Abstract
The rates of infection with Fusarium molds are increasing, and a diverse number of Fusarium spp. belonging to different species complexes can cause infection. Conventional species identification in the clinical laboratory is time-consuming and prone to errors. We therefore evaluated whether matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a useful alternative. The 289 Fusarium strains from the Belgian Coordinated Collections of Microorganisms (BCCM)/Institute of Hygiene and Epidemiology Mycology (IHEM) culture collection with validated sequence-based identities and comprising 40 species were used in this study. An identification strategy was developed, applying a standardized MALDI-TOF MS assay and an in-house reference spectrum database. In vitro antifungal testing was performed to assess important differences in susceptibility between clinically relevant species/species complexes. We observed that no incorrect species complex identifications were made by MALDI-TOF MS, and 82.8% of the identifications were correct to the species level. This success rate was increased to 91% by lowering the cutoff for identification. Although the identification of the correct species complex member was not always guaranteed, antifungal susceptibility testing showed that discriminating between Fusarium species complexes can be important for treatment but is not necessarily required between members of a species complex. With this perspective, some Fusarium species complexes with closely related members can be considered as a whole, increasing the success rate of correct identifications to 97%. The application of our user-friendly MALDI-TOF MS identification approach resulted in a dramatic improvement in both time and accuracy compared to identification with the conventional method. A proof of principle of our MALDI-TOF MS approach in the clinical setting using recently isolated Fusarium strains demonstrated its validity.
INTRODUCTION
Fusarium is a widely distributed fungal genus of soil inhabitants and plant pathogens that are important in agriculture (1). At least 70 Fusarium species have also been reported as opportunistic human pathogens, and infection rates have increased over the past years (2). Fusarium spp. can cause superficial infections, such as keratitis and onychomycosis, as well as locally invasive and disseminated infections (1, 2). These disseminated infections particularly affect immunosuppressed patients and are associated with a high mortality rate (2).
Fusarium species are grouped into several species complexes (3). Molecular evolutionary analysis has been performed for most species complexes and revealed many additional phylogenetically distinct species (4–12). These closely related species are often morphologically indistinguishable, and multilocus gene sequencing is needed to attempt species delimitation (13, 14). The most important clinically relevant Fusarium species complexes include the F. solani species complex (FSSC), F. oxysporum species complex (FOSC), F. fujikuroi species complex (FFSC), F. dimerum species complex (FDSC), F. incarnatum-F. equiseti species complex (FIESC), and F. chlamydosporum species complex (FCSC). Members of the FSSC are estimated to cause the majority of fusarioses (50%), followed by members of the FOSC and FFSC (both 20%) (1, 12, 15).
Compared to other molds, Fusarium spp., especially isolates of the FSSC, are considered relatively resistant to most antifungals. However, according to in vitro testing, antifungal susceptibility profiles have been shown to vary between Fusarium species (16–26). Correct identification to the species or species complex level may thus be crucial for patient treatment, although no in vitro-in vivo correlations have been demonstrated, and treatment is dependent on the type of infection (27). Identification to the species level is also essential for epidemiological purposes.
A differential diagnosis of fusariosis from other mycoses is not that straightforward, and identification of the infecting Fusarium sp. is even more difficult. Classically, in the clinical laboratory, the identification of a mold infection relies on a morphological examination of an isolate by expert mycologists. This approach is challenging, especially with Fusarium, which contains many cryptic species, and for some isolates, specific morphological traits are lacking and appear atypical in culture. Inaccurate species identifications or identifications to only the genus level are thus not uncommon. DNA sequencing, the gold standard, can complement the morphological identification but is costly, and different DNA markers may need to be evaluated. Moreover, routine conventional identification is a time-consuming process. It is widely accepted that the early onset of appropriate treatment for infected patients decreases the mortality rates of invasive infections and increases the chance of success. With this perspective, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has received major interest.
MALDI-TOF MS has improved the identification of bacteria and yeasts in the clinical setting, increasingly replacing the conventional methods (28–32). It has emerged as a rapid, simple, cost-effective, reliable, and reproducible identification tool in which a spectrum is generated based on the proteomic content of a microorganism and compared against a reference spectrum database for identification. Recently, the MALDI-TOF MS approach was also applied to mold identification and was shown to be able to accurately identify a wide array of species from various genera or groups of clinical interest, including Aspergillus spp. and dermatophytes (33–44). Species identification by MALDI-TOF MS within the Fusarium genus has not yet been thoroughly investigated. Studies have either focused on the comparison of different sample preparation procedures (45, 46) or analyzed only a limited number of Fusarium spp. and strains (47, 48). Moreover, it has been shown that for some mold species, the data obtained by MALDI-TOF MS are correlated with their phylogenetic data (49).
The main objective of the present study was to investigate whether MALDI-TOF MS is a useful alternative for the species identification of Fusarium isolates compared to identification with conventional methods. Therefore, we aimed to take into account a substantial part of the species diversity in the Fusarium genus. We focused on the performance of MALDI-TOF MS to accurately discriminate between species complexes and especially the members showing important differences in their antifungal susceptibility profiles.
MATERIALS AND METHODS
Taxon sampling.
A total of 323 Fusarium strains preserved and referenced in the Belgian Coordinated Collections of Microorganisms (BCCM)/Institute of Hygiene and Epidemiology Mycology (IHEM) culture collection were used in this study (see Table S1 in the supplemental material). The strains were gathered over the previous 30 years from different origins, mostly clinical. Since many taxonomical changes have occurred during this time period and the strains were identified only by morphological examination or assigned a Fusarium sp. upon the time of their deposit, we reidentified all strains.
Multilocus sequencing.
Our reidentification relied on BLAST analysis in GenBank and FUSARIUM-ID (50), using sequences of the internal transcribed spacer (ITS) region and part of the ribosomal large subunit (LSU), as well as sequences of a partial fragment of the beta-tubulin (BT) and translation elongation factor 1-alpha (TEF1α) gene. The choice of DNA markers was based on previous studies (11, 51). DNA extraction, PCR amplification, and sequencing were performed according to the protocol applied by Beguin et al. (52). The following primers were described previously: those for ITS by White et al. (53), for LSU by Hopple and Vilgalys (54), for BT by Glass and Donaldson (55), and for TEF1α by Carbone and Kohn (56).
Morphological characterization.
The strains were also reanalyzed morphologically. This was done after cultivation on potato dextrose agar (PDA) and nutrient-poor agar at 25°C in an incubator for ≥7 days, according to The Fusarium laboratory manual of Leslie, Summerell, and Bullock (57).
Phylogenetic analysis.
In order to determine the evolutionary species relationships in our Fusarium data set, phylogenetic analysis was performed. The sequences were first aligned using the Clustal W algorithm in MEGA4 (58) and edited manually. Subsequently, Bayesian inference (BI) analysis was executed on the combined sequence data set with MrBayes3.2 (59), using the Monte Carlo Markov chain method with runs of 1 million generations and sampling a tree every 500 generations. The first 25% of the sampled trees were discarded (i.e., burn-in). A consensus tree with posterior probabilities was assessed from the remaining trees. Our combined sequence data set was partitioned according to the different gene regions, and the GTR+I+Γ model of evolution was applied with parameters estimated separately for each partition. Tracer version 1.5 (60) was used to check the convergence of the likelihood scores, showing acceptable mixing of the runs and sufficient sampling. An Acremonium sp. (strain IHEM 7465) was selected as the outgroup.
In vitro antifungal susceptibility testing.
