of cells reveals chiral morphogenesis. Stem Cell Res Ther 4, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. Q. et al. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci U S A 108, 12295–12300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. Q. et al. Geometric control of human stem cell morphology and differentiation. Integr Biol 2, 346–353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. Q. & Vunjak-Novakovic G. Micropatterning chiral morphogenesis. Commun Integr Biol 4, 745–748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu A. et al. Superimposed topographic and chemical cues synergistically guide neurite outgrowth. Lab Chip 13, 3070–3081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlock T., Elisseeff J., Langer R., Vacanti J. & Cheney M. A tissue-engineered conduit for peripheral nerve repair. Archives of Otolaryngology–Head & Neck Surgery 124, 1081–1086 (1998). [DOI] [PubMed] [Google Scholar]
- Silver J. & Miller J. H. Regeneration beyond the glial scar. Nat Rev Neurosci 5, 146–156 (2004). [DOI] [PubMed] [Google Scholar]
- Wang H. B., Mullins M. E., Cregg J. M., McCarthy C. W. & Gilbert R. J. Varying the diameter of aligned electrospun fibers alters neurite outgrowth and Schwann cell migration. Acta Biomater 6, 2970–2978 (2010). [DOI] [PubMed] [Google Scholar]
- Mitragotri S. & Lahann J. Physical approaches to biomaterial design. Nat Mater 8, 15–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pêgo A. P. et al. Regenerative medicine for the treatment of spinal cord injury: more than just promises? J Cell Mol Med 16, 2564–2582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A. et al. Robust CNS regeneration after complete spinal cord transection using aligned poly-l-lactic acid microfibers. Biomaterials 32, 6068–6079 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereifej E. S. et al. Nanopatterning effects on astrocyte reactivity. J Biomed Mater Res A 101A, 1743–1757 (2013). [DOI] [PubMed] [Google Scholar]
- Meng F., Hlady V. & Tresco P. A. Inducing alignment in astrocyte tissue constructs by surface ligands patterned on biomaterials. Biomaterials 33, 1323–1335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S. K. et al. Effect of topography of an electrospun nanofiber on modulation of activity of primary rat astrocytes. Neurosci Lett 534, 80–84 (2013). [DOI] [PubMed] [Google Scholar]
- Mattotti M. et al. Inducing functional radial glia-like progenitors from cortical astrocyte cultures using micropatterned PMMA. Biomaterials 33, 1759–1770 (2012). [DOI] [PubMed] [Google Scholar]
- Hadjiargyrou M. & Chiu J. B. Enhanced composite electrospun nanofiber scaffolds for use in drug delivery. Expert Opin Drug Deliv 5, 1093–1106 (2008). [DOI] [PubMed] [Google Scholar]
- Schaub N. J. & Gilbert R. J. Controlled release of 6-aminonicotinamide from aligned, electrospun fibers alters astrocyte metabolism and dorsal root ganglia neurite outgrowth. J Neural Eng 8, 046026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. V. et al. Rapid prototyping of nano- and micro-patterned substrates for the control of cell neuritogenesis by topographic and chemical cues. Mater Sci Eng C 31, 892–899 (2011). [Google Scholar]
- Singh A. et al. Bottom-up engineering of the surface roughness of nanostructured cubic zirconia to control cell adhesion. Nanotechnology 23, 475101 (2012). [DOI] [PubMed] [Google Scholar]
- Takano H. et al. Micropatterned Substrates: Approach to Probing Intercellular Communication Pathways. Anal Chem 74, 4640–4646 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. Regulated ATP release from astrocytes through lysosome exocytosis. Nat Cell Biol 9, 945–953 (2007). [DOI] [PubMed] [Google Scholar]
- Lee W. & Parpura V. Micropatterned substrates for studying astrocytes in culture. Front Neurosci 3, 381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch A. S., Miller D., Luebke K. J. & Hu W. Correlation of anisotropic cell behaviors with topographic aspect ratio. Biomaterials 30, 1560–1567 (2009). [DOI] [PubMed] [Google Scholar]
- Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A. & Horwitz A. F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385, 537–540 (1997). [DOI] [PubMed] [Google Scholar]
- East E., de Oliveira D. B., Golding J. P. & Phillips J. B. Alignment of astrocytes increases neuronal growth in three-dimensional collagen gels and is maintained following plastic compression to form a spinal cord repair conduit. Tissue Eng Part A 16, 3173–3184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Koito H., Li J. & Han A. Multi-compartment neuron–glia co-culture platform for localized CNS axon–glia interaction study. Lab Chip 12, 3296–3304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M., Grevesse T. & Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun 3, 671 (2012). [DOI] [PubMed] [Google Scholar]
- Liazoghli D., Roth A. D., Thostrup P. & Colman D. R. Substrate Micropatterning as a New in Vitro Cell Culture System to Study Myelination. ACS Chem Neurosci 3, 90–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badique F. et al. Directing nuclear deformation on micropillared surfaces by substrate geometry and cytoskeleton organization. Biomaterials 34, 2991–3001 (2013). [DOI] [PubMed] [Google Scholar]
- Lu X. L., Huo B., Chiang V. & Guo X. E. Osteocytic network is more responsive in calcium signaling than osteoblastic network under fluid flow. J Bone Miner Res 27, 563–574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields R. D. Visualizing Calcium Signaling in Astrocytes. Sci Signal 3, tr5–tr5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti B., Parpura V. & Haydon P. G. Imaging Extracellular Waves of Glutamate during Calcium Signaling in Cultured Astrocytes. J Neurosci 20, 1800–1808 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco S. et al. Storage and Release of ATP from Astrocytes in Culture. J Biol Chem 278, 1354–1362 (2003). [DOI] [PubMed] [Google Scholar]
- Guthrie P. B. et al. ATP Released from Astrocytes Mediates Glial Calcium Waves. J Neurosci 19, 520–528 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaepel J. & Blum R. Capturing ER calcium dynamics. Eur J Cell Biol 90, 613–619 (2011). [DOI] [PubMed] [Google Scholar]
- Zuidema J. M. et al. Enhanced GLT-1 mediated glutamate uptake and migration of primary astrocytes directed by fibronectin-coated electrospun poly-l-lactic acid fibers. Biomaterials 35, 1439–1449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongin A. A., Hyzinski-García M. C., Vincent M. Y. & Keller R. W. A simple method for measuring intracellular activities of glutamine synthetase and glutaminase in glial cells. Am J Physiol Cell Physiol 301, C814–C822 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci 114, 797–809 (2001). [DOI] [PubMed] [Google Scholar]
- Schnetkamp P. P., Basu D. K., Li X. B. & Szerencsei R. T. Regulation of intracellular free Ca2+ concentration in the outer segments of bovine retinal rods by Na-Ca-K exchange measured with fluo-3. II. Thermodynamic competence of transmembrane Na+ and K+ gradients and inactivation of Na(+)-dependent Ca2+ extrusion. J Biol Chem 266, 22983–22990 (1991). [PubMed] [Google Scholar]
- Parpura V. & Haydon P. G. Physiological astrocytic calcium levels stimulate glutamate release to modulate adjacent neurons. Proc Natl Acad Sci U S A 97, 8629–8634 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee K. R. et al. Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell Calcium 27, 97–106 (2000). [DOI] [PubMed] [Google Scholar]
- Cotrina M. L. et al. Connexins regulate calcium signaling by controlling ATP release. Proc Natl Acad Sci U S A 95, 15735–15740 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary info
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3