Abstract
In reptiles, sex-determining mechanisms have evolved repeatedly and reversibly between genotypic and temperature-dependent sex determination. The gene Dmrt1 directs male determination in chicken (and presumably other birds), and regulates sex differentiation in animals as distantly related as fruit flies, nematodes and humans. Here, we show a consistent molecular difference in Dmrt1 between reptiles with genotypic and temperature-dependent sex determination. Among 34 non-avian reptiles, a convergently evolved pair of amino acids encoded by sequence within exon 2 near the DM-binding domain of Dmrt1 distinguishes species with either type of sex determination. We suggest that this amino acid shift accompanied the evolution of genotypic sex determination from an ancestral condition of temperature-dependent sex determination at least three times among reptiles, as evident in turtles, birds and squamates. This novel hypothesis describes the evolution of sex-determining mechanisms as turnover events accompanied by one or two small mutations.
Keywords: convergence, sex determination, ancestral reconstruction, squamate, archosaur
1. Introduction
Genotypic (GSD) and temperature-dependent sex determination (TSD) have evolved reversibly and repeatedly in reptiles [1,2]. At least four studies have reported reptile phylogenies with varying frequencies of change between GSD and TSD [1–4]. Although sex chromosomes have been retained throughout birds and snakes, lizards exhibit abundant turnover of sex chromosomes and sex-determining systems [5–8]. TSD is attributed to all crocodilians, most turtles and some lizards, and GSD has been inferred for extinct marine reptiles [9].
In this study, we describe an amino acid shift within the protein-coding region of the gene Dmrt1 that largely distinguishes reptiles characterized as exhibiting either TSD or GSD. DMRT1 is essential in double dosage for male development in humans, despite its location on an autosome [10] and its functional activation after Sry, the master sex-determining gene [11]. Dmrt1 is directly responsible for the initiation of male sex differentiation in chicken, Gallus gallus [12]. This gene is considered the sex-determining gene in chicken, because among genes that govern sexual development, Dmrt1 resides on the sex chromosomes, acts in a sex-specific pattern in the gonad earliest among those genes, and sex is reversed when Dmrt1 expression is suppressed [12]. In chicken, Dmrt1 knockdown is followed by decreased activity of Sox9, a gene responsible for testis differentiation, resulting in a female phenotype [12]. Here, we show that two mutated amino acids in Dmrt1 accompanied a convergent phenotype of GSD at least three times in reptiles.
2. Material and methods
Novel Dmrt1 sequences were obtained for fourteen reptiles by sequencing targeted PCR products from RNAs and for tuatara, Sphenodon punctatus, by searching an unpublished genome database. Other Dmrt1 sequences were downloaded from GenBank at the National Center for Biotechnology Information [13] or from Ensembl [14] (electronic supplementary material). The amino acid alignment was curated using BioEdit [15] and aligned in Seaview v. 4.5.1 using the Muscle algorithm [16]. The program Prank was used to align DNA sequences, using the codon option [17].
PhyML v. 3.0 [18] was used to construct the phylogeny of Dmrt1 using DNA sequences (less than or equal to 733 nt). Topology and branch lengths of the tree were inferred from the Dmrt1 sequence data using the HKY model. We specified a general time reversible model with gamma-distributed rate variation and invariant sites. We used MEGA v. 6.0 [19] to infer ancestral amino acid sequences at selected nodes using maximum-likelihood and the LG model [20] of amino acid evolution. A time-calibrated tree was created in Mesquite v. 3.01 [21] with the topology from our phylogenetic inference and branch lengths manually adjusted by Date-a-clade [22], using previously published divergence times [23–25].
The BayesTraits program (http://www.evolution.rdg.ac.uk/BayesTraits.html) was used to run a reversible-jump MCMC algorithm on character states for sex-determining mechanism in extant species [9] and ancestral reconstructions of sex-determining mechanisms in the ancestral archosaur, diapsid and lepidosaur [1] to test for correlated evolution between TSD and the combination of threonine at position no. 54 and serine at position no. 57. We used a gamma-distributed hyperprior with mean and variance randomly sampled from 0 to 10. The MCMC ran for 2 100 000 iterations with a 100 000 burn-in and a sampling frequency of 1000. Sex-determining mechanisms were coded as 0 for GSD and 1 for TSD. Sequence data were coded as presence (1) or absence (0) of threonine at position no. 54 and presence (1) or absence (0) of serine at position no. 57.
