Abstract
Objective
Huntington’s disease (HD) is a rare neurological disorder, and its current status in Korea is not well investigated. This study aims to determine the prevalence and incidence of HD and to investigate the clinical features of HD patients in Korea.
Methods
We estimated the crude prevalence and annual incidence of HD based on the databases of the Rare Diseases Registry (RDR) and the National Health Insurance (NHI). The clinical data of genetically confirmed HD patients was collected from 10 referral hospitals and analyzed.
Results
The mean calculated annual incidence was 0.06 cases per 100,000 persons, and the mean calculated prevalence was 0.38 based on the NHI database. The estimated crude prevalence based on the RDR was 0.41. Of the sixty-eight HD patients recruited, the mean age of onset was 44.16 ± 14.08 years and chorea was most frequently reported as the initial symptom and chief complaint. The mean CAG repeat number of the expanded allele was 44.7 ± 4.8 and correlated inversely with the age of onset (p < 0.001). About two-thirds of the patients have a positive family history, and HD patients without positive family history showed a delay in onset of initial symptoms, a prolonged interval between initial symptom onset and genetic diagnosis and a delay in the age of genetic diagnosis.
Conclusions
To the best of our knowledge, this is the first study to estimate the prevalence and incidence of HD in Korea and the largest HD series in the Asian population. Our analyses might be useful for further studies and large-scale investigations in HD patients.
Keywords: Huntington’s disease, Phenotype, Database, Prevalence, Incidence
Huntington’s disease (HD) is a well-known neurological disorder of inherited genetic cause, the incurable nature of which has made patients and doctors helpless. Even after the discovery of the CAG repeat expansion of the IT-15 gene in 1993, no satisfactory treatment has emerged [1]. In Korea, studies involving genetically confirmed HD patients were first published from 1996, and the first clinical analysis was presented in 1997 [2–5]. These studies showed that there were a number of genetically confirmed HD patients without a family history of the disease, and also revealed that caudate atrophy might be ambiguous even after several years of disease duration. Recently, decreased cortical glucose metabolism was suggested as a predictor of disease progression in Korean patients with HD [6]. However, the current status of HD in Korea requires additional investigation.
Huntington’s disease is an example of a rare disease, which are also referred to as rare and intractable diseases or orphan diseases, and are defined as affecting fewer than 20,000 people in Korea (Korean Centers for Disease Control and Prevention, 2006) [7]. The Korean National Health Insurance (NHI) is a quasi-governmental organization supplying medical services for the entire Korean population, and its database contains a comprehensive dataset of its users. In July 2009, a nationwide registry for rare and intractable diseases (Rare Diseases Registry, RDR), was established by the Korean NHI in cooperation with the Ministry of Health and Welfare, and patients with HD have been registered in the RDR. The RDR database differs from the NHI database in that it gathers identifying information on each patient with a physician-certified diagnosis, making it possible to know the cumulative number of patients regardless of the usage of the NHI during a given period. Moreover, the annual incidence of diagnosis could be estimated by the number of newly registered patients with a given disease. To investigate the current status of HD in Korea, we collected data from 10 nationwide referral hospitals and used open data from the databases of the RDR and NHI.
MATERIALS & METHODS
Data collection from the Korean Rare Diseases Registry and the National Health Insurance databases
We collected the data on HD patients who registered in the RDR between July 2009 and December 2013. During this period, the total number of registered HD cases and the number of new registrations each year were identified. To estimate the prevalence of HD, the size of the Korean population in December 2013 was ascertained from resident registration data with respect to population data gathered by the Korean Ministry of Security and Public. The annual incidence of HD from 2010 to 2013 was calculated based on the number of newly registered HD patients and the number of registered residents in December of the corresponding year. The Korean NHI database was used to crosscheck the prevalence of HD, based on the number of patients receiving medical care.
Data collection from 10 referral hospitals around the country
For analysis of the clinical data of individual HD patients, medical records of 68 HD patients from 10 referral hospitals around the country were reviewed. All patients had been diagnosed by a genetic test, which confirmed pathologically expanded CAG repeats at the HTT locus. There were no duplicate cases. The collected clinical data included patient demographics, the number of expanded and normal CAG repeats, initial symptoms and age of onset, the cause of the hospital visit (i.e., the chief complaints), the date of the hospital visit for neurological examination, and the age at genetic diagnosis. There were sometimes multiple initial symptoms and chief complaints for a single patient. Depending on the age of initial symptoms, patients were categorized into juvenile-onset (i.e., HD before the age of 21 years), adult-onset (21–59) and elderly (after 60 years) according to the definition of juvenile-onset HD and the characteristic onset age of typical HD [8,9]. This research was approved by the Institutional Review Board of CHA University.
