Abstract
One unreported case of extended-spectrum-beta-lactamase (ESBL)-producing Salmonella enterica serovar Typhi was identified, whole-genome sequence typed, among other analyses, and compared to other available genomes of S. Typhi. The reported strain was similar to a previously published strain harboring blaSHV-12 from the Philippines and likely part of an undetected outbreak, the first of ESBL-producing S. Typhi.
TEXT
The occurrence of extended-spectrum-beta-lactamase (ESBL)-producing Salmonella enterica serovar Typhi is an alarming development which may significantly complicate the treatment of typhoid fever. To date, ESBL-producing S. enterica serovar Typhi has only been reported from Bangladesh, Egypt, India, Iran, Iraq, Pakistan, and the Philippines (1). A subset of strains (from India, Iraq, and the Philippines) has been independently confirmed, and an assortment of ESBL genes (blaCTX-M-15, blaSHV-12, and blaCMY-2) have been identified and sequenced.
The purpose of the present study was to identify potentially unreported cases of typhoid fever caused by ESBL-producing S. Typhi at a global level, to confirm ESBL production phenotypically, and to identify the responsible ESBL genes. Furthermore, we wanted to investigate the genetic relatedness to other available ESBL-producing S. Typhi isolates using whole-genome sequence typing (WGST) and a variety of molecular and genomic studies and to test the hypothesis that impaired restriction modification (RM) systems could be a factor for the development of ESBL resistance.
On 4 May 2012, an electronic message requesting information about any confirmed or suspected ESBL-producing S. Typhi isolates was sent to the members of the World Health Organization (WHO) Global Foodborne Infections Network (GFN). Of 1,062 recipients, 3 members (0.28%) responded. We believe that the low response rate reflects a true low prevalence as the members frequently receive and respond to messages.
One strain (strain 1107-3567) confirmed as an ESBL-producing S. Typhi strain was submitted by The Norwegian Institute of Public Health. The Norwegian patient in question and a previously published case from the Netherlands (strain TY5359) (2) had travel histories to the Philippines in late 2007 that were almost identical. The Norwegian patient had gastroenteritis, whereas the Dutch patient was admitted to a hospital with typhoid fever caused by an ESBL-producing S. Typhi containing the blaSHV-12 gene. The Dutch patient was treated successfully with ciprofloxacin; the treatment of the Norwegian patient was unknown.
The isolates were sequenced using the MiSeq platform (Illumina, Inc., San Diego, CA) (see Methods in the supplemental material). The raw reads were assembled using the Assembler pipeline (version 1.0) available from the Center for Genomic Epidemiology (CGE) (http://cge.cbs.dtu.dk/services/all.php) and submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/PRJEB6961) (accession no. ERS525820 and ERS525821). A complete list of genomic sequence data is available in Table S1A in the supplemental material.
MIC determination (3–5; see Methods in the supplementary material) and acquired antimicrobial resistance genes using the CGE pipeline ResFinder (version 2.1, 80% threshold for % infective dose [ID]/60% minimum length) (6) revealed resistance to the following antimicrobials and genes: ampicillin, blaTEM-1; cefotaxime, cefpodoxime, ceftazidime, ceftiofur, and ceftriaxone, blaSHV-12; gentamicin, aac(6′)IIc; streptomycin, strA/strB; tetracycline, tetD; and trimethoprim, dfrA18 (Table 1). The isolates were susceptible to the following antimicrobials: apramycin, azithromycin, cefepime, cefoxitin, chloramphenicol, ciprofloxacin, colistin, florfenicol, imipenem, meropenem, nalidixic acid, neomycin, spectinomycin, and sulfamethoxazole (Table 1).
TABLE 1.
