Abstract
Objectives/Hypothesis
The role of fungi in chronic rhinosinusitis (CRS) is still controversial. The present study was conducted to detect and identify fungal species from the nasal polyp tissues of eosinophilic and noneosinophilic CRS, and to determine the role of fungal antigens in cytokine production.
Study Design
Prospective study.
Methods
Thirty-five specimens of nasal polyps were collected from patients with CRS and examined for fungus using culture, histology, and polymerase chain reaction analysis. The secretion of 14 cytokines stimulated by fungal extracts using dispersed nasal polyp cells (DNPCs) was determined by multiplex immunoassay.
Results
There was no microbiological growth (including fungus) in the cultures of homogenized nasal polyps. Furthermore, Grocott methanamine silver staining for all nasal polyps showed no fungal bodies. Sixteen of 35 samples of the nasal polyps showed amplification of fungal DNA. In none of the mucosa of the sphenoid sinus was fungal DNA detected. The number of eosinophils in the nasal polyps in which fungal DNA was detected was significantly higher than in the nasal polyps in which fungal DNA was not detected (P < .01). The extract of fungus enhanced the secretion of eosinophil-associated cytokines such as interleukine (IL)-5, IL-13, IL-17A, and RANTES (regulated on activation normal T-cell expressed and secreted), and proinflammatory cytokines such as IL-6, IL-8, tumor necrosis factor-α, and granulocyte-macrophage colony-stimulating factor from DNPCs.
Conclusions
The present study offers direct evidence supporting that fungal elements modify the inflammatory response in the nasal polyps of eosinophilic CRS.
Level of Evidence: NA
Keywords: Fungus, nasal polyp, polymerase chain reaction, cytokine, eosinophil
INTRODUCTION
Chronic rhinosinusitis (CRS) is considered to be a multifactorial disease within a heterogeneous group of diseases, with different underlying etiologies and pathophysiologies. CRS has been classified broadly as CRS with nasal polyps (CRSwNP), CRS without nasal polyps, and allergic fungal rhinosinusitis (AFRS). In particular, CRSwNP may have a complex pathogenesis and is thought to arise from multiple factors including allergy. Microorganisms have always been popularly suspected in the pathology of CRSwNP. There is recent evidence suggesting that 1) Staphylococcus aureus enterotoxins, 2) type I hypersensitivity to fungus, and 3) non-immunoglobulin E (IgE)-mediated hypersensitivity to fungus may play a role in the pathogenesis of eosinophilic inflammation.1 However, the role of the microorganisms, particularly fungal pathogens, in the etiology of CRS remains largely unknown.
The role of fungi in CRS is still controversial. Conflicting with the prevailing belief that fungi were responsible for CRS in a selected group of patients with distinct pathophysiology, Ponikau et al.2 and Braun et al.3 observed that fungus is a ubiquitous intranasal presence, identified in close to 100% of both CRS patients and controls. The former group also detected fungi along with eosinophil and eosinophil-degraded products with mucus. Shin et al.4 exposed peripheral blood mononuclear cells to fungal antigens in vitro and reported increased interleukin (IL)−5 and IL-13 production in 89% of CRS patients but not in controls. These observations formed the basis of the “fungal hypothesis of CRS.” As further evidence, nasal mucus or tissue from CRS patients triggered eosinophil migration,5 and Alternaria fungus in particular can directly induce eosinophil degranulation mediated by protease-activated receptor (PAR) activation.6
However, other investigators reported the absence of a universal hyper-responsiveness to fungal antigens in CRS patients.7,8 Furthermore, a multicenter, randomized clinical trial of topical antifungal agents for CRS eventually failed to show any evidence of efficacy,9 and a meta-analysis did not support the routine use of topical antifungals for CRS.10 Thus, the precise roles of fungi in the etiopathology of CRS remain unknown.
The present study was conducted to detect and identify fungal species from the nasal polyp tissues of eosinophilic and noneosinophilic CRS using Grocott methanamine silver staining and polymerase chain reaction (PCR) methods. Moreover, the effects of fungal extracts identified in the nasal polyps were examined by the ex vivo cellular responses of dispersed nasal polyp cells (DNPCs).