Statistical analysis
The relationship between CAG repeat length and age at initial symptom onset was determined by nonlinear regression analysis. The differences associated with family history and the affected parental sex were compared by independent t test. All of the statistical analyses were performed using Predictive Analytics Software (version 21.0, IBM, New York, NY, USA).
RESULTS
Crude prevalence and incidence of HD based on RDR and National Health Insurance databases (Table 1)
Table 1.
Number of HD patients according to the Korean RDR and NHI from 2009 to 2013
| Year | Total Korean population | RDR | NHI | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| New HD patients | Cumulative # of patients | Estimated incidence | Total # (male %) | Estimated prevalence | ||
| 2009 | 49,773,145 | 99 | 99 | NA | 222 (50.5) | 0.45 |
|
| ||||||
| 2010 | 50,515,666 | 36 | 135 | 0.07 | 182 (44.5) | 0.36 |
|
| ||||||
| 2011 | 50,734,284 | 19 | 154 | 0.04 | 171 (42.7) | 0.34 |
|
| ||||||
| 2012 | 50,948,272 | 25 | 179 | 0.05 | 181 (40.3) | 0.36 |
|
| ||||||
| 2013 | 51,141,463 | 29 | 208 | 0.06 | 197 (39.1) | 0.39 |
Total Korean population taken from resident registration data (December of each year). Estimated annual incidence = (incident cases per year / total Korean population) × 105, Estimated prevalence = (total cases per year / total Korean population) × 105. HD: Huntington’s disease, NA: not available, NHI: National Health Insurance, RDR: Rare Diseases Registry.
Excluding the HD patients registered in 2009 when the RDR program began, the mean number of new patients registered annually between 2010 and 2013 was 27.25 ± 7.14. Based on the resident registration data, the mean of calculated crude annual incidences was 0.06 ± 0.01 cases per 100,000 persons. The estimated crude prevalence of HD was 0.41 cases per 100,000 persons, based on the cumulative number of HD cases in the RDR database (n = 208) and the size of the Korean population (n = 51,141,463) as of December 2013. According to the NHI database, the mean number of HD patients receiving medical services annually between 2009 and 2013 was 190.60 ± 19.86 (ranges from 171 to 222), with the mean percentage of males being 43.42 ± 4.48%. The crude prevalence estimated based on the NHI database was 0.38 ± 0.04.
Analyses of HD patient data collected from 10 referral hospitals around the country
Genetic analysis and clinical features (Table 2)
Table 2.
Demographics and clinical characteristics of the patients, based on the collective data from 10 referral hospitals
| Clinical findings | Number of patients or means ± SD | Range |
|---|---|---|
| Sex (M:F) | 41:27 | |
| Number of expanded CAG repeats | 44.7 ± 4.8 | 37–60 |
| Number of normal CAG repeats | 17.4 ± 3.2 | 7–27 |
| Family history (present:possible:absent) | 43:4:21 | |
| Paternal:maternal | 26:17 | |
| Age at initial symptom onset (year old) | 44.16 ± 14.08 | 12–77 |
| Age at genetic diagnosis (year old) | 47.91 ± 14.42 | 17–80 |
| Juvenile/adult/elderly | 1/53/8 | |
| Interval from age at initial symptom onset to age at genetic diagnosis (years) | 4.30 ± 2.96 | 1–10 |
| Interval from chief complaints appearance to hospital visit (months) | 33.53 ± 28.99 | 1–120 |
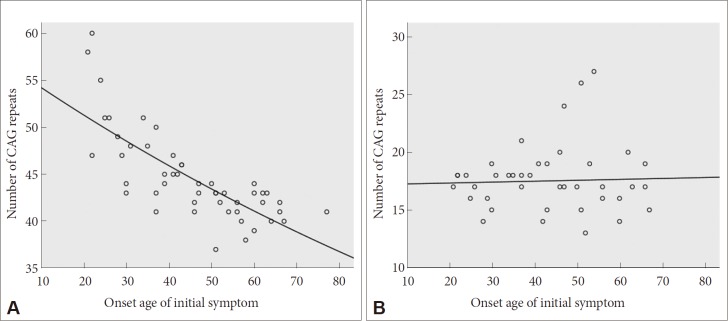