Antimicrobial resistance profiles of the two ESBL-producing Salmonella serovar Typhi isolates from the Norwegian and Dutch patients
| Antimicrobial class | Antimicrobial(s) | CLSI clinical resistance (R) breakpoint (mg/liter) | MIC (mg/liter) for isolatea: |
|
|---|---|---|---|---|
| TY5359 (NL) | 1107-3567 (N) | |||
| β-Lactam/inhibitor | Amoxicillin + clavulanic acid | ≥32 | 8 | 4 |
| Aminocyclitol | Spectinomycinb | 64 | ≤16 | ≤16 |
| Aminoglycoside | Apramycinc | 32 | ≤4 | ≤4 |
| Gentamicin | ≥16 | >16 | >16 | |
| Neomycinb | 4 | ≤2 | ≤2 | |
| Streptomycinb | 16 | >128 | >128 | |
| Carbapenem | Imipenem | ≥4 | ≤0.5 | ≤0.5 |
| Meropenem | ≥4 | ≤1 | ≤1 | |
| Cephalosporin | ||||
| Second generation | Cefoxitin | ≥32 | ≤4 | 8 |
| Third generation | Cefotaxime | ≥4 | 32 | 8 |
| Cefpodoxime | ≥8 | >32 | >32 | |
| Ceftazidime | ≥16 | 64 | 64 | |
| Ceftiofurb | 2 | >8 | >8 | |
| Ceftriaxone | ≥4 | 32 | 16 | |
| Fourth generation | Cefepime | ≥32 | 4 | 2 |
| Macrolide | Azithromycin | ≥32 | 8 | 16 |
| Penicillin | Ampicillin | ≥32 | >32 | >32 |
| Phenicol | Chloramphenicol | ≥32 | 4 | 4 |
| Florfenicolb | 16 | 4 | ≤2 | |
| Polypeptide | Colistinb | 2 | ≤1 | ≤1 |
| Quinolone | Ciprofloxacin | ≥1 | ≤0.016 | ≤0.016 |
| Nalidixic acid | ≥32 | ≤4 | ≤4 | |
| Sulfonamide | Sulfamethoxazole | ≥512 | ≤64 | 256 |
| Tetracycline | Tetracycline | ≥16 | >32 | >32 |
| Trimethoprim | Trimethoprim | ≥16 | >32 | >32 |
NL, the Netherlands, N: Norway. MICs in bold type indicate resistance toward the antimicrobial.
EUCAST epidemiological cutoff values.
According to Technical University of Denmark (DTU) food research.
Pulsed-field gel electrophoresis (PFGE) typing (7) with XbaI showed that both isolates clustered together but displayed a three-band difference; TY5359 produced a unique XbaI pattern. Among the 5,968 S. Typhi PFGE profiles uploaded to the U.S. and global databases, isolate 1107-3567 was indistinguishable from pattern JPPX01.0621, and this pattern had been seen six times before; two of the matches were from the Philippines (unknown susceptibility status), submitted in 2007 (see Methods in the supplemental material).
Multilocus sequence typing (MLST) based on the CGE pipeline MLST (version 1.7) (8) and haplotypes based on a previously described WGS-based method (9) (see Table S1B in the supplemental material) showed that the two S. Typhi strains belonged to MLST sequence type 1 (ST1) and the relatively infrequent haplotype H13, defined by the biallelic polymorphisms (BiPs) 40, 53, 56, and 81.
Single nucleotide polymorphisms (SNPs) were determined using the CGE pipeline snpTree (version 1.1, default settings) (10) (see Methods in the supplemental material). Assembled genomes or contigs were aligned against the reference genome, S. Typhi strain CT18. The qualified SNPs were selected once they met the following criteria: (i) a minimum coverage (number of reads mapped to reference positions) of 10, (ii) a minimum distance of 15 bp between each SNP, and (iii) a minimum quality score for each SNP at 30; all indels were excluded. Prior to creating the phylogenetic SNP tree, all SNPs were visually curated. The final phylogenetic SNP tree was computed using the maximum likelihood method (see Methods in the supplemental material).
The tree exhibited 2,776 high-quality whole-genome SNPs overall. No SNP differences were observed between the two isolates (TY5359 and 1107-3567). The minor differences between the PFGE profiles compared to the detection of no SNPs between the isolates can be explained by possible DNA recombination or the exclusion of indels, islands, and accessory genes containing possible XbaI cleavage sites in the SNP analysis. All SNPs identified between the two isolates and the public available genomes included are listed in Table S1C in the supplemental material.
The reference-rooted tree showed that the closest neighbors to the two Philippine S. Typhi genomes were BL196 and CR0044, which were both acquired in Malaysia and separated by 149 and 182 SNPs, respectively (Fig. 1). Interestingly, both of the Malaysian S. Typhi isolates were pan-susceptible, belonged to the same haplotype (H13) as the two Philippine strains, and were isolated within the same time period (2005 and 2007, respectively), thus suggesting that they were not part of the Filipino cluster but may share an ancestor.
FIG 1.