MATERIALS AND METHODS
Patients
Thirty-five patients with CRS with nasal polyps (21 males and 14 females, ranging in age from 23–77 years, mean age of 49 years) were consecutively recruited from the Department of Otorhinolaryngology of Juntendo University Hospital from April 2011 to March 2012. CRS with nasal polyps was diagnosed based on the criteria of the European position paper.11 None of the patients was treated with antibiotics, systemic or topical corticosteroids, or other immune-modulating drugs for at least 1 month before the surgery. Subjects with AFRS were excluded from the present study. The criteria of AFRS of two positive findings, 1) specific IgE antibodies against fungi, and 2) the presence of fungi in the sinus effusion using Grocott methanamine cytological silver staining or microbiological examination. Serum fungus-specific IgE concentrations against Alternaria, Aspergillus, Candida, Penicillium, Mucor, Cladosporium, and Pityrosporium were measured. Patients with CRSwNP associated with current signs of purulent nasal discharge, chronic obstructive pulmonary disease, diffuse panbronchiolitis, fungal sinus disease, congenital mucociliary disease, or cystic fibrosis were excluded from this study. The control group consisted of 15 patients with pituitary tumor surgery (four males and 11 females, age range from 36 to 73 years, mean age of 55 years). The study was approved by the ethics committee of the Juntendo University Faculty of Medicine.
Sampling of Tissue and Pretreatment
Surgically removed human nasal polyps located in the middle meatus were obtained from the patients with CRSwNP, and the mucosa of the sphenoid sinus as a control were procured from patients with pituitary tumor. They were treated with 70% ethanol and physiologic saline to eliminate microorganisms outside of the nasal polyps. The samples were placed immediately in a 50% glycerol, then transferred to a −80°C freezer for storage until DNA extraction could be performed. At the same time, some of these samples were fixed in 10% formalin, embedded in paraffin wax, processed routinely, and stained with hematoxylin-eosin. According to our previous studies,12,13 the eosinophilic and noneosinophilic groups were defined as eosinophil counts of the nasal polyps pf more and less 100/microacopic field (magnification ×400) using three fields, respectively. Eosinophils were quantified in the foci of the densest cellular infiltrate. The total number of eosinophils present with a 10 × 10-mm reticulate present in the eyepiece was determined as the count per high-power field. The samples for the DNPCs were treated within 30 minutes postoperatively.
Culture and Histology of Fungi
After the nasal polyps were disrupted with glass beads and 100 μL of Tris-EDTA (10 mM Tris-Cl, 1 mM ethylenediaminetetraacetic acid pH 8.0) using a homogenizer (BioMasher) (Takara Bio Inc., Otsu, Japan), and 1 μL of homogenized polyp samples were spread onto 5% sheep blood agar, chocolate agar, Columbia anaerobic blood agar, Drigalski agar, and Sabouraud agar, and incubated for 2 weeks at 35°C. All specimens were stained with Grocott methanamine silver staining for confirmation of the existence of fungal organisms.
PCR of Fungi in the Nasal Polyps
DNA was extracted from the tissue of homogenized nasal polyps. PCR amplification of fungal specific ribosomal DNA was performed using ITS-1F and ITS-4R primer, the PCR product covering the end of the 18SrDNA gene to the start of 26SrDNA gene and NL-1 and NL-4 primer, the PCR product covering D1/D2 26SrDNA (Table1). A universal fungal primer set designed by the National Institute for Occupational Safety and Health, FF2 and FR1 primers specific for the amplification of 18S rDNA, was also used. Amplification was performed in a TaKaRa PCR Thermal Cycler Dice Gradient (Takara Bio Inc., Otsu, Japan) according to the manufacturer's specifications.
Table 1.
Primers Used in the Study.
| Primer | 5′ → 3′ |
|---|---|
| ITS-IF | GTC GTA ACA AGG |
| TTA ACC TGC GG | |
| ITS-4R | TCC TCC GCT TAT |
| TGA TAT GC | |
| NL-1 | GCA TAT CAA TAA |
| GCG GAG GAA AAG | |
| NL-4 | GGT CCG TGT TTC |
| AAG ACG G | |
| FF-2 | GGT TCT ATT TTG TTG |
| GTT TCT A | |
| FR-1 | CTC TCA ATC TGT |
| CAA TCC TTA TT |
Species Identification
PCR products were purified by the High Pure PCR Product Purification Kit (Roche, Indianapolis, IN). These amplifications were sequenced using ITS1F, ITS4R, NL1, NL4, FF2, and FR1 primers by a Big Dye Terminator V3.1 Cycle sequencing kit and ABI sequence analyzer 3730×I (Applied Biosystems Inc., Carlsbad, CA). Species identification was determined by a BLAST search(DNA Data Bank of Japan, Mishima, Japan).