The Fusarium strains in the collection, which were isolated from patients, were subjected to in vitro antifungal susceptibility testing, according to the EUCAST E.DEF 9.1 broth microdilution method, as described previously (61). Inoculum suspensions were prepared from cultures grown on PDA for 2 days at 35°C and subsequently for 5 days at 25°C in a humidified incubator. A panel of eight commonly used antifungal agents was evaluated. Two-fold serial drug dilutions were prepared in 96-well plates, with concentrations ranging from 64 to 0.032 μg/ml for fluconazole and 5-fluorocytosine and from 16 to 0.008 μg/ml for amphotericin B, voriconazole, itraconazole, ketoconazole, posaconazole, and terbinafine. The minimum 100% inhibitory concentrations (MIC100) were assessed for each strain, and internal control strains with known MIC100s were included. An antifungal susceptibility profile was constructed for each species tested in the data set.
MALDI-TOF MS assay.
Our MALDI-TOF MS assay was based on the optimized procedure developed by Cassagne et al. (33). This assay has already been applied in several studies for MALDI-TOF MS identification of molds from different genera (40, 42–44). Some adaptations were made to the protocol. The strains were cultivated on Sabouraud-chloramphenicol agar plates at 25°C in a humidified incubator for 3 days. The incubation times of the sample in formic acid and acetonitrile for protein extraction were both augmented to 15 min, and the volume administered was increased to 50 μl. The acquisition of mass spectra was performed on a microflex LT mass spectrometer (Bruker Daltonics, Bremen, Germany), using the default settings of the manufacturer. Instrument calibration was achieved with a Bacterial Test Standard (Bruker Daltonics). The time of flight measurements were converted to m/z values, and all raw spectra were automatically processed by the Flexcontrol version 2.4 AutoXecute software (Bruker Daltonics). The resulting peak lists were exported to the MALDI Biotyper 3.0 software (Bruker Daltonics).
Construction of reference spectrum database.
A reference spectrum was constructed for each Fusarium strain, as described by Cassagne et al. (33), taking into account 4 culture replicates per strain for separate protein extraction and spotting each extract 10 times onto a MALDI 96 polished steel target plate (Bruker Daltonics). The resulting Fusarium reference spectra were implemented in an in-house database, which already contained the reference spectra of 1,021 other validated strains comprising 152 different fungal genera from the BCCM/IHEM collection.
Identification strategy.
In order to make unbiased MALDI-TOF MS identifications for our Fusarium strains, we applied the following strategy. For each strain, 4 additional raw spectra (i.e., the identification spectra) were collected from a separate culture. These identification spectra were subsequently challenged against the reference spectrum database using the MALDI Biotyper 3.0 software and excluding the reference spectrum of the analyzed strain to determine its identity. The second step was done to ensure that the strain would not be recognized by its own reference spectrum. The results of the matching process were expressed as logarithmic scores from 0 (no spectrum match) to 3 (a perfect match). For identification in the present study we took into account the reference strain associated with the highest logarithmic score (i.e., the top score). Analogous to the methodology of Cassagne et al. (33), the identification of a strain by MALDI-TOF MS was considered interpretable when two criteria were fulfilled: (i) at least three out of four identification spectra were identified by the reference spectrum of the same species, and (ii) at least one of their top scores had a value above the standard manufacturer-defined 2.0 cutoff for reliable species identification. The MALDI-TOF MS identification of a strain was found to be correct when the obtained identity matched that determined from the gold standard multilocus sequence analysis. Correct species complex identifications were also taken into account. When the criteria for identification were not fulfilled, it was evaluated whether lowering the cutoff value for reliable identification would allow the strain to be identified; otherwise, the strain remained unidentified by MALDI-TOF MS. Per Fusarium species in the data set for which more than one strain could be analyzed, we calculated the percentages of correct species identifications, correct species complex identifications, erroneous identifications, and identifications in which the criteria were not fulfilled (for both the standard 2.0 and a lowered cutoff value). Species represented by only one strain in the data set were not included in the calculations, since, in our setup, they were always misidentified (seeing as their own reference spectrum is not taken into account). For these one-strain species, it was checked whether they were identified by MALDI-TOF MS as a phylogenetically closely related species (i.e., within the correct species complex).
Comparison.
In order to visualize the species relationships in the Fusarium reference spectrum data set, a dendrogram was constructed. Therefore, the distance matrix of the MALDI Biotyper 3.0 software was used, in which distance values are relative and normalized to a maximum value of 1,000. The topology of the dendrogram was compared with that of the BI consensus tree. In this way, we evaluated if there was a correlation between the MALDI-TOF MS data and the phylogenetic data.
Proof of principle.
The validity of our MALDI-TOF MS identification approach was assessed in a clinical routine by screening 20 recent clinical isolates with a presumable Fusarium identity. The isolates were recovered from patient samples between 2012 and 2013 in the medical mycology laboratories of the Universitair Ziekenhuis Brussel (Belgium) and the Universitair Medisch Centrum Sint-Pieter (Belgium). We subjected them to MALDI-TOF MS identification, according to the criteria described above, and in parallel to a DNA sequence-based identification using ITS or, when ITS was not found to be sufficiently discriminatory, TEF1α.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank with the accession numbers given in Table 1; see also the accession numbers in Table S1 in the supplemental material.
TABLE 1.
Assessment of the validity of our MALDI-TOF MS approach by screening recent clinical isolates, with a presumable Fusarium identity, against our in-house reference spectrum database, using 4 identification spectra, and comparison of the MALDI-TOF MS identification with the one obtained by DNA sequence analysis, using ITS or, when ITS was not sufficiently discriminatory, TEF1α
| Reference no. | Source of isolation of the strain (substrate), patient pathology, hospital | Identification by ITS | ITS GenBank accession no. | Identification by TEF1α | TEF1α GenBank accession no. | Identification spectrum 1 |
Identification spectrum 2 |
Identification spectrum 3 |
Identification spectrum 4 |
MALDI-TOF MS identification with cutoff ofa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Top score | Associated IHEM strain in database | Identified as: | Top score | Associated IHEM strain in database | Identified as: | Top score | Associated IHEM strain in database | Identified as: | Top score | Associated IHEM strain in database | Identified as: | >2.0 | >1.8 | ||||||
| 12/0930 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. petroliphilum | KJ173886 | 1.288 | IHEM 2813 | F. petroliphilum | 1.649 | IHEM 18411 | F. petroliphilum | 1.704 | IHEM 18411 | F. petroliphilum | 1.844 | IHEM 18411 | F. petroliphilum | Criteria not fulfilled | F. petroliphilum | ||