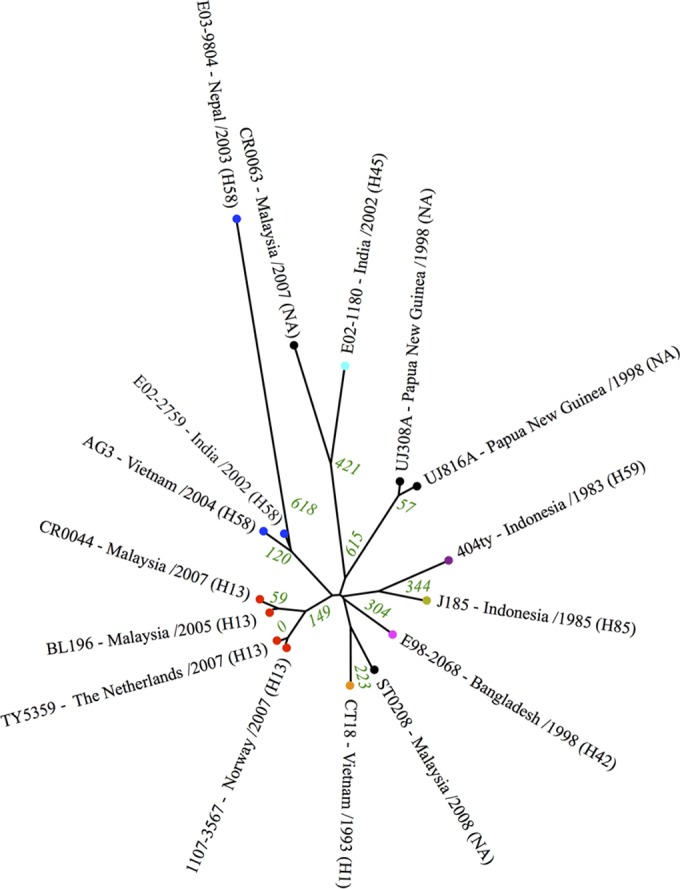
Phylogenetic SNP analysis of the Salmonella serovar Typhi genomes. The green numbers indicate the total SNP differences between isolates. The colored dots represent identical haplotypes, NA, haplotype not assessed.
The ESBL-producing S. Typhi contained a single plasmid with an approximate size of 280 kb confirmed by S1 PFGE (11; see Methods in the supplemental material). Two closely related incHI2 and incHI2A plasmid replicon subgroups were identified, and both incHI2 plasmids belonged to pMLST ST1 by using the CGE pipelines PlasmidFinder (version 1.2, 80% threshold for % ID) and pMLST (version 1.2, default settings) (12; see Methods in the supplemental material).
Plate mating was performed with the two S. Typhi isolates as donors and plasmid-free, rifampin- and nalidixic acid-resistant Escherichia coli MT102RN strains as recipients (see Methods in the supplemental material). A transformant was subjected to plasmid purification followed by whole-genome sequencing, which revealed the transfer of the blaSHV-12 gene on a plasmid with a size of approximately 280 kb. Additionally, the antimicrobial resistance genes, strA/B, aac(6′)IIc, blaTEM-1, tetD, and dfrA18, were successfully cotransferred. In contrast, conjugation of strain 1107-3567 was unsuccessful under the laboratory conditions used. In addition, subsequent SNP analysis covering 83% of the pEC_IMPQ reference sequence produced the same 17 SNPs for the two isolates.
A BLAST atlas was used to detect putative deletions in a comparison of the genomes against the reference genome CT18. One large deletion of 144 kb between sequence positions 4358279 (STY4472) and 4502238 (STY4635) was absent in isolate 1107-3567 relative to the CT18 reference genome (see Table S1D in the supplemental material). The deletion was equivalent to a 144-kb DNA fragment carrying the 120-kb Salmonella pathogenicity island SPI-7 encoding the synthesis of the Vi capsule exopolysaccharide previously described (13). The 144-kb deletion consists of various distinct regions, including the conjugal DNA transfer region, the type IVB pilus system, the DNA conjugational transfer region, and a partial sopE bacteriophage region (see Table S1D in the supplemental material). Deletions of different sizes of SPI-7 have previously been described (13–17) and related to plasmid- or chromosome-borne integrative and conjugative elements (ICEs) (17, 18). ICEs, the conjugative system, allow the exchange of new genetic material by, e.g., horizontal gene transfer (18). In this study, we did not succeed in performing conjugation mating experiments with S. Typhi isolate 1107-3567 harboring the deletion in contrast to the isolate harboring a fully intact SPI-7. We speculate whether the lack of the conjugal DNA transfer region, the type IVB pilus system, and the DNA conjugational transfer region that is part of the ICE may have prevented successful conjugation.
The integrity of the RM systems present in the two S. Typhi isolates were analyzed and compared to those of 11 publicly available S. Typhi genomes using the CGE Restriction-ModificationFinder (version 1.0, 80% threshold for % ID/80% minimum length) The outcome was subsequently blasted against the Conserved Domain Database at NCBI (see Methods in the supplemental material).