Cell Cultures and Multiplex Immunoassay
DNPCs were prepared from nasal polyps of eosinophilic CRSwNPs in five randomly selected patients by enzymatic digestion, as described by Okano et al.14 In flat-bottomed 24-well culture plates (Nunc, Roskiide, Denmark), 500 μL of 1 × 106/mL DNPCs were stimulated with serial concentrations (2 and 200 μg/mL) of Candida parapsilosis, Rhodotorula mucilaginosa. The crude extracts of C parapsilosis and R mucilaginosa were provided by Teikyo University Japanese Society for Medical Mycology Research Center. After the incubation, freeze-dried fungi were dissolved and sonicated in phosphate-buffered saline (PBS) with 0.2% NaN3. Afterward, the extract was subjected to sterilizing filtration and lyophilization. The antigen extracts of fungus were dissolved with PBS (Sigma-Aldrich Inc., St. Louis, MO). As a control, DNPCs were cultured without antigen stimulation. The culture supernatant was collected after 72 hours and stored at −80°C, after which the levels of IL-4, IL-5, IL-6, IL-8, IL-13, IL-17A, IL-23, IL-25, IL-33, eotaxin, RANTES (regulated on activation normal T-cell expressed and secreted), tumor necrosis factor (TNF)-α, interferon (IFN)-γ, and granulocyte-macrophage colony-stimulating factor (GM-CSF) were measured by Bio-Plex Suspension Array System (Bio-Rad Laboratories, Inc., Hercules, CA). Data were expressed as the fold change relative to that without stimulus of five cell cultures. Viability was assessed by the exclusion of trypan blue stain.
Statistical Analyses
Values are given as means ± standard errors for the multiplex immunoassay. Differences between the values were determined using the Student t test. Tukey's hinge was used in the comparison of eosinophils within nasal polyps. P values <.05 were considered significant. All analyses were conducted using Statmate IV for windows (ATMS Co., Ltd., Tokyo, Japan).
RESULTS
Culture and Histology of Fungi
There was no microbiological growth (including fungus) in the culture media of the homogenized nasal polyps. Furthermore, Grocott methanamine silver staining for the all nasal polyps showed no fungal bodies.
Detection and Identification of Fungal DNA
Sixteen of 35 samples of the nasal polyps showed amplification of fungal DNA. Using the D1/D2 domain of the large subunit (26S) ribosomal DNA primer, NL1 /NL4, we found that 16 cellular tissues of the homogenized nasal polyps (100%) were positive for fungal DNA, whereas using the 18S rDNA primer, ITS1F/ITS4R amplified sequences four of the 16 (25%) were positive. On the other hand, using the universal fungal primer set, FF2 and FR1, no fungal DNA was detected. Incidentally, in none of the mucosa of the sphenoid sinus was fungal DNA detected. The amplification products obtained from 16 patients were sequenced to identify the detected species in the tissue of the homogenized nasal polyps, such as C parapsilosis, R mucilaginosa, and Aspergillus sp. Table2 summarizes the data for all patients. The average number of eosinophils within the nasal polyps in which fungal DNA was detected was significantly higher than that in the absence of fungal DNA (P < .01, Fig. 1).
Table 2.
Clinical Characteristics of Patients and Identified Fungal Species.