| 12/1188 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173887 | 1.768 | IHEM 15652 | F. oxysporum | 1.940 | IHEM 15652 | F. oxysporum | 1.817 | IHEM 19994 | F. oxysporum | 1.839 | IHEM 23185 | F. oxysporum | Criteria not fulfilled | F. oxysporum | ||
| 12/1342 | Human nail, contaminant ?, Universitair Ziekenhuis Brussel | F. equiseti | KJ173888 | F. equiseti | KJ173883 | 1.330 | IHEM 3571 | F. equiseti | 1.384 | IHEM 20883 | F. incarnatum | 1.344 | IHEM 15929 | F. sporotrichioides | 1.332 | IHEM 20883 | F. incarnatum | Criteria not fulfilled | Criteria not fulfilled |
| 12/1573 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173889 | 1.929 | IHEM 22005 | F. oxysporum | 1.643 | IHEM 15895 | F. oxysporum | 1.658 | IHEM 22005 | F. oxysporum | 1.709 | IHEM 22005 | F. oxysporum | Criteria not fulfilled | F. oxysporum | ||
| 13/0077 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173890 | 1.682 | IHEM 17811 | F. oxysporum | 1.701 | IHEM 18037 | F. oxysporum | 1.803 | IHEM 19992 | F. oxysporum | 1.747 | IHEM 19992 | F. oxysporum | Criteria not fulfilled | F. oxysporum | ||
| 13/0080 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173891 | 1.891 | IHEM 21643 | F. oxysporum | 1.971 | IHEM 21643 | F. oxysporum | 1.948 | IHEM 21643 | F. oxysporum | 1.844 | IHEM 21643 | F. oxysporum | Criteria not fulfilled | F. oxysporum | ||
| 13/0091 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173892 | 1.949 | IHEM 18448 | F. oxysporum | 1.930 | IHEM 25352 | F. oxysporum | 2.080 | IHEM 18448 | F. oxysporum | 2.047 | IHEM 18448 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/0093 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173893 | 1.872 | IHEM 25352 | F. oxysporum | 1.984 | IHEM 25352 | F. oxysporum | 2.081 | IHEM 19994 | F. oxysporum | 2.079 | IHEM 19994 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/0158 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. petroliphilum | KJ173894 | 2.104 | IHEM 22467 | F. petroliphilum | 1.586 | IHEM 18410 | F. petroliphilum | 1.959 | IHEM 18410 | F. petroliphilum | 1.594 | IHEM 18528 | F. solani | F. petroliphilum | F. petroliphilum | ||
| 13/0585 | Human tracheal secretions, cardiac arrest and prolonged resuscitation at intensive care unit, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173895 | 2.181 | IHEM 15652 | F. oxysporum | 2.102 | IHEM 19994 | F. oxysporum | 1.936 | IHEM 25352 | F. oxysporum | 2.246 | IHEM 15652 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/0638 | Human tracheal secretions, cardiac arrest and prolonged resuscitation at intensive care unit, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173896 | 2.099 | IHEM 18037 | F. oxysporum | 2.021 | IHEM 18037 | F. oxysporum | 2.091 | IHEM 18037 | F. oxysporum | 2.042 | IHEM 18037 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/0882 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173897 | 2.001 | IHEM 18037 | F. oxysporum | 2.093 | IHEM 18037 | F. oxysporum | 2.041 | IHEM 18037 | F. oxysporum | 2.084 | IHEM 18037 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/0889 | Human tracheal secretions, cardiac arrest, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173898 | 2.042 | IHEM 18037 | F. oxysporum | 2.077 | IHEM 18037 | F. oxysporum | 2.022 | IHEM 18037 | F. oxysporum | 2.091 | IHEM 18037 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/1134 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173899 | 2.026 | IHEM 15652 | F. oxysporum | 1.793 | IHEM 15652 | F. oxysporum | 1.918 | IHEM 15652 | F. oxysporum | 2.070 | IHEM 15652 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/1149 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. oxysporum | KJ173900 | 2.450 | IHEM 23153 | F. oxysporum | 2.381 | IHEM 13316 | F. oxysporum | 2.396 | IHEM 13316 | F. oxysporum | 2.378 | IHEM 13316 | F. oxysporum | F. oxysporum | F. oxysporum | ||
| 13/1159 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. proliferatum | KJ173901 | F. proliferatum | KJ173884 | 1.688 | IHEM 25354 | F. proliferatum | 1.905 | IHEM 25671 | F. proliferatum | 1.708 | IHEM 9534 | F. verticillioides | 1.790 | IHEM 25671 | F. proliferatum | Criteria not fulfilled | F. proliferatum |
| 13/1161 | Human nail, onychomycosis, Universitair Ziekenhuis Brussel | F. petroliphilum | KJ173902 | 2.353 | IHEM 18411 | F. petroliphilum | 2.312 | IHEM 18411 | F. petroliphilum | 2.123 | IHEM 18411 | F. petroliphilum | 2.311 | IHEM 18411 | F. petroliphilum | F. petroliphilum | F. petroliphilum | ||
| 12/0547 | Human toe, lymphoblastic lymphoma, Universitair Medisch Centrum Sint-Pieter | F. solani | KJ173903 | 2.387 | IHEM 19488 | F. solani | 2.270 | IHEM 19488 | F. solani | 2.316 | IHEM 19488 | F. solani | 2.263 | IHEM 19488 | F. solani | F. solani | F. solani | ||
| 13/0176 | Human brain biopsy specimen, transplant patient, Universitair Medisch Centrum Sint-Pieter | F. oxysporum | KJ173904 | 1.885 | IHEM 17811 | F. oxysporum | 1.861 | IHEM 17890 | F. oxysporum | 1.547 | IHEM 23189 | F. oxysporum | 1.955 | IHEM 9896 | F. oxysporum | Criteria not fulfilled | F. oxysporum | ||
| 13/0678 | Human blood, acute leukemia and chemotherapy, Universitair Medisch Centrum Sint-Pieter | F. verticillioides | KJ173905 | F. musae | KJ173885 | 2.080 | IHEM 18495 | F. musae | 2.137 | IHEM 19881 | F. musae | 2.170 | IHEM 18495 | F. musae | 2.104 | IHEM 19667 | F. musae | F. musae | F. musae |
MALDI-TOF MS identification of a strain was considered interpretable when two criteria were fulfilled: (i) at least three out of four identification spectra are identified by the reference spectrum of the same species, and (ii) at least one of their top scores meets the cutoff listed here (i.e., >2.0 or >1.8).
RESULTS
Taxon sampling, multilocus sequencing, and morphological characterization.
While performing multilocus sequencing, we encountered difficulties or inconsistencies with 61 of the 323 Fusarium strains (see Table S1 in the supplemental material). For 4 strains, no viable or pure culture was obtained. Sequencing failed for one of the DNA markers in 4 strains, and 5 strains were reclassified in a Fusarium-like genus that previously belonged to Fusarium sensu stricto. Eight other strains were identified as a species not belonging to Fusarium or a Fusarium-like genus (mostly Acremonium). The remaining 40 strains showed a different Fusarium identity than the one under which it was preserved in the collection. Twenty-seven of these 40 strains were retained in the collection and this study, with a corrected identity, since the species concerned had not yet been defined upon the time of deposit of the strain or a misidentification on the basis of morphology seemed likely. For the other 13 strains, a misidentification seemed unlikely, and they were rejected from this study together with the abovementioned 21 unviable, impure, incompletely sequenced, or reclassified strains. A total of 289 strains were thus available for further analysis. These 289 validated strains contained 40 species of Fusarium sensu stricto divided across 9 species complexes: the FSSC, FOSC, FFSC, FDSC, FIESC, FCSC, F. sambucinum species complex (FSAMSC), F. tricinctum species complex (FTSC), and the F. lateritium species complex (FLSC). There were 21 one-strain species, of which five remained as Fusarium due to phylogenetic and morphological considerations (i.e., insufficient sequence similarity and morphological similarity with known formally described Fusarium species). Nineteen species were represented by more than one strain (i.e., a total of 268 strains). In summary, 8.2% of these strains were initially misidentified by classical morphological examination and 35.1% were classified as Fusarium sp. only. The other 56.7% of the strains had been correctly identified to the species level.
Phylogenetic analysis.