Sixty-eight HD patients, including 12 patients from six families, were recruited for this study. The mean number of CAG repeats of the expanded allele was 44.7 ± 4.8. In normal alleles, the mean number of CAG repeats was 17.4 ± 3.2 without skewness in the distribution (data not shown). The number of CAG expansion showed an inverse correlation with the onset age of initial symptoms (R2 = 0.574, p-value < 0.001), and there was no correlation between the onset age of initial symptoms and the number of CAG repeats in normal allele (Figure 1). The mean age at initial symptom onset was 44.16 ± 14.08 years old, and the mean interval from initial symptom onset to the diagnosis of HD was 4.30 ± 2.96 years. Chorea (n = 41, 60.3%) was the most frequent initial symptom followed by psychiatric symptoms (n = 19), gait disturbance (n = 13), dysarthria (n = 9), cognitive impairment (n = 8), parkinsonism (n = 2), and urinary symptom (n = 1). Causes of hospital visit (chief complaints) were chorea (n = 58, 85.3%), gait disturbance (n = 18), psychiatric symptoms (n = 16), cognitive impairment (n = 12), dysarthria (n = 10), parkinsonism (n = 4), tremor (n = 3), limb ataxia (n = 2), dysphagia (n = 1), and dystonia (n = 1). The age at initial symptom onset was identified in 62 patients, among whom there was 1 juvenile-onset patient (1.6%) and 8 elderly-onset patients (12.8%). The occupations of 50 patients (73.5%) were classified based on the Korean standard classification of occupations with the addition of several categories such as “no employment” and “housewife”. There was no significant correlation between HD diagnosis and occupation (Figure 2).
Figure 1.
Scatter plot for age of initial symptom onset and for the number of CAG repeats in expanded allele (A) and in normal allele (B). There is a significant decrease of age at initial symptom onset in accordance with the number of expanded CAG repeats based on a nonlinear regression model. The number of CAG repeats in normal allele has no correlation with the age at the initial symptom onset.
Figure 2.
Occupation of the HD patients categorized based on the Korean standard classification of occupations, with modifications. The occupations of HD patients were evenly distributed across the category. Patient #: number of patients, HD: Huntington’s disease.
Comparisons according to the family history (Table 3)
Table 3.
Differences according to the presence of family history
| Clinical findings | Family history | p-value | |
|---|---|---|---|
| Presence | Absence | ||
| Age at initial symptom onset | 41.81 ± 13.40 | 49.47 ± 14.49 | 0.047* |
| Interval between the age at initial symptom onset to the genetic diagnosis (years) | 3.80 ± 2.71 | 5.58 ± 3.21 | 0.031* |
| Age at genetic diagnosis (years) | 45.40 ± 13.64 | 54.11 ± 14.79 | 0.025* |
| Interval between the onset of chief complaint to hospital visit (months) | 29.45 ± 23.76 | 43.59 ± 38.08 | 0.17 |
| Number of expanded CAG repeat | 45.30 ± 5.14 | 43.41 ± 3.64 | 0.18 |
| Number of normal CAG repeat | 17.28 ± 3.49 | 17.83 ± 2.48 | 0.62 |
independent t test.
Positive family history of HD was found in 43 patients (63.2%), with paternal transmission occurring in 26 patients (60.5%) and maternal transmission in 17 patients (39.5%). In 21 patients (30.9%), there was no identifiable family history. Four patients had possible family histories, having had family members with early or unexpected mortality associated with psychiatric symptoms and abnormal movements. The comparison of clinical variables between HD patients with and without family histories showed a delay in the onset of initial symptoms (p = 0.047), a prolonged interval between initial symptom onset and genetic diagnosis (p = 0.031) and a delay of the age at genetic diagnosis (p = 0.025) in those without a positive family history (Table 3). Neither the number of expanded CAG repeats nor the interval between the onset of chief complaints and hospital visit was different between the HD patients with and without family histories.