| Patient | Sex/Age, yr | Blood Eosinophils | Blood IgE | RAST Fungus | Asthma | Eosinophilic Mucin | Tissue Eosinophils | Identified Fungal Species |
|---|---|---|---|---|---|---|---|---|
| 1 | M/47 | 420 | 38 | − | + | + | 58 | Candida parapsilosis |
| 2 | M/36 | 508 | 332 | + | + | + | 148 | Candida parapsilosis |
| 3 | F/74 | 301 | 65 | − | + | − | 111 | Rhodotorula mucilaginosa |
| 4 | F/35 | 644 | 77 | − | + | − | 410 | Rhodotorula mucilaginosa |
| 5 | M/45 | 328 | 162 | − | + | − | 184 | Rhodotorula mucilaginosa |
| 6 | M/67 | 809 | 301 | − | + | − | 275 | Malassezzia restricta |
| 7 | M/39 | 462 | 118 | − | + | − | 164 | Aspergillus s/o |
| 8 | F/46 | 381 | 1,990 | − | + | − | 194 | Candida parapsilosis |
| 9 | M/42 | 418 | 86 | − | + | − | 291 | Aspergillus gracillus |
| 10 | M/44 | 675 | 51 | − | + | − | 650 | Candida parapsilosis |
| 11 | M/49 | 428 | 517 | − | − | − | 198 | Rhodotorula mucilaginosa |
| 12 | F/49 | 731 | 430 | + | − | − | 189 | Candida parapsilosis |
| 13 | M/39 | 412 | 46 | − | − | − | 341 | Candida parapsilosis |
| 14 | F/51 | 353 | 89 | − | − | − | 184 | Candida glabrata |
| 15 | F/62 | 192 | 38 | − | − | − | 176 | Candida tropicalis |
| 16 | F/55 | 280 | 1,882 | + | − | − | 266 | Candida parapsilosis |
| 17 | M/49 | 420 | 104 | − | + | + | 232 | None |
| 18 | M/45 | 328 | 162 | − | + | − | 124 | None |
| 19 | F/60 | 468 | 191 | − | + | − | 158 | None |
| 20 | M/32 | 538 | 25 | − | + | − | 175 | None |
| 21 | M/66 | 271 | 83 | − | − | − | 18 | None |
| 22 | F/34 | 795 | 5 | − | − | − | 91 | None |
| 23 | F/56 | 99 | 24 | − | − | − | 14 | None |
| 24 | M/25 | 152 | 118 | − | − | − | 12 | None |
| 25 | F/60 | 1,372 | 191 | − | − | + | 158 | None |
| 26 | F/58 | 413 | 65 | − | − | + | 186 | None |
| 27 | M/63 | 567 | 515 | + | − | + | 58 | None |
| 28 | F/57 | 413 | 104 | − | − | − | 153 | None |
| 29 | M/42 | 246 | 250 | − | − | − | 111 | None |
| 30 | M/44 | 292 | 91 | − | − | − | 128 | None |
| 31 | F/52 | 270 | 287 | − | − | − | 36 | None |
| 32 | M/76 | 7 | 11 | − | − | − | 27 | None |
| 33 | F/63 | 384 | 528 | + | − | − | 146 | None |
| 34 | F/34 | 30 | 1,409 | + | − | − | 148 | None |
| 35 | M/43 | 150 | 500 | + | − | − | 66 | None |
F = female; IgE = immunoglobulin E; M = male; RAST = radioallergosorbent test.
Figure 1.
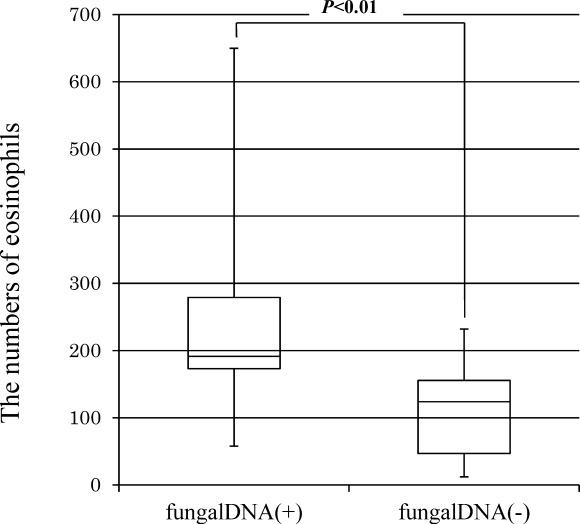
Comparison of the numbers of tissue eosinophils with nasal polyps between positive samples and those negative for fungal DNA. The number of eosinophils in three fields with cell clusters were counted using light microscopy (×400 magnification). The box and whisker plots show the median and the interquartile range of the number. The whiskers extend to the maximum and the minimum data points.