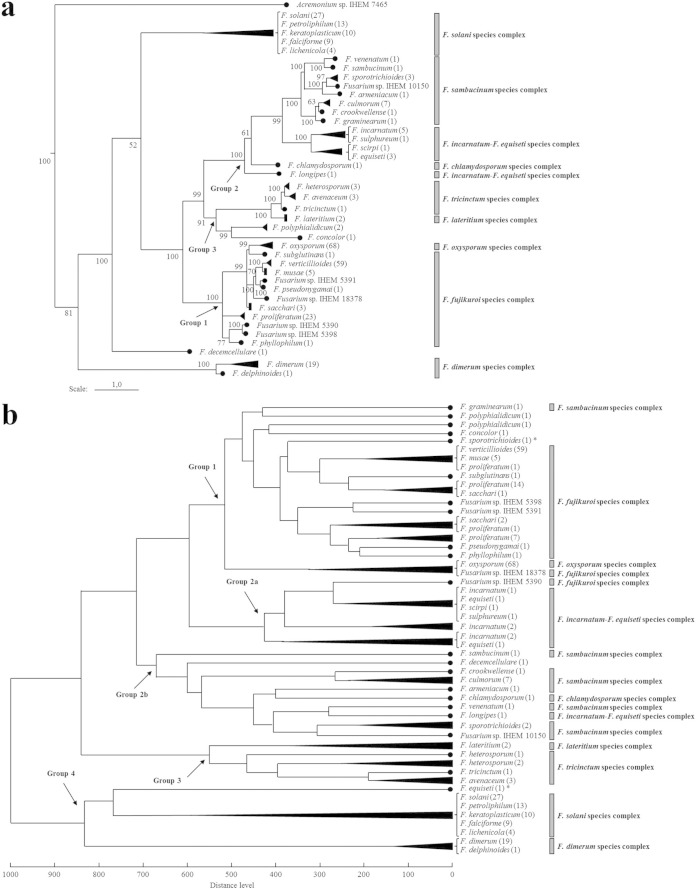
A representation of the obtained BI consensus tree, showing the evolutionary relationships between the species clades in our Fusarium data set, is given in Fig. 1a. The effective sample sizes were >100 for most parameters of the BI analysis, and the consensus tree was inferred from the 3,002 remaining trees after burn-in.
FIG 1.
(a) Representation of our BI consensus tree of combined sequences (ITS, BT, TEF1α, and LSU). The different species clades are shown. Our Fusarium data set consisted of 289 validated strains, containing 40 species (the number of strains per species is indicated in parentheses) divided across 9 species complexes. Species with an unresolved phylogenetic relationship are combined with a bracket. Posterior probabilities (in percentages) are indicated at the nodes of the tree. An Acremonium sp. was applied to root the tree. (b) Representation of the distance matrix dendrogram of our Fusarium reference spectra. The different species clades are shown. Most strains of a same species fall into a single clade or closely related clades. The topology of this dendrogram looks similar to that of the BI consensus tree, as the major species groups in phylogeny (indicated by an arrow and with group 4 comprising the species at the basis of the tree) were also found to be distinguishable and with few inconsistencies in the dendrogram. Most members of a same species complex also clustered together in the dendrogram. *, misidentified strain.
In vitro antifungal susceptibility testing.
Of the 289 validated Fusarium strains, 180 had been isolated from patients (blood samples or biopsy specimens from infected sites). This included 15 species and 4 species complexes. An antifungal susceptibility profile was constructed for each species represented (Table 2). We found that for the azoles, except for voriconazole and ketoconazole, as well as for 5-fluorocytosine, all strains had MIC100s higher than the maximum concentration tested (i.e., 16 μg/ml or 64 μg/ml). For ketoconazole, most strains also displayed a high MIC100. In the FSSC and FOSC as well as for Fusarium verticillioides and F. dimerum, voriconazole had no or only a limited effect compared to the effects of other antifungals. Amphotericin B was the only drug active against all Fusarium spp., except for some strains, especially F. verticillioides, which showed a high MIC100. For these F. verticillioides strains, terbinafine generally displayed the lowest MIC100. This was also the case in the FDSC and with Fusarium sacchari and the 3 species for which only one strain was analyzed. In the FSSC and FOSC, terbinafine had little or no activity. For Fusarium proliferatum and Fusarium musae, similar MIC100s were observed with terbinafine, amphotericin B, and voriconazole.
TABLE 2.
In vitro antifungal susceptibility profiles (distribution of the MIC100 per antifungal tested) of the Fusarium spp. represented in the 180 patient-isolated strains
| Antifungal agenta | Fusarium species (no. of isolates) | Species complex | No. of strains with MIC100 (μg/ml) ofb: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | >16 | |||
| Amphotericin B | F. solani (21) | F. solani | 3 | 3 | 10 | 4 | 1 | |||
| F. falciforme (6) | F. solani | 2 | 3 | 1 | ||||||
| F. petroliphilum (10) | F. solani | 5 | 5 | |||||||
| F. keratoplasticum (10) | F. solani | 1 | 5 | 3 | 1 | |||||
| F. lichenicola (3) | F. solani | 1 | 2 | |||||||
| F. oxysporum (47) | F. oxysporum | 1 | 25 | 19 | 1 | 1 | ||||
| F. verticillioides (39) | F. fujikuroi | 1 | 3 | 20 | 14 | 1 | ||||
| F. proliferatum (16) | F. fujikuroi | 8 | 7 | 1 | ||||||
| F. musae (4) | F. fujikuroi | 1 | 3 | |||||||
| F. sacchari (3) | F. fujikuroi | 2 | 1 | |||||||
| F. pseudonygamai (1) | F. fujikuroi | 1 | ||||||||
| F. dimerum (17) | F. dimerum | 3 | 7 | 7 | ||||||
| F. delphinoides (1) | F. dimerum | 1 | ||||||||
| F. concolor (1) | 1 | |||||||||
| F. decemcellulare (1) | 1 | |||||||||
| Voriconazole | F. solani (21) | F. solani | 1 | 1 | 5 | 14 | ||||
| F. falciforme (6) | F. solani | 2 | 2 | 2 | ||||||
| F. petroliphilum (10) | F. solani | 3 | 7 | |||||||
| F. keratoplasticum (10) | F. solani | 1 | 4 | 5 | ||||||
| F. lichenicola (3) | F. solani | 1 | 1 | 1 | ||||||
| F. oxysporum (47) | F. oxysporum | 8 | 20 | 10 | 9 | |||||
| F. verticillioides (39) | F. fujikuroi | 6 | 3 | 2 | 1 | 27 | ||||
| F. proliferatum (16) | F. fujikuroi | 1 | 4 | 6 | 4 | 1 | ||||
| F. musae (4) | F. fujikuroi | 2 | 2 | |||||||
| F. sacchari (3) | F. fujikuroi | 2 | 1 | |||||||
| F. pseudonygamai (1) | F. fujikuroi | 1 | ||||||||
| F. dimerum (17) | F. dimerum | 2 | 10 | 5 | ||||||
| F. delphinoides (1) | F. dimerum | 1 | ||||||||
| F. concolor (1) | 1 | |||||||||
| F. decemcellulare (1) | 1 | |||||||||
| Terbinafine | F. solani (21) | F. solani | 21 | |||||||
| F. falciforme (6) | F. solani | 6 | ||||||||
| F. petroliphilum (10) | F. solani | 10 | ||||||||
| F. keratoplasticum (10) | F. solani | 10 | ||||||||
| F. lichenicola (3) | F. solani | 3 | ||||||||
| F. oxysporum (47) | F. oxysporum | 6 | 4 | 4 | 33 | |||||
| F. verticillioides (39) | F. fujikuroi | 2 | 3 | 13 | 12 | 2 | 3 | 4 | ||
| F. proliferatum (16) | F. fujikuroi | 5 | 5 | 5 | 1 | |||||
| F. musae (4) | F. fujikuroi | 1 | 2 | 1 | ||||||
| F. sacchari (3) | F. fujikuroi | 2 | 1 | |||||||
| F. pseudonygamai (1) | F. fujikuroi | 1 | ||||||||
| F. dimerum (17) | F. dimerum | 3 | 12 | 2 | ||||||
| F. delphinoides (1) | F. dimerum | 1 | ||||||||
| F. concolor (1) | 1 | |||||||||
| F. decemcellulare (1) | 1 | |||||||||
| Ketoconazole | F. solani (21) | F. solani | 1 | 20 | ||||||
| F. falciforme (6) | F. solani | 6 | ||||||||
| F. petroliphilum (10) | F. solani | 10 | ||||||||
| F. keratoplasticum (10) | F. solani | 10 | ||||||||
| F. lichenicola (3) | F. solani | 3 | ||||||||
| F. oxysporum (47) | F. oxysporum | 47 | ||||||||
| F. verticillioides (39) | F. fujikuroi | 3 | 3 | 4 | 29 | |||||
| F. proliferatum (16) | F. fujikuroi | 2 | 1 | 13 | ||||||
| F. musae (4) | F. fujikuroi | 3 | 1 | |||||||
| F. sacchari (3) | F. fujikuroi | 3 | ||||||||
| F. pseudonygamai (1) | F. fujikuroi | 1 | ||||||||
| F. dimerum (17) | F. dimerum | 17 | ||||||||
| F. delphinoides (1) | F. dimerum | 1 | ||||||||
| F. concolor (1) | 1 | |||||||||
| F. decemcellulare (1) | 1 | |||||||||
For itraconazole, posaconazole, fluconazole, and 5-fluorocytosine, all strains had a MIC100 higher than the maximum concentration tested.