3. Results
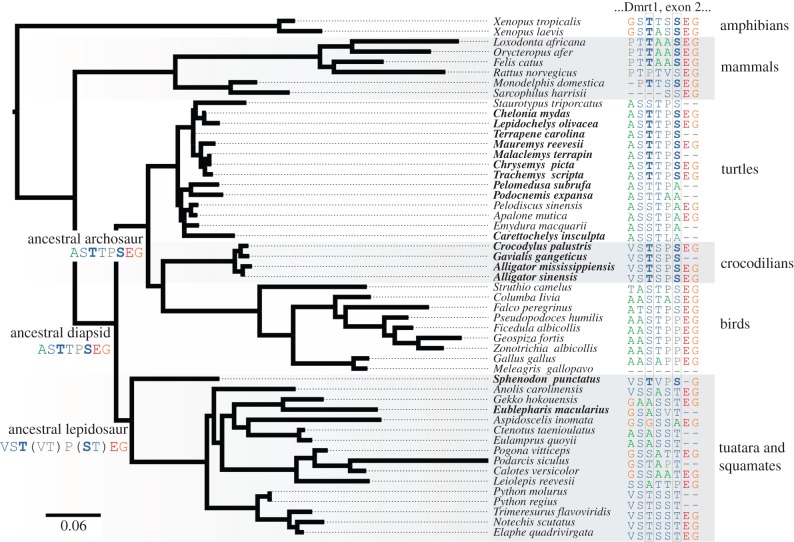
Using the time-calibrated tree, we found evidence that changes from an ancestral TSD state with threonine at position no. 54 (T54) and serine at position no. 57 (S57) of Dmrt1 (posterior probability (PP) = 95%) are associated with GSD evolution in reptiles. Further, we found 89% posterior support using the tree with branch lengths in units of substitutions per site of Dmrt1. Our results also suggest that evolution of T54 was dependent on evolution of S57 (PP = 99.9% using the time-calibrated tree and PP = 95% using the tree with branch lengths in substitutions). Sites 54 and 57 are found within exon 2 near the DM-binding domain of Dmrt1. Maximum-likelihood phylogenetic inference supports T54 and S57 in the ancestral archosaur and diapsid and T54 and T57 in the ancestral lepidosaur with probabilities greater than 90% (figure 1). The covariation of these two amino acids and sex-determining mechanism was originally identified by visual inspection of full-length Dmrt1 sequences across a subset of reptiles. The T54–S57 condition is found in all available Dmrt1 sequences for extant reptiles with TSD, with the exception of Pelomedusa subrufa, Podocnemis expansa, Carettochelys insculpta and Eublepharis macularius. All 18 extant reptiles in our dataset that exhibit GSD, and all nine sampled birds differ in one or both of these sites, suggesting a conformational difference between Dmrt1 in avian and non-avian reptiles with different sex-determining mechanisms. Low levels of documented homology and partial coverage for Dmrt1 protein prevented us from characterizing the effect of the two AA shift on three-dimensional conformation of the protein.
Figure 1.
Maximum-likelihood analysis of an ancestral Dmrt1 protein for diapsids, archosaurs and lepidosaurs. A consistent amino acid shift at positions 54 and 57 in the alignment largely distinguishes GSD and TSD species. Most TSD species (bold Latin binomials) have T and S in positions 54 and 57 of this sequence, suggesting mutation at these positions in the gene that codes for Dmrt1 accompanies a change in sex-determining mechanism. Grey outlined columns indicate positions 54 and 57. Bold amino acids indicate species with the T54–S57 amino acid state. The set of states at each node is ordered from most to least likely, excluding states with probabilities below 5%. Amino acid states in parentheses are ambiguous.