Cytokine Secretion Stimulated by Fungal Extracts
To determine the biological and immunological roles of the identified fungi in CRSwNP in the present study, five samples of DNPCs were stimulated by extracts of C parapsilosis and R mucilaginosa. The expression levels of each cytokine and chemokine are summarized and compared with those of the unstimulated group in Figure 2). The secretion levels of the cytokines were set to the maximal level of each standard curve because the levels were above the maximal levels of detection.
Figure 2.
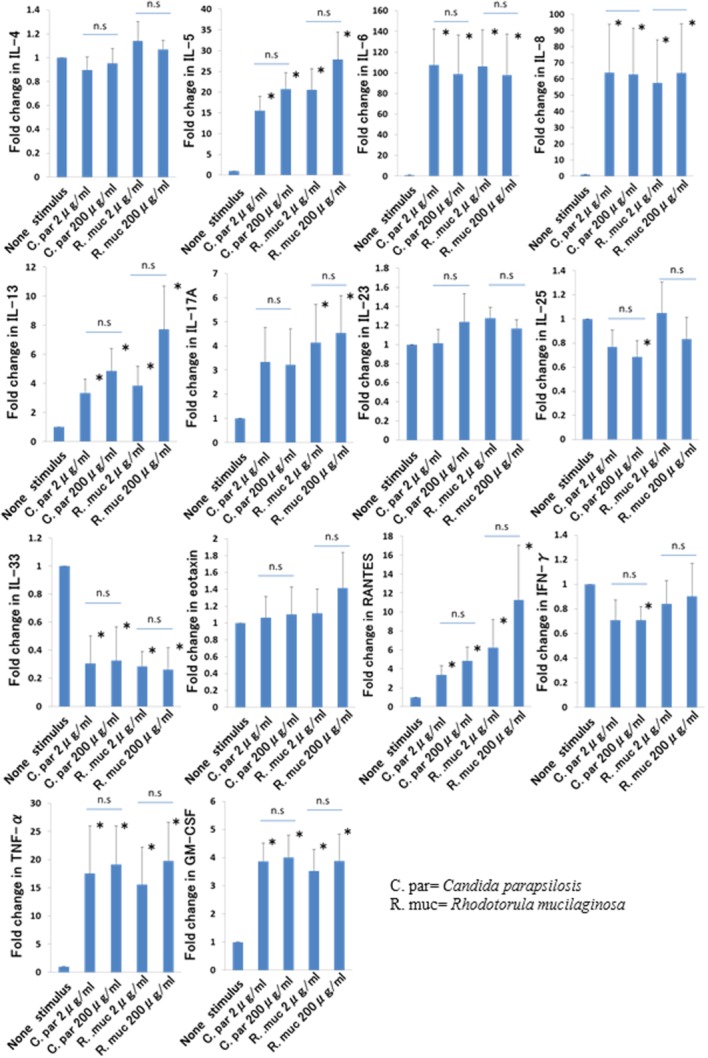
Effect of extracts of Candida parapsilosis and Rhodotorula mucilaginosa on interleukin (IL)−4, IL-5, IL-6, IL-8, IL-13, IL-17A, IL-23, IL-25, IL-33, eotaxin, RANTES, interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and granulocyte-macrophage colony-stimulating factor (GM-CSF) production by dispersed nasal polyp cells. Five hundred microliters of 1 × 106/mL dispersed nasal polyp cells were collected after 72 hours, and the levels of the cytokines were determined by multiplex immunoassay. Data were expressed as the fold change relative to that without stimulus of five cell cultures (the means ± standard errors). Significant differences (*) were considered if P values were <.05. RANTES = regulated on activation normal T-cell expressed and secreted. [Color figure can be viewed in the online issue, which is available at www.laryngoscope.com.]
C parapsilosis stimulation of DNPCs significantly induced the secretion of IL-5, IL-6, IL-8, IL-13, RANTES, TNF-α, and GM-CSF. The most remarkable upregulation was observed in IL-6 (approximately 100-fold) and IL-8 (approximately 60-fold), followed by IL-5, TNF-α, RANTES, IL-13, and GM-CSF. IL-4, IL-17A, IL-23, and eotaxin showed no significant increase from stimulation by C parapsilosis extracts. The expression of IL-25, IL-33 and IFN-γ showed a decreasing trend. No significant differences were seen in the responses between 2 and 200 μg/mL of any cytokines.