MIC100s of <0.25 μg/ml were not observed.
MALDI-TOF MS assay, construction of reference spectrum database, and identification strategy.
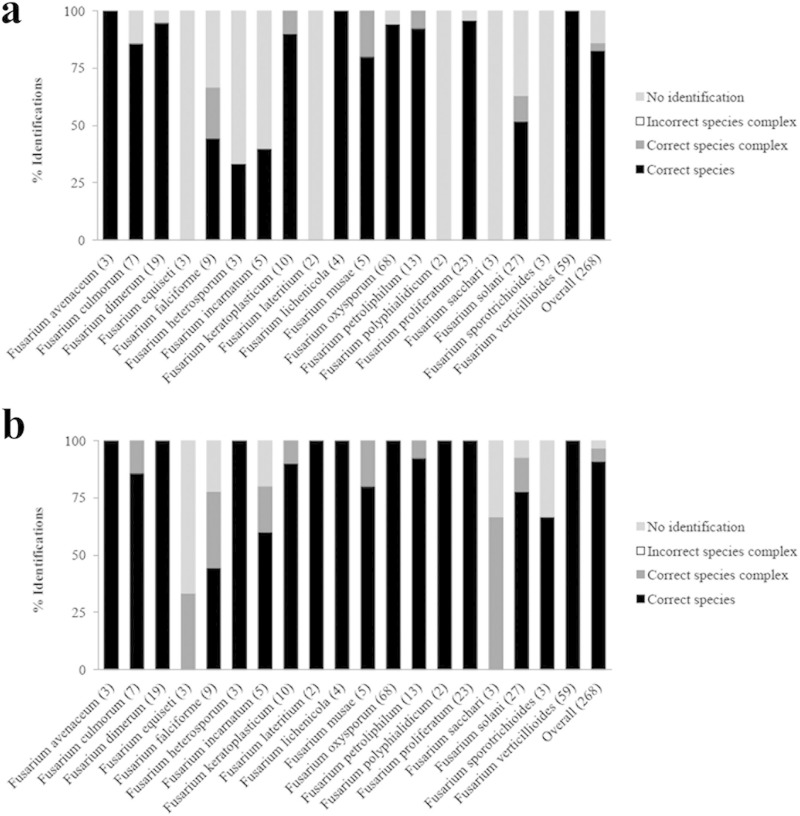
MALDI-TOF MS reference spectra were created for the 289 validated Fusarium strains and implemented in our in-house database. Each strain was subsequently challenged against this database according to well-defined criteria, and the individual identification results are shown in Table S1 in the supplemental material. It should be noted that the identification spectra of each strain matched their own reference spectrum with the highest logarithmic scores observed, always well above the standard 2.0 cutoff value. For the 19 species represented by more than one strain, the percentages of strains with a correct species identification, correct species complex identification, and incorrect species complex identification, as well as the percentage of unidentified strains were calculated (Fig. 2). Using 2.0 as a cutoff, we found that 82.8% of the MALDI-TOF MS-based identifications were correct to the species level, whereas 3% of the strains were incorrectly identified, although always within the correct species complex. The incorrectly identified strains include 8 strains (see Table S2 in the supplemental material), with seven of them belonging to the FSSC and the other to the FFSC. For 14.2% of the strains (i.e., 38 strains), the criteria for identification were not fulfilled. Lowering the cutoff value to 1.4 would allow 22 of these strains to be identified correctly to the species level and 7 to the species complex level (see Table S2), resulting in an overall success rate of 91% correct species identifications and 5.6% correct species complex identifications. When we also consider the FSSC and the FIESC as a whole, an additional 5 strains would have been identified correctly, leading to an overall success rate of 97% correct identifications. The inability of MALDI-TOF MS to achieve identification would then concern 4 strains: one F. incarnatum strain, one F. equiseti strain, one Fusarium sporotrichioides strain, and one F. sacchari strain. No incorrect species complex or non-Fusarium spp. identifications occurred by MALDI-TOF MS. Thirteen of the 21 species represented by only one strain in the data set were identified as a phylogenetically closely related species with our MALDI-TOF MS approach when using 1.4 as the cutoff (see Table S2). The other one-strain species remained unidentified or were misidentified.
FIG 2.
Summary of the identification results with the MALDI-TOF MS assay. The validated Fusarium strains were screened against our in-house reference spectrum database, according to well-defined identification criteria using a cutoff of either 2.0 (a) or 1.4 (b). The MALDI-TOF MS identifications were compared with those obtained by the multilocus sequence analysis. For the 19 species represented by more than one strain in the data set, the percentages of strains with a correct species identification, correct species complex identification, and incorrect species complex identification, as well as the percentage of unidentified strains were assessed. The number of strains per species is indicated in parentheses.
Comparison.
A distance matrix dendrogram of the 289 Fusarium reference spectra was constructed, showing the relationships between the different strains and species (Fig. 1b). We observed that with exception of a F. equiseti (IHEM 3571) and a F. sporotrichioides (IHEM 3235) strain, all strains of a same species clustered together into a single clade or closely related clades. The two aberrant strains are also the ones that were not identified by MALDI-TOF MS. The topology of the dendrogram appeared to be similar to that of the BI consensus tree (Fig. 1a). To emphasize this, we defined 4 groups of species forming a well-supported lineage in the phylogeny (group 4, comprising the species at the basis of the tree). These groups of species were also distinguished, with few inconsistencies, in the dendrogram. Differences with the BI consensus tree were noticeable for the species Fusarium graminearum (in group 1 instead of group 2), Fusarium polyphialidicum (in group 1 instead of group 3), Fusarium concolor (in group 1 instead of group 3), Fusarium sp. strain IHEM 5390 (in group 2a instead of group 1), Fusarium decemcellulare (in group 2b instead of group 4), and the aberrant strains IHEM 3235 and IHEM 3571 (in groups 1 and 4, respectively, instead of group 2). In addition, we found that most members of the same species complex also clustered together in the dendrogram, with the exception of Fusarium longipes, F. graminearum, Fusarium sp. strain IHEM 5390, Fusarium sp. strain IHEM 18378, and the two aberrant strains.
Proof of principle.