DISCUSSION
To the best of our knowledge, this is the first study to estimate the prevalence and incidence of HD in Korea and the largest HD series of multi-center study in the Asian population. Most of the previous Asian epidemiologic studies of HD were performed on a regional basis, such as in Hong Kong, China or the San-in area of Japan [10,11]. In Taiwan, the average annual incidence rate was estimated at 0.1 and the prevalence at 0.42 per 100,000 persons, based on the National Health Insurance database [12]. The annual incidence estimated in our study is about half of the average annual incidence of Taiwan; however, our estimate is similar to the annual incidence in China (0.046 per 100,000) [10]. Our estimation of the prevalence of HD in Korea based on two data sources was 0.41 per 100,000 in 2013 (Korean RDR) and 0.38 per 100,000 bewteen 2009 to 2013 (National Health Insurance database), which are comparable to Taiwanese data and a meta-analysis [11,13]. It is known that HD is less prevalent in Asian countries compared with Western countries, in which the overall prevalence is 5.70 according to a meta-analysis [13]. Recent data suggests that the prevalence of HD may be over 10 per 100,000 in the UK [14]. These racial differences in prevalence are accounted for by the differences in CAG tract size, HTT haplotype and CCG polymorphism [15–17]. The mean CAG repeat length of the normal allele was reported to be significantly longer in the European (18.4 ± 3.7) population compared with the Chinese, Japanese, and Thai (16.4 ± 1.5, 16.6 ± 1.5, and 16.5 ± 1.9, respectively) population [16,18]. Furthermore, the distribution of CAG length was positively skewed in Europeans, which represents a length-dependent mutational bias towards longer alleles [19]. In our collected dataset, we did not observe skewness in CAG length of normal alleles. The mean CAG repeat length of the normal allele in our HD patients was 17.4 ± 3.2, which is longer than that of other Asian populations, but shorter than that reported by a previous single center study in Korea [20] and than those of Western populations. Recently, HTT haplogroups (types A, B, and C) were identified, and CAG instability was more likely to occur in the cis-acting haplogroup A [15]. HTT haplogroups are known to show a regional difference, and the higher risk haplotype was infrequent in China, Japan, and Thailand [18,21,22]. The CCG polymorphic region is located adjacent to the CAG triplet repeat of the HD gene, and there is a linkage disequilibrium between CCG polymorphism and the pathological CAG expansion according to the race [17,23]. HTT haplogroup and CCG polymorphism have not yet been studied in Korean HD patients.
The mean age at initial symptom onset (44.16 ± 14.08) and the inverse correlation between age at initial symptom onset and CAG repeat expansion size were compatible with previous reports [20,24–26]. The mean length of CAG repeats were similar to the previously reported range of 44.7 to 49.0 (range: 36–70) in Korea and Western countries [5–7,20,27]. Moreover, the length of the expanded CAG repeat determines the age of onset in approximately 50–70% of patients in the 40 to 55 CAG repeat range, but possible roles of other genetic and environmental modifiers are currently being investigated [28,29].
Chorea is the most frequent initial symptom (60.3%) in our collected dataset as well as the chief complaint (85.3%), and the latter is compatible to the previously reported frequency (89%) at first visit to hospital [20]. However, this finding also showed that approximately 40% of HD patients had initial symptoms other than chorea. Among the symptomatic triad of problems involving movement, cognition, and mood, the most prominent deficit may differ between patients, and recent studies have recognized that non-motor symptoms occur before overt motor features in a fraction of patients [30,31].
About one third of the patients comprising our collected dataset had no family history of HD, which is higher than reported in a study from British Columbia (29.8%) [32] and in a previous report from a single center in Korea (20%).20 Given that the probability of de novo expansion of CAG repeats through paternal transmission was estimated to range from 1/6,241 to 1/951 [33], the higher rate of HD patients with a negative family history might not be explained solely by this mechanism. In addition to de novo expansion of CAG repeats, unstable familial transmission, reduced penetrance or underreported family history may be another cause of negative family history among HD patients [34–36]. Interestingly, our HD patients without family history showed a delayed age at onset of initial symptoms, a delayed age at genetic diagnosis and a prolonged interval between the age of initial symptom onset to the age of genetic diagnosis compared with the HD patients with a positive family history. Lower CAG repeat number in negative family history groups may be related with these findings; however, a clinician’s delay in suspicion of HD in choreic patients apparently lacking family history may have also contributed to this observation. Our study has some limitations. Because this research is based on retrospective chart review, extensive data gathering was impossible. Even after the diagnosis of HD, some patients may have refused to be registered into RDR because of social stigma and discrimination, which may have resulted in the underestimation of the real incidence and prevalence of HD [37].
In conclusion, our estimated incidence and prevalence of HD were comparable with previous reports of Asian populations and were relatively low compared with estimates for Western populations. The clinical features showed similar phenotypes, and the effect of CAG repeat expansion on the age of disease onset was comparable. Our analyses using national registries and multi-center clinical data could be useful for future studies and large-scale investigations in HD patients.
Footnotes
Conflicts of Interest
The authors have no financial conflicts of interest.