The stimulation of DNPCs by R mucilaginosa resulted in a significant induction of IL-5, IL-8, IL-13, IL-17A, RANTES, TNF-α, and GM-CSF. Similar to C parapsilosis, R mucilaginosa most strongly upregulated the production of both IL-6 and IL-8, which was followed by IL-5, TNF-α, RANTES, IL-13, IL-17A, and GM-CSF. IL-4, IL-23, IL-25, IFN-γ, and eotaxin showed no significant production with stimulation of R mucilaginosa extract. On the other hand, R mucilaginosa inhibited IL-33 production. There was no significant difference in the cytokine releases induced by 2 and 200 μg/mL.
DISCUSSION
In the present study, we found fungal DNA in the nasal polyps in spite of the lack of detection of fungus using histology and culture. A variety of studies agree that PCR is superior to both culture and Grocott methanamine silver staining for detecting fungal elements.15 Because the PCR studies showed the ubiquitous presence of fungi such as Aspergillus, Penicillium, Cladosporium, Candida, Aureobasidium, and Alternaria in the nose and paranasal sinuses of both CRS patients and healthy controls,16–18 it is unlikely that fungal species and fungal load play a role in disease development.19 Gosepath et al.20 reported that fungal DNA was detected in all 27 CRS specimens with universal PCR primers. The differences in the detection rates of fungal DNA between Gospath et al.'s report and our study may be due to the sampling method, surgical specimens, or environmental factors. On the other hand, Rao et al.21 reported that the proper design of PCR primers for fungi and the meticulous harvesting of nasal mucosa resulted in a low but significant incidence of fungi in CRS. The primer pairs NL1/NL4, which encode the D1/D2 domain of large subunit (26S) ribosomal DNA, have been widely used for the detection of fungal DNA and the identification of fungal species, as compared with two other primers, ITS1F/ITS4R and FF2/FR1.22,23 No PCR products could be obtained from normal mucosa as negative controls. Positive and negative controls were correctly amplified in every experiment. Therefore, the three PCR amplified systems used here were appropriately operated. The exact reason why the detection rates of three PCR systems showed different values is not known. The large amount of human DNA extracted from the nasal polyps might have inhibited several fungus-specific PCR products, because fungi share the same class of ribosomal RNA with humans. We detected PCR products derived from four species of fungi including Candida, Rhodotorula, Malasezzia, and Aspergillus in the nasal polyps, which were never detected in the normal sinus mucosa. The Rhodotorula species are ubiquitous saprophytic yeast that can be recovered from many environmental sources. Previously reported as nonpathogenic, Rhodotorula species have emerged as opportunistic pathogens with the ability to colonize and infect susceptible patients.24 Malasezzia is a monophyletic genus of fungi detected as a ubiquitous component of the human skin microbiome and is associated with a myriad of skin problems.25 Although we attempted to observe the localization of the DNA of Candida and Rhodotorula using in situ hybridization in the nasal polyps, the experimental results failed to detect the fungal element, presumably due to the use of inappropriate probes. The fungal DNA would be identified within bacterial biofilms26 and/or phagocytic vesicles of antigen-presenting cells such as macrophages and neutrophils in the nasal polyps.27
The detection rate of fugal DNA was significantly elevated in the eosinophilic CRS as compared with noneosinophilic CRS, suggesting that the presence of fungi is closely related with eosinophil accumulation. Eosinophils are known to be prominent in the reaction to parasitic infections. A concentration-dependent increase in eosinophil migration toward both CRS nasal mucin and CRS nasal tissue extract was augmented as compared with the mucin of healthy controls.5 Exposure of peripheral blood mononuclear cells to fungal antigen in vitro increased IL-5 and IL-13 production, whereas cells from normal controls did not respond.4 A component of Alternaria was shown to degranulate eosinophils from CRS patients by acting on PARs. Moreover, activation of nasal epithelial cells with fungi resulted in the upregulation of PAR2 and PAR3 mRNA.28 These findings may lead to a hypothesis that fungi on the sinus mucosal surface induce the production of cytokines, which promote eosinophil migration through the epithelial cells and other constitutive cells of the nasal polyps. Although we did not employ quantitative assays of PCR products, it was considered that the patients with more fungal DNA would have significantly higher eosinophil counts.