RM system analysis revealed three potential RM systems (types I, III, and IV), all identical in the 11 strains. In addition to the three potential systems, both isolates also revealed identity to a type II methyltransferase which has not been linked to restriction endonuclease but is required for the viability of E. coli cells. We were not able to obtain genetic evidence for the loss of integrity of the RM systems in the two ESBL-producing isolates included in this study. Therefore, either the RM systems may not be restricting this plasmid in S. Typhi or the plasmid may have been initially introduced into an intermediate S. Typhi host with an impaired restriction gene but a functional methylation gene and subsequently disseminated by horizontal transfer to the present S. Typhi host.
Overall, the ESBL-producing S. Typhi isolates were strikingly similar based on a variety of analyses. Thus, put into context with only a few reported cases worldwide of ESBL-producing S. Typhi, the occurrence of two nonrelated travelers being infected with these low-prevalence strains during the same time period in the same country supports the possibility that both ESBL-producing S. Typhi have the same clonal origin. When one considers that additional strains with indistinguishable PFGE profiles were reported in the Philippines during the same time period, it is highly likely that these strains were associated with an undetected outbreak according to the WHO definition “A single case of a communicable disease long absent from a population, or caused by an agent not previously recognized in that community or area”; to our knowledge, this is the first outbreak of ESBL-producing S. Typhi.
There is a vital need to establish a well-functioning surveillance system with the capability of conducting reliable antimicrobial susceptibility testing in regions where typhoid is endemic and the likelihood of ESBL-producing S. Typhi emerging is high and in areas where confirmed cases of ESBL-producing S. Typhi have already been identified. This surveillance will be crucial for determining if these strains persist and implementing interventions should they reemerge.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Mark Achtman and Zhemin Zhou, Warwick Medical School University of Warwick Coventry, Ireland, for providing biallelic polymorphisms positions.
This work was supported by the Danish Council for Strategic Research (grant 09-067103), the Center for Genomic Epidemiology (www.genomicepidemiology.org), and the World Health Organization Global Foodborne Infections Network (www.who.int/gfn).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03104-14.
REFERENCES
- 1.Wain J, Hendriksen RS, Mikoleit M, Keddy KH, Ochiai RL. Typhoid fever. Lancet, in press. doi: 10.1016/S0140-6736(13)62708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Naiemi N, Zwart B, Rijnsburger MC, Roosendaal R, Debets-Ossenkopp YJ, Mulder JA, Fijen CA, Maten W, Vandenbroucke-Grauls CM, Savelkoul PH. 2008. Extended-spectrum-beta-lactamase production in a Salmonella enterica serotype Typhi strain from the Philippines. J Clin Microbiol 46:2794–2795. doi: 10.1128/JCM.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing, 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Hendriksen RS, Le HS, Bortolaia V, Pulsrikarn C, Nielsen EM, Pornruangmong S, Chaichana P, Svendsen CA, Weill FX, Aarestrup FM. 2012. Characterization of isolates of Salmonella enterica serovar Stanley, a serovar endemic to Asia and associated with travel. J Clin Microbiol 50:709–720. doi: 10.1128/JCM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 8.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, Achtman M. 2006. Evolutionary history of Salmonella typhi. Science 314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leekitcharoenphon P, Kaas RS, Thomsen MC, Friis C, Rasmussen S, Aarestrup FM. 2012. snpTree—a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13(Suppl 7):S6. doi: 10.1186/1471-2164-13-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirichote P, Hasman H, Pulsrikarn C, Schonheyder HC, Samulioniene J, Pornruangmong S, Bangtrakulnonth A, Aarestrup FM, Hendriksen RS. 2010. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol 48:883–888. doi: 10.1128/JCM.01792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Zankari E, Garcia-Fernandez A, Volby LM, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickard D, Wain J, Baker S, Line A, Chohan S, Fookes M, Barron A, Gaora PO, Chabalgoity JA, Thanky N, Scholes C, Thomson N, Quail M, Parkhill J, Dougan G. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J Bacteriol 185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker S, Dougan G. 2007. The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 45(Suppl 1):S29–S33. doi: 10.1086/518143. [DOI] [PubMed] [Google Scholar]
- 15.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat Genet 40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair S, Alokam S, Kothapalli S, Porwollik S, Proctor E, Choy C, McClelland M, Liu SL, Sanderson KE. 2004. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J Bacteriol 186:3214–3223. doi: 10.1128/JB.186.10.3214-3223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth-Smith HM, Fookes MC, Okoro CK, Baker S, Harris SR, Scott P, Pickard D, Quail MA, Churcher C, Sanders M, Harmse J, Dougan G, Parkhill J, Thomson NR. 2012. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J Bacteriol 194:1494–1504. doi: 10.1128/JB.06403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


