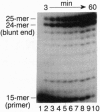
Abstract
Human immunodeficiency virus type 1 (HIV-1) is genetically highly variable. This is attributed to the error-prone nature of HIV-1 replication and its proclivity for recombination. During replication and recombination, reverse transcriptase (RT) must polymerize DNA to the 5' ends of multiple RNA and DNA template termini while converting HIV-1 RNA to double-stranded DNA. We have determined the fidelity of HIV-1 RT in vitro during polymerization to the 5' ends of HIV-1 long terminal repeat DNA template sequences and to the end of a partial HIV-1 genomic RNA template that mimics a recombination intermediate. HIV-1 RT readily extended recessed DNA primers to form full-length blunt-end DNA-DNA and DNA-RNA duplexes. In addition, HIV-1 RT catalyzed high yields of products with one to four extra nucleotides at the 3' ends of the nascent DNAs. These products were formed processively via a nontemplated mechanism that is highly specific for the addition of purine nucleotides (A > G >> T > or = C). Thus, HIV-1 RT is extremely unfaithful at both DNA and RNA template ends, introducing errors (extra nucleotides) in one out of every two or three nascent strands processively polymerized. This error rate is 1000 times higher than for HIV-1 RT-catalyzed errors at internal template positions. Blunt-end additions were also catalyzed by other retroviral RTs at relative rates of HIV-1 approximately Moloney murine leukemia virus > avian myeloblastosis virus. These data suggest a potentially important mechanism for retroviral mutation mediated by nontemplated blunt-end addition of purines prior to forced copy-choice recombination.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bebenek K., Abbotts J., Roberts J. D., Wilson S. H., Kunkel T. A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989 Oct 5;264(28):16948–16956. [PubMed] [Google Scholar]
- Clark J. M. DNA synthesis on discontinuous templates by DNA polymerase I of Escherichia coli. Gene. 1991 Jul 31;104(1):75–80. doi: 10.1016/0378-1119(91)90467-p. [DOI] [PubMed] [Google Scholar]
- Clark J. M., Joyce C. M., Beardsley G. P. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol. 1987 Nov 5;198(1):123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- Clark J. M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988 Oct 25;16(20):9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- DeStefano J. J., Mallaber L. M., Rodriguez-Rodriguez L., Fay P. J., Bambara R. A. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J Virol. 1992 Nov;66(11):6370–6378. doi: 10.1128/jvi.66.11.6370-6378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A. S. The human immunodeficiency virus: infectivity and mechanisms of pathogenesis. Science. 1988 Feb 5;239(4840):617–622. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. The origin of extrachromosomal circular copia elements. Cell. 1983 Sep;34(2):415–419. doi: 10.1016/0092-8674(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Klarmann G. J., Schauber C. A., Preston B. D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993 May 5;268(13):9793–9802. [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman L. V., Petruska J., Goodman M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990 Feb 5;265(4):2338–2346. [PubMed] [Google Scholar]
- Olsen J. C., Swanstrom R. A new pathway in the generation of defective retrovirus DNA. J Virol. 1985 Dec;56(3):779–789. doi: 10.1128/jvi.56.3.779-789.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauza C. D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990 Dec;179(2):886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Preston B. D., Sandell L. L., Loeb L. A. Extension of mismatched 3' termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. D., Poiesz B. J., Loeb L. A. Fidelity of HIV-1 reverse transcriptase. Science. 1988 Nov 25;242(4882):1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Ratner L., Fisher A., Jagodzinski L. L., Mitsuya H., Liou R. S., Gallo R. C., Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987 Spring;3(1):57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- Richman D. D. AZT resistance in isolates of HIV. Immunodefic Rev. 1991;2(4):315–318. [PubMed] [Google Scholar]
- Roberts J. D., Bebenek K., Kunkel T. A. The accuracy of reverse transcriptase from HIV-1. Science. 1988 Nov 25;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Roberts J. D., Preston B. D., Johnston L. A., Soni A., Loeb L. A., Kunkel T. A. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989 Feb;9(2):469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P. L., Goff S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989 Jul 14;58(1):47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Kim S. Y., Roth M. J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J Virol. 1990 Dec;64(12):6286–6290. doi: 10.1128/jvi.64.12.6286-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Varela-Echavarría A., Garvey N., Preston B. D., Dougherty J. P. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992 Dec 5;267(34):24681–24688. [PubMed] [Google Scholar]
- Varela-Echavarría A., Prorock C. M., Ron Y., Dougherty J. P. High rate of genetic rearrangement during replication of a Moloney murine leukemia virus-based vector. J Virol. 1993 Nov;67(11):6357–6364. doi: 10.1128/jvi.67.11.6357-6364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb J. M., Kumar R., Hughes S. H. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990 Oct;64(10):4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]