REFERENCES
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Lyoo CH, Lee MS, Kim YJ, Suk SH, Yang KH, Song KS. Huntington’s disease confirmed by genetic and pathological study. J Korean Neurol Assoc. 1996;14:725–737. [Google Scholar]
- 3.Kim BJ, Park KW, Lee DH. Diagnosis of Huntington’s disease with polymerase chain reaction. J Korean Neurol Assoc. 1996;14:502–510. [Google Scholar]
- 4.Jeon BS, Choi SH, Kim MH, Joo SI, Park SS. Gene analysis in Huntington disease. J Korean Neurol Assoc. 1996;14:494–501. [Google Scholar]
- 5.Kim SY, Park SS, Joo SI, Lee DH, Wie BA, Kwon HY, et al. Clinical analysis of Huntington’s disease in Korea. J Korean Neurol Assoc. 1997;15:1256–1264. [Google Scholar]
- 6.Shin H, Kim MH, Lee SJ, Lee KH, Kim MJ, Kim JS, et al. Decreased metabolism in the cerebral cortex in early-stage Huntington’s disease: a possible biomarker of disease progression? J Clin Neurol. 2013;9:21–25. doi: 10.3988/jcn.2013.9.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates G, Tabrizi S, Jones L. Huntington’s Disease. 4th ed. New York: Oxford University Press; 2014. [Google Scholar]
- 8.Hille ET, Siesling S, Vegter-van der Vlis M, Vandenbroucke JP, Roos RA, Rosendaal FR. Two centuries of mortality in ten large families with Huntington disease: a rising impact of gene carriership. Epidemiology. 1999;10:706–710. [PubMed] [Google Scholar]
- 9.Telenius H, Kremer HP, Theilmann J, Andrew SE, Almqvist E, Anvret M, et al. Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet. 1993;2:1535–1540. doi: 10.1093/hmg/2.10.1535. [DOI] [PubMed] [Google Scholar]
- 10.Chang CM, Yu YL, Fong KY, Wong MT, Chan YW, Ng TH, et al. Huntington’s disease in Hong Kong Chinese: epidemiology and clinical picture. Clin Exp Neurol. 1994;31:43–51. [PubMed] [Google Scholar]
- 11.Nakashima K, Watanabe Y, Kusumi M, Nanba E, Maeoka Y, Nakagawa M, et al. Epidemiological and genetic studies of Huntington’s disease in the San-in area of Japan. Neuroepidemiology. 1996;15:126–131. doi: 10.1159/000109899. [DOI] [PubMed] [Google Scholar]
- 12.Chen YY, Lai CH. Nationwide population-based epidemiologic study of Huntington’s Disease in Taiwan. Neuroepidemiology. 2010;35:250–254. doi: 10.1159/000319462. [DOI] [PubMed] [Google Scholar]
- 13.Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. The incidence and prevalence of Huntington’s disease: a systematic review and meta-analysis. Mov Disord. 2012;27:1083–1091. doi: 10.1002/mds.25075. [DOI] [PubMed] [Google Scholar]
- 14.Evans SJ, Douglas I, Rawlins MD, Wexler NS, Tabrizi SJ, Smeeth L. Prevalence of adult Huntington’s disease in the UK based on diagnoses recorded in general practice records. J Neurol Neurosurg Psychiatry. 2013;84:1156–1160. doi: 10.1136/jnnp-2012-304636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warby SC, Visscher H, Collins JA, Doty CN, Carter C, Butland SL, et al. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet. 2011;19:561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squitieri F, Andrew SE, Goldberg YP, Kremer B, Spence N, Zeisler J, et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994;3:2103–2114. doi: 10.1093/hmg/3.12.2103. [DOI] [PubMed] [Google Scholar]
- 17.Pêcheux C, Mouret JF, Dürr A, Agid Y, Feingold J, Brice A, et al. Sequence analysis of the CCG polymorphic region adjacent to the CAG triplet repeat of the HD gene in normal and HD chromosomes. J Med Genet. 1995;32:399–400. doi: 10.1136/jmg.32.5.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulkes T, Papsing C, Wattanapokayakit S, Mahasirimongkol S. CAG-Expansion Haplotype Analysis in a Population with a Low Prevalence of Huntington’s Disease. J Clin Neurol. 2014;10:32–36. doi: 10.3988/jcn.2014.10.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinsztein DC, Amos W, Leggo J, Goodburn S, Ramesar RS, Old J, et al. Mutational bias provides a model for the evolution of Huntington’s disease and predicts a general increase in disease prevalence. Nat Genet. 1994;7:525–530. doi: 10.1038/ng0894-525. [DOI] [PubMed] [Google Scholar]