A screening of the 20 recent clinical isolates against our in-house database revealed a Fusarium identity for each of them (Table 1). Nineteen isolates received a species identity by MALDI-TOF MS, though only when a cutoff of 1.8 instead of the standard 2.0 was used for accepting identification; otherwise, eight isolates could not be identified. For isolate 12/1342, 3 of the 4 identification spectra corresponded to a member of the FIESC (i.e., F. incarnatum or F. equiseti) but with top scores of <1.4 (Table 1). The MALDI-TOF MS identifications were all confirmed by the identifications made with DNA sequencing. In three cases, TEF1α sequencing was deemed necessary for reliable DNA sequence-based identification. For isolate 13/0678, this resulted in an identification of F. musae instead of F. verticillioides using ITS only, which was concordant with the obtained MALDI-TOF MS identity. A total of 6 distinct Fusarium spp. were identified: F. petroliphilum (3 strains), F. oxysporum (13 strains), F. proliferatum (1 strain), F. solani (1 strain), F. musae (1 strain), and a member of the FIESC (i.e., F. equiseti).
DISCUSSION
The present study demonstrates the validity of the standardized MALDI-TOF MS assay for the identification of molds developed by Cassagne et al. (33), to identify Fusarium isolates in the clinical setting. No incorrect species complex or non-Fusarium identifications were made by MALDI-TOF MS and our in-house reference spectrum database. However, identification of the correct species complex member was not always guaranteed, and some strains remained unidentified. The problem of unidentified strains can be resolved without increasing the number of misidentifications by lowering the cutoff value for accepting identification, the feasibility of which has been demonstrated in previous studies (42). Moreover, taking into account the FSSC and the FIESC as a whole and not discriminating between its members would increase the percentage of correct identifications even more. This leaves only 4 unidentified strains of species rarely or not yet encountered as human pathogen (1).
In vitro antifungal susceptibility testing showed that discriminating between Fusarium species complexes can indeed be important for appropriate patient treatment, as previously indicated (27). Although amphotericin B was active against all strains analyzed and was the drug of choice for all members of the FSSC and FOSC (i.e., the other antifungals tested had no or only a limited effect), our results suggest alternatives for members of the FFSC and FDSC. For F. verticillioides, F. sacchari, F. dimerum, and Fusarium delphinoides, terbinafine generally showed the lowest MIC100, and for F. sacchari and F. delphinoides, voriconazole might also be a good alternative. Similar MIC100s for amphotericin B, terbinafine, and voriconazole were observed with F. proliferatum and F. musae strains. Discriminating between members of the same species complex thus appeared not to be that clinically relevant, with the exception of the FFSC.
Since our MALDI-TOF MS approach did not make any incorrect species complex identifications, and most incorrect species identifications occurred within the FSSC, the most resistant species complex for which only amphotericin B seems effective, these misidentifications are likely to have no clinical implications (i.e., no repercussion on the treatment of the patient). Moreover, in the FSSC, many new species formerly identified as F. solani sensu stricto are defined or may be defined in the near future based upon the phylogenetic species concept (13). The species Fusarium keratoplasticum and F. petroliphilum are examples of this. They were described by Short et al. (13) as phylogenetically distinct species, despite the fact that they cannot be discriminated from each other or from other FSSC members on the basis of morphology, niche, or clinical appearance. It can be questioned whether identification to the species level is clinically relevant in this species complex, though from an epidemiological point of view, it is always recommended. When we consider the FSSC as a whole and no distinction is made between its members, a success ratio of 100% correct identifications would be obtained with MALDI-TOF MS for this species complex, comparable to that of F. oxysporum.
In the FFSC, for which correct species identification seems important due to differences in antifungal susceptibility profiles, the applied MALDI-TOF MS assay works well. Only for the F. sacchari strains and one F. musae strain we were unable to obtain a correct species identification by MALDI-TOF MS. Yet, the clinical significance of these species is rather limited, and they are recognized as a phylogenetically closely related species (i.e., F. proliferatum and F. verticillioides, respectively) (1, 62). Other incorrect species identifications occurred with members of species complexes that are of less clinical importance (i.e., the FIESC) or that have not been found to be associated with human pathogenesis (i.e., the FSAMSC).
With respect to the complexity of the Fusarium genus and its often closely related species, our identification results are comparable to those obtained in other studies using the same standardized MALDI-TOF MS procedure. Cassagne et al. (33), L'Ollivier et al. (40), Gautier et al. (43), and Ranque et al. (44) correctly identified 87%, 97.8%, 98.8%, and 89% of their clinical mold isolates, respectively. Although the assay was optimized on a large panel of clinically relevant molds and several genera have been analyzed, its feasibility for identification within the Fusarium genus was not yet established in detail. To our knowledge, Marinach-Patrice et al. (48) was the only study to focus on the capacity of MALDI-TOF MS to identify Fusarium isolates. However, the reference spectrum database used in this study contained only 5 Fusarium spp., represented by a few strains, and species other than Fusarium were not included. In contrast, our applied in-house database contained 40 different Fusarium spp., of which 19 had more than one strain, as well as various other species from 152 different fungal genera. Moreover, it is well known that the currently available Bruker database contains only a limited number of mold reference spectra, insufficiently capturing the diversity in clinically relevant species. Using the Bruker database, identification within Fusarium seems extremely problematic, since only one species (i.e., F. proliferatum) is represented.
The robustness of our reference spectrum database and principle of our MALDI-TOF MS approach were proven by successfully screening recent clinical Fusarium isolates. All isolates except one were correctly identified to the species level by MALDI-TOF MS. For one isolate, MALDI-TOF MS even outperformed ITS sequencing. Another isolate appeared to be F. equiseti, but MALDI-TOF MS identified it only as a member of the FIESC, with low identification scores. The identification of FIESC members seems difficult with the applied MALDI-TOF MS approach. Also, when screening our collection material, only 37.5% of the FIESC strains were identified correctly to the species level, even when the cutoff value for accepting identification was lowered. When the FIESC was taken as a whole and no distinction was made between its members, the success rate was increased to 75%. Nevertheless, the identification scores were often low, especially with F. equiseti strains, indicating that the extraction procedure is not that stable for members of this species complex.
Accurate identification by MALDI-TOF MS is largely limited by the species diversity in the reference spectrum database. This is emphasized by the one-strain species in our data set, which could not be correctly identified according to the identification strategy. These species were generally identified as phylogenetically closely related species. This observation triggered a comparison of the available phylogenetic and MALDI-TOF MS data. We saw an interesting similarity in the topology of a BI phylogenetic tree and that of a dendrogram outlining the relationships between the Fusarium reference spectra. Such a correlation was also demonstrated in the study of Packeu et al. (49) with molds of the Trichophyton mentagrophytes species complex and indicates that phylogenetically closely related species also have similar protein spectra.
In the routine clinical laboratory, the advantages of MALDI-TOF MS for the identification of Fusarium isolates and molds in general are that it is easy to perform, requires no expert mycologists, and is faster than the conventional identification methods. Furthermore, it allows an objective identification not prone to interpretation by the examiner. With the MALDI-TOF MS assay of Cassagne et al. (33), an identification was generated within 1 h after the 3 days of culture, whereas with DNA sequence analysis, ≥2 additional days are needed after culturing. Moreover, after 3 days of culture, certain characteristics needed for morphological identification may not yet be developed. Reducing the time frame for mold identification is of major clinical interest, especially with invasive infections for which patient prognosis depends on a timely onset of appropriate treatment. Even more important is that MALDI-TOF MS outperforms the conventional identification of molds. Ranque et al. (44) found a 31% to 61% increase in correct identifications with MALDI-TOF MS compared to the conventional method for their non-Aspergillus isolates. After the implementation of MALDI-TOF MS in a clinical routine, Gautier et al. (43) observed a dramatic increase in mold identifications at the species level and a decrease in the rate of misidentifications. In our study, the morphological examination of the Fusarium strains upon the time of deposit in the collection showed a correct identification in only 56.7% of the cases to the species level. This percentage was dramatically increased by using MALDI-TOF MS technology, rendering a success rate of 82.8% and even 97% when the cutoff value for accepting identification was lowered and some species complexes were taken as a whole.