4. Discussion
This is the first report, to the best of our knowledge, of a molecular difference in a sex-determining gene between reptiles with GSD or TSD. Several lines of evidence support the idea that this amino acid shift accompanies turnover of sex-determining mechanisms. Dmrt1 is known to direct sexual development in chicken [12], a species found within Reptilia, a clade known for frequent and reversible changes in sex-determining mechanism [1]. Dmrt1 is expressed more in gonadal tissues of males than of females, even in a turtle that has TSD [26]. In other species, including mice and rats, Dmrt1 contributes to male sexual development but does not act first among sex-differentiating genes [27]. Capture or influence by one gene on sexual development could occur at any point in the gene cascade [28]. Therefore, we are not surprised to find exceptions to the two AA pattern across sampled reptiles. Sex-determining mechanisms have changed frequently, suggesting more than one cause. GSD, in extant therian mammals, is driven predominantly by Sry. Amino acid shifts in Dmrt1 are not likely to alter sex-determining mechanism in those species or in amphibians, all of which have GSD. Nonetheless, the relatively tight correlation of T54-S57 or other amino acid states and sex determination phenotypes, despite shifts in key driver genes, is intriguing. Even as a midpoint in the cascade, change in function of Dmrt1 could change overall function and thermal sensitivity of the cascade. By this model, in most sampled reptiles with TSD, Dmrt1 has the T54–S57 condition but still functions as part of the cascade of genes responsible for sexual development, enabling function and avoiding loss by gene conversion.
By using the amino acid shift as a proxy for sex-determining mechanism in a phylogenetic analysis, we replicated ancestral reconstructions that were previously inferred based on characterizations of mechanisms in a family-level analysis [1]. The ancestral lepidosaur is reconstructed as potentially exhibiting T54–T57, predicted to accompany GSD, as inferred using a different criterion. Likewise, the ancestral archosaur and diapsid are reconstructed as exhibiting T54–S57, indicative of TSD, also shown by previous reconstructions [1]. The ancestral amniote was also reconstructed as exhibiting T54–S57 despite lack of robust reconstruction of that ancestor as exhibiting TSD. Staurotypus triporcatus, a turtle with XY sex chromosomes homologous to chicken ZW sex chromosomes [29], exhibits an S54–S57 pattern also found in birds (figure 1). Finally, Carettochelys inscupta, a turtle with the only inferred reversal from GSD to TSD in chelonians [4], retains the S54–A57 pattern found in its GSD sister family Trionychidae, suggesting that TSD may have re-evolved by a different path from the TS amino acid state in Carettochelys. Thus, the TS amino acid state appears to allow TSD, but TSD is likely to be possible in the absence of the TS state, as GSD is possible in the presence of TS seen in amphibians and mammals. The relationship between our described amino acid shift and sex determination is only part of a complex evolutionary history of sex-determining mechanisms. This discovery suggests a new model for turnover of reptiles' sex-determining mechanisms in which one or two amino acid mutations accompanied a series of changes that altered the mechanisms' thermal sensitivity.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank G. Rivera and M. Penko for donating chelonian samples and L. Mickelson and N. Nelson for donating tuatara samples. Nagtiwai iwi and the New Zealand Department of Conservation permitted tuatara sampling.
Funding statement
Funding was provided by NSF grant no. MCB0817687 to N.V. and S.V.E, MCB 1244355 to N.V. The Harvard Club of Australia Foundation provided additional support for collaboration between D.E.J. and colleagues in Australia. T.E. is supported by an ARC Future Fellowship (FT110100733). The tuatara genome project is a strategic initiative supported by the Allan Wilson Centre.