- 20.Shin CW, Choi YJ, Kim M, Jeon BS. Preliminary analysis of Huntington’s Disease in South Korea. J Huntingtons Dis. 2013;2:83–87. doi: 10.3233/JHD-120040. [DOI] [PubMed] [Google Scholar]
- 21.Masuda N, Goto J, Murayama N, Watanabe M, Kondo I, Kanazawa I. Analysis of triplet repeats in the huntingtin gene in Japanese families affected with Huntington’s disease. J Med Genet. 1995;32:701–705. doi: 10.1136/jmg.32.9.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma M, Yang Y, Shang H, Su D, Zhang H, Ma Y, et al. Evidence for a predisposing background for CAG expansion leading to HTT mutation in a Chinese population. J Neurol Sci. 2010;298:57–60. doi: 10.1016/j.jns.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Morovvati S, Nakagawa M, Osame M, Karami A. Analysis of CCG repeats in Huntingtin gene among HD patients and normal populations in Japan. Arch Med Res. 2008;39:131–133. doi: 10.1016/j.arcmed.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Aziz NA, Jurgens CK, Landwehrmeyer GB EHDN Registry Study Group. van Roon-Mom WM, van Ommen GJ, et al. Normal and mutant HTT interact to affect clinical severity and progression in Huntington disease. Neurology. 2009;73:1280–1285. doi: 10.1212/WNL.0b013e3181bd1121. [DOI] [PubMed] [Google Scholar]
- 25.Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–1210. [PMC free article] [PubMed] [Google Scholar]
- 26.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR International Huntington’s Disease Collaborative Group. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 27.Orth M, Schwenke C. Age-at-onset in Huntington disease. PLoS Curr. 2011;3:RRN1258. doi: 10.1371/currents.RRN1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gusella JF, MacDonald ME, Lee JM. Genetic modifiers of Huntington’s disease. Mov Disord. 2014;29:1359–1365. doi: 10.1002/mds.26001. [DOI] [PubMed] [Google Scholar]
- 29.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen JS, Long JD. Onset of Huntington’s disease: can it be purely cognitive? Mov Disord. 2014;29:1342–1350. doi: 10.1002/mds.25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabrizi SJ, Scahill RI, Durr A, Roos RA, Leavitt BR, Jones R, et al. Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol. 2011;10:31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 32.Almqvist EW, Elterman DS, MacLeod PM, Hayden MR. High incidence rate and absent family histories in one quarter of patients newly diagnosed with Huntington disease in British Columbia. Clin Genet. 2001;60:198–205. doi: 10.1034/j.1399-0004.2001.600305.x. [DOI] [PubMed] [Google Scholar]
- 33.Hendricks AE, Latourelle JC, Lunetta KL, Cupples LA, Wheeler V, MacDonald ME, et al. Estimating the probability of de novo HD cases from transmissions of expanded penetrant CAG alleles in the Huntington disease gene from male carriers of high normal alleles (27–35 CAG) Am J Med Genet A. 2009;149A:1375–1381. doi: 10.1002/ajmg.a.32901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semaka A, Collins JA, Hayden MR. Unstable familial transmissions of Huntington disease alleles with 27–35 CAG repeats (intermediate alleles) Am J Med Genet B Neuropsychiatr Genet. 2010;153B:314–320. doi: 10.1002/ajmg.b.30970. [DOI] [PubMed] [Google Scholar]
- 35.Houge G, Bruland O, Bjørnevoll I, Hayden MR, Semaka A. De novo Huntington disease caused by 26–44 CAG repeat expansion on a low-risk haplotype. Neurology. 2013;81:1099–1100. doi: 10.1212/WNL.0b013e3182a4a4af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sequeiros J, Ramos EM, Cerqueira J, Costa MC, Sousa A, Pinto-Basto J, et al. Large normal and reduced penetrance alleles in Huntington disease: instability in families and frequency at the laboratory, at the clinic and in the population. Clin Genet. 2010;78:381–387. doi: 10.1111/j.1399-0004.2010.01388.x. [DOI] [PubMed] [Google Scholar]
- 37.Dispelling the stigma of Huntington’s disease. Lancet Neurol. 2010;9:751. doi: 10.1016/S1474-4422(10)70170-8. [DOI] [PubMed] [Google Scholar]