In conclusion, our study highlights once again the usefulness of MALDI-TOF MS and in particular the user-friendly standardized procedure of Cassagne et al. (33) for mold identification in the clinical setting. In combination with the constructed in-house reference spectrum database, identification within the Fusarium genus was found to be highly accurate, taking into account the complexity of the genus. Indeed, the availability of an extended database of reference spectra is indispensable for routine use, and the database currently provided by the manufacturer might be too limited for mold identification. Therefore, we are currently working on creating an online portal that will allow MALDI-TOF MS identifications to be performed by querying our reference spectrum database.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Belgian Science Policy (Belspo) for funding, Annelies De Bel for sending us clinical isolates from the Universitair Ziekenhuis Brussel, and Delphine Martiny for sending us clinical isolates from the Universitair Medisch Centrum Sint-Pieter.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02213-14.
REFERENCES
- 1.Nucci M, Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin Microbiol Rev 20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miceli MH, Lee SA. 2011. Emerging moulds: epidemiological trends and antifungal resistance. Mycoses 54:666–678. doi: 10.1111/j.1439-0507.2011.02032.x. [DOI] [PubMed] [Google Scholar]
- 3.Geiser DM, Aoki T, Bacon CW, Baker SE, Bhattacharyya MK, Brandt ME, Brown DW, Burgess LW, Chulze S, Coleman JJ, Correll JC, Covert SF, Crous PW, Cuomo CA, De Hoog GS, Di Pietro A, Elmer WH, Epstein L, Frandsen RJ, Freeman S, Gagkaeva T, Glenn AE, Gordon TR, Gregory NF, Hammond-Kosack KE, Hanson LE, Jímenez-Gasco Mdel M, Kang S, Kistler HC, Kuldau GA, Leslie JF, Logrieco A, Lu GZ, Lysøe E, Ma LJ, McCormick SP, Migheli Q, Moretti A, Munaut F, O'Donnell K, Pfenning L, Ploetz RC, Proctor RH, Rehner SA, Robert VA, Rooney AP, Bin Salleh B, Scandiani MM, Scauflaire J, Short DP, et al. 2013. One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves the longstanding use. Phytopathology 103:400–408. doi: 10.1094/PHYTO-07-12-0150-LE. [DOI] [PubMed] [Google Scholar]
- 4.O'Donnell K, Cigelnik E, Nirenberg HI. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493. doi: 10.2307/3761407. [DOI] [Google Scholar]
- 5.O'Donnell K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919–938. doi: 10.2307/3761588. [DOI] [Google Scholar]
- 6.O'Donnell K, Nirenberg HI, Aoki T, Cigelnik E. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41:61–78. doi: 10.1007/BF02464387. [DOI] [Google Scholar]
- 7.Summerbell RC, Schroers HJ. 2002. Analysis of phylogenetic relationship of Cylindrocarpon lichenicola and Acremonium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic infections caused by these fungi. J Clin Microbiol 40:2866–2875. doi: 10.1128/JCM.40.8.2866-2875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell K, Sutton DA, Rinaldi MG, Magnon KC, Cox PA, Revankar SG, Sanche S, Geiser DM, Juba JH, van Burik JA, Padhye A, Anaissie E, Francesconi A, Walsh TJ, Robinson JS. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J Clin Microbiol 42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N, O'Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye A, Geiser DM. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol 44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol 46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroers HJ, O'Donnell K, Lamprecht SC, Kammeyer PL, Johnson S, Sutton DA, Rinaldi MG, Geiser DM, Summerbell RC. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101:44–70. doi: 10.3852/08-002. [DOI] [PubMed] [Google Scholar]
- 12.O'Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. 2009. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol 47:3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Short DP, O'Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM. 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet Biol 53:59–70. doi: 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Laurence MH, Summerell BA, Burgess LW, Liew EC. 2014. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biol 118:374–384. doi: 10.1016/j.funbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell K, Sarver BA, Brandt M, Chang DC, Noble-Wang J, Park BJ, Sutton DA, Benjamin L, Lindsley M, Padhye A, Geiser DM, Ward TJ. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J Clin Microbiol 45:2235–2248. doi: 10.1128/JCM.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azor M, Gené J, Cano J, Guarro J. 2007. Universal in vitro antifungal resistance of genetic clades of the Fusarium solani species complex. Antimicrob Agents Chemother 51:1500–1503. doi: 10.1128/AAC.01618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azor M, Gené J, Cano J, Sutton DA, Fothergill AW, Rinaldi MG, Guarro J. 2008. In vitro antifungal susceptibility and molecular characterization of clinical isolates of Fusarium verticillioides (F. moniliforme) and Fusarium thapsinum. Antimicrob Agents Chemother 52:2228–2231. doi: 10.1128/AAC.00176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alastruey-Izquierdo A, Cuenca-Estrella M, Monzón A, Mellado E, Rodríguez-Tudela JL. 2008. Antifungal susceptibility profile of clinical Fusarium spp. isolates identified by molecular methods. J Antimicrob Chemother 61:805–809. doi: 10.1093/jac/dkn022. [DOI] [PubMed] [Google Scholar]
- 19.Iqbal NJ, Boey A, Park BJ, Brandt ME. 2008. Determination of in vitro susceptibility of ocular Fusarium spp. isolates from keratitis cases and comparison of Clinical and Laboratory Standards Institute M38-A2 and E test methods. Diagn Microbiol Infect Dis 62:348–350. doi: 10.1016/j.diagmicrobio.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Tortorano AM, Prigitano A, Dho G, Esposto MC, Gianni C, Grancini A, Ossi C, Viviani MA. 2008. Species distribution and in vitro antifungal susceptibility patterns of 75 clinical isolates of Fusarium spp. from northern Italy. Antimicrob Agents Chemother 52:2683–2685. doi: 10.1128/AAC.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie L, Zhai H, Zhao J, Sun S, Shi W, Dong X. 2008. Antifungal susceptibility for common pathogens of fungal keratitis in Shandong Province, China. Am J Ophthalmol 146:260–265. doi: 10.1016/j.ajo.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Azor M, Gené J, Cano J, Manikandan P, Venkatapathy N, Guarro J. 2009. Less-frequent Fusarium species of clinical interest: correlation between morphological and molecular identification and antifungal susceptibility. J Clin Microbiol 47:1463–1468. doi: 10.1128/JCM.02467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azor M, Cano J, Gené J, Guarro J. 2009. High genetic diversity and poor in vitro response to antifungals of clinical strains of Fusarium oxysporum. J Antimicrob Chemother 63:1152–1155. doi: 10.1093/jac/dkp095. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Xiao M, Kong F, Chen S, Dou HT, Sorrell T, Li RY, Xu YC. 2011. Accurate and practical identification of 20 Fusarium species by seven-locus sequence analysis and reverse line blot hybridization, and an in vitro antifungal susceptibility study. J Clin Microbiol 49:1890–1898. doi: 10.1128/JCM.02415-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homa M, Shobana CS, Singh YR, Manikandan P, Selvam KP, Kredics L, Narendran V, Vágvölgyi C, Galgóczy L. 2013. Fusarium keratitis in South India: causative agents, their antifungal susceptibilities and a rapid identification method for the Fusarium solani species complex. Mycoses 56:501–511. doi: 10.1111/myc.12062. [DOI] [PubMed] [Google Scholar]