References
- 1.Organ CL, Janes DE. 2008. Evolution of sex chromosomes in Sauropsida. Integr. Comp. Biol. 48, 512–519. ( 10.1093/icb/icn041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarre SD, Ezaz T, Georges A. 2011. Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genomics Hum. Genet. 12, 18.1–18.16. ( 10.1146/annurev-genom-082410-101518) [DOI] [PubMed] [Google Scholar]
- 3.Janzen FJ, Phillips PC. 2006. Exploring the evolution of environmental sex determination in reptiles: ecology, evolution, and experimental design. Q. Rev. Biol. 66, 149–170. ( 10.1111/j.1420-9101.2006.01138.x) [DOI] [PubMed] [Google Scholar]
- 4.Valenzuela N, Adams DC. 2011. Chromosome number and sex determination co-evolve in turtles. Evolution 65, 1808–1813. ( 10.1111/j.1558-5646.2011.01258.x) [DOI] [PubMed] [Google Scholar]
- 5.Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara CH, Agata K, Matsuda Y. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and stepwise differentiation of snake sex chromosomes. Proc. Natl Acad. Sci. USA 103, 18 190–18 195. ( 10.1073/pnas.0605274103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezaz T, Sarre SD, O'Meally D, Graves JAM. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet. Genome Res. 127, 249–260. ( 10.1159/000300507) [DOI] [PubMed] [Google Scholar]
- 7.O'Meally D, Patel HR, Stiglec R, Sarre SD, Georges A, Graves JAM, Ezaz T. 2010. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 18, 787–800. ( 10.1007/s10577-010-9152-9) [DOI] [PubMed] [Google Scholar]
- 8.Pokorna M, et al. 2011. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma 120, 455–468. ( 10.1007/s00412-011-0322-0) [DOI] [PubMed] [Google Scholar]
- 9.Organ CL, Janes DE, Meade A, Pagel M. 2009. Genotypic sex determination enabled adaptive radiations of extinct marine reptiles. Nature 461, 389–392. ( 10.1038/nature08350) [DOI] [PubMed] [Google Scholar]
- 10.Bennett CP, Docherty Z, Robb SA, Ramani P, Hawkins JR, Grant D. 1993. Deletion 9p and sex reversal. J. Med. Genet. 30, 518–520. ( 10.1136/jmg.30.6.518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair AH, et al. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. ( 10.1038/346240a0) [DOI] [PubMed] [Google Scholar]
- 12.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Fairlie PG, Doran TJ, Sinclair AH. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271. ( 10.1038/nature08298) [DOI] [PubMed] [Google Scholar]
- 13.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res. 33, D34–D38. ( 10.1093/nar/gki063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flicek P, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42, D749–D755. ( 10.1093/nar/gkt1196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 16.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. ( 10.1093/molbev/msp259) [DOI] [PubMed] [Google Scholar]
- 17.Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170. ( 10.1007/978-1-62703-646-7_10) [DOI] [PubMed] [Google Scholar]
- 18.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6 molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le SQ, Gascuel O. 1993. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. ( 10.1093/molbev/msn067) [DOI] [PubMed] [Google Scholar]
- 21.Maddision WP, Maddison DR. 2014. Mesquite: a modular system for evolutionary analysis. Version 3.01. See http://mesquiteproject.org.
- 22.Benton MJ, Donoghue PCJ. 2007. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26–53. ( 10.1093/molbev/msl150) [DOI] [PubMed] [Google Scholar]
- 23.Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base. See www.timetree.net Pennsylvania and Arizona State Universities. [Google Scholar]
- 24.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444–448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 25.Dornburg A, Beaulieu JM, Oliver JC, Near TJ. 2011. Integrating fossil preservation biases in the selection of calibrations for molecular divergence time estimation. Syst. Biol. 60, 519–527. ( 10.1093/sysbio/syr019) [DOI] [PubMed] [Google Scholar]
- 26.Murdock C, Wibbels T. 2003. Expression of Dmrt1 in a turtle with temperature-dependent sex determination. Cytogenet. Genome Res. 101, 302–308. ( 10.1159/000074353) [DOI] [PubMed] [Google Scholar]
- 27.Lei N, Heckert LL. 2004. Gata4 regulates testis expression of Dmrt1. Mol. Cell. Biol. 24, 377–388. ( 10.1128/MCB.24.1.377-388.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georges A, Ezaz T, Quinn AE, Sarre SD. 2010. Are reptiles predisposed to temperature-dependent sex determination? Sex Dev. 4, 7–15. ( 10.1159/000279441) [DOI] [PubMed] [Google Scholar]
- 29.Kawagoshi T, Uno Y, Nishida C, Matsuda Y. 2014. The Staurotypus turtles and aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS ONE 9, e105315 ( 10.1371/journal.pone.0105315) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.