- 26.Oechsler RA, Feilmeier MR, Miller D, Shi W, Hofling-Lima AL, Alfonso EC. 2013. Fusarium keratitis: genotyping, in vitro susceptibility and clinical outcomes. Cornea 32:667–673. doi: 10.1097/ICO.0b013e318277ac74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortorano AM, Prigitano A, Esposto MC, Arsic Arsenijevic V, Kolarovic J, Ivanovic D, Paripovic L, Klingspor L, Nordøy I, Hamal P, Arikan Akdagli S, Ossi C, Grancini A, Cavanna C, Lo Cascio G, Scarparo C, Candoni A, Caira M, Drogari Apiranthitou M, ECMM Working Group . 2014. European Confederation of Medical Mycology (ECMM) epidemiological survey on invasive infections due to Fusarium species in Europe. Eur J Clin Microbiol Infect Dis 33:1623–1630. doi: 10.1007/s10096-014-2111-1. [DOI] [PubMed] [Google Scholar]
- 28.Marklein G, Josten M, Klanke U, Müller E, Horré R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol 47:2912–2917. doi: 10.1128/JCM.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 30.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 49:1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. 2012. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:1313–1325. doi: 10.1128/JCM.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt V, Jarosch A, März P, Sander C, Vacata V, Kalka-Moll W. 2012. Rapid identification of bacteria in positive blood culture by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis 31:311–317. doi: 10.1007/s10096-011-1312-0. [DOI] [PubMed] [Google Scholar]
- 33.Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6:e28425. doi: 10.1371/journal.pone.0028425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos C, Paterson RR, Venâncio A, Lima N. 2010. Filamentous fungal characterizations by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Appl Microbiol 108:375–385. doi: 10.1111/j.1365-2672.2009.04448.x. [DOI] [PubMed] [Google Scholar]
- 35.Alanio A, Beretti JL, Dauphin B, Mellado E, Quesne G, Lacroix C, Amara A, Berche P, Nassif X, Bougnoux ME. 2011. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin Microbiol Infect 17:750–755. doi: 10.1111/j.1469-0691.2010.03323.x. [DOI] [PubMed] [Google Scholar]
- 36.Theel ES, Hall L, Mandrekar J, Wengenack NL. 2011. Dermatophyte identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49:4067–4071. doi: 10.1128/JCM.01280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alshawa K, Beretti JL, Lacroix C, Feuilhade M, Dauphin B, Quesne G, Hassouni N, Nassif X, Bougnoux ME. 2012. Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 50:2277–2281. doi: 10.1128/JCM.06634-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Carolis E, Posteraro B, Lass-Flörl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. 2012. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect 18:475–484. doi: 10.1111/j.1469-0691.2011.03599.x. [DOI] [PubMed] [Google Scholar]
- 39.Chalupová J, Raus M, Sedlářová M, Sebela M. 2014. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol Adv 32:230–241. doi: 10.1016/j.biotechadv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 40.L'Ollivier C, Cassagne C, Normand AC, Bouchara JP, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. 2013. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol 51:713–720. doi: 10.3109/13693786.2013.781691. [DOI] [PubMed] [Google Scholar]
- 41.Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Gräser Y. 2013. MALDI-TOF mass spectrometry—a rapid method for the identification of dermatophyte species. Med Mycol 51:17–24. doi: 10.3109/13693786.2012.685186. [DOI] [PubMed] [Google Scholar]
- 42.Normand AC, Cassagne C, Ranque S, L'Ollivier C, Fourquet P, Roesems S, Hendrickx M, Piarroux R. 2013. Assessment of various parameters to improve MALDI-TOF MS reference spectra libraries constructed for the routine identification of filamentous fungi. BMC Microbiol 13:76. doi: 10.1186/1471-2180-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gautier M, Ranque S, Normand AC, Becker P, Packeu A, Cassagne C, L'Ollivier C, Hendrickx M, Piarroux R. 2014. MALDI-TOF mass spectrometry: revolutionising clinical laboratory diagnosis of mould infections. Clin Microbiol Infect doi: 10.1111/1469-0691.12750. [DOI] [PubMed] [Google Scholar]
- 44.Ranque S, Normand AC, Cassagne C, Murat JB, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M, Piarroux R. 2014. MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses 57:135–140. doi: 10.1111/myc.12115. [DOI] [PubMed] [Google Scholar]
- 45.Kemptner J, Marchetti-Deschmann M, Mach R, Druzhinina IS, Kubicek CP, Allmaier G. 2009. Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact Fusarium spores by MALDI linear time-of flight mass spectrometry. Rapid Commun Mass Spectrom 23:877–884. doi: 10.1002/rcm.3949. [DOI] [PubMed] [Google Scholar]
- 46.Dong HJ, Kemptner J, Marchetti-Deschmann M, Kubicek CP, Allmaier G. 2009. Development of a MALDI two-layer volume sample preparation technique for analysis of colored conidia spores of Fusarium by MALDI linear TOF mass spectrometry. Anal Bioanal Chem 395:1373–1383. doi: 10.1007/s00216-009-3067-3. [DOI] [PubMed] [Google Scholar]
- 47.Seyfarth F, Ziemer M, Sayer HG, Burmester A, Erhard M, Welker M, Schliemann S, Straube E, Hipler UC. 2008. The use of ITS DNA sequence analysis and MALDI-TOF mass spectrometry in diagnosing an infection with Fusarium proliferatum. Exp Dermatol 17:965–971. doi: 10.1111/j.1600-0625.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 48.Marinach-Patrice C, Lethuillier A, Marly A, Brossas JY, Gené J, Symoens F, Datry A, Guarro J, Mazier D, Hennequin C. 2009. Use of mass spectrometry to identify clinical Fusarium isolates. Clin Microbiol Infect 15:634–642. doi: 10.1111/j.1469-0691.2009.02758.x. [DOI] [PubMed] [Google Scholar]
- 49.Packeu A, Hendrickx M, Beguin H, Martiny D, Vandenberg O, Detandt M. 2013. Identification of the Trichophyton mentagrophytes complex species using MALDI-TOF mass spectrometry. Med Mycol 51:580–585. doi: 10.3109/13693786.2013.770605. [DOI] [PubMed] [Google Scholar]
- 50.O'Donnell K, Humber RA, Geiser DM, Kang S, Park B, Robert VA, Crous PW, Johnston PR, Aoki T, Rooney AP, Rehner SA. 2012. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 104:427–445. doi: 10.3852/11-179. [DOI] [PubMed] [Google Scholar]
- 51.Gräfenhan T, Schroers HJ, Nirenberg HI, Seifert KA. 2011. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol 68:79–113. doi: 10.3114/sim.2011.68.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beguin H, Pyck N, Hendrickx M, Planard C, Stubbe D, Detandt M. 2012. The taxonomic status of Trichophyton quinckeanum and T. interdigitale revisited: a multigene phylogenetic approach. Med Mycol 50:871–882. doi: 10.3109/13693786.2012.684153. [DOI] [PubMed] [Google Scholar]
- 53.White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 54.Hopple JS Jr, Vilgalys R. 1999. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol Phylogenet Evol 13:1–19. doi: 10.1006/mpev.1999.0634. [DOI] [PubMed] [Google Scholar]
- 55.Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in ascomycetes. Mycologia 91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 57.Leslie JF, Summerell BA, Bullock S. 2006. The Fusarium laboratory manual. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 58.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A, Suchard M, Drummond AJ. 2007. Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer/.
- 61.Rodriguez-Tudela JL, Arendrup MC, Arikan S, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Denning DW, Donnelly JP, Fegeler W, Lass-Flörl C, Moore C, Richardson M, Gaustad P, Schmalreck A, Velegraki A, Verweij P. 2008. EUCAST DEFINITIVE DOCUMENT E.DEF 9.1: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST), London, United Kingdom. [Google Scholar]
- 62.Van Hove F, Waalwijk C, Logrieco A, Munaut F, Moretti A. 2011. Gibberella musae (Fusarium musae) sp. nov., a recently discovered species from banana is sister to F. verticillioides. Mycologia 103:570–585. doi: 10.3852/10-038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.