The final series of experiments aimed to evaluate immunological aspects of the detected fungi in the induction of eosinophilic inflammation. The fungal extracts derived from Candida and Rhodotorula, which were detected from the nasal polyps, apparently and remarkably upregulated a Th1/Th2 and Th1Th2/Th17 cytokine profile, respectively, in the ex vivo models of the nasal polyp in the present study. It would be desirable to have stimulated the cytokine-producing cells from healthy sphenoid control tissue. However, it is well known that healthy tissue does not contain eosinophils and very minimal amounts of lymphocytes if any. Thus, no quantitative comparison would have been useful. Protease activity contained in fungi can activate epithelial cells via their PARs. Kaufman et al.29 showed that the interaction of protease present in fungal extracts from the inferior nasal conchae of nonatopic subjects led to morphologic change, cell desquamation, and the induction of proinflammatory Th1 cytokines. In addition, PAR2 stimulation in CRS patients did not induce the release of eosinophil attracting cytokines like eotaxins or RANTES.30 Recent investigations are focusing on the contribution of Th17 cells to the pathology of and resistance to fungi. Zelante et al.31 found that by affecting fungal clearance and by promoting chronic inflammation and tissue damage, Th17 responses at the mucosal surface had a detrimental effect on the course of fungal infections. Our previous study revealed that the infiltration of cells positive for both CD4 and IL-17A (Th17 cells) showed a significant correlation with the numbers of eosinophils and mucosal remodeling in Japanese CRSwNP.12
Another underlying mechanism that might explain the fungus-mediated secretion of cytokines and chemokines, such as IL-6 and IL-8, from DNPCs may involve toll-like receptors (TLRs) expressed on the sinonasal mucosa. The recognition of various pathogen-molecular patterns by the TLRs, including mannan, Candida and CpG DNA induces a cascade of downstream signaling that provokes an inflammatory cytokine profile.32 A quantitative increase in TLR2 mRNA was seen in cystic fibrosis polyps as well as in CRS,33,34 whereas in some studies35,36 a decrease in mucosal TLR2 and TLR9 mRNA in samples from CRSwNP occurred. Despite inconsistencies in previous data, TLR signaling appears to play an important role in mediating host inflammation, with potential derangements contributing to the development of CRS.37 Moreover, nucleotide-binding oligomerization domain-like receptors (NLRs) are newly discovered cytosolic receptors belonging to the pattern-recognition receptor family. NLR mRNA was found to be higher in nasal polyps than in normal nasal mucosa.38 Further studies are required to elucidate the role of innate immunity in fungus-related CRS.
CONCLUSION
The present study offers direct evidence to support the notion that fungal elements modify the inflammatory responses in the nasal polyps of eosinophilic CRS, though the underlying mechanisms responsible for the eosinophil-based inflammation require further examination.
Acknowledgments
The authors thank Kazusaku Kamiya for advice on the experiment and Mayumi Sakuraba for assistance with the experiment.
BIBLIOGRAPHY
- Sok JC, Ferguson BJ. Differential diagnosis of eosinophilic chronic rhinosinusitis. Curr Allergy Asthma Rep. 2006;6:203–214. doi: 10.1007/s11882-006-0036-1. [DOI] [PubMed] [Google Scholar]
- Ponikau JU, Sherris DA, Kern EB, et al. The diagnosis and incidence of allergic fungal sinusitis. Mayo Clin Proc. 1999;74:877–884. doi: 10.4065/74.9.877. [DOI] [PubMed] [Google Scholar]
- Braun H, Buzina W, Freudenschuss K, Beham A, Stammberger H. Eosinophilic fungal rhinosinusitis: a common disorder in Europe? Laryngoscope. 2003;113:264–269. doi: 10.1097/00005537-200302000-00013. [DOI] [PubMed] [Google Scholar]
- Shin SH, Ponikau JU, Sherris DA, et al. Chronic rhinosinusitis: an enhanced immune response to ubiquitous airborne fungi. J Allergy Clin Immunol. 2004;114:1369–1375. doi: 10.1016/j.jaci.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Wei JL, Kita H, Sherris DA, Kern EB, Weaver A, Ponikau JU. The chemotactic behavior of eosinophils in patients with chronic rhinosinusitis. Laryngoscope. 2003;113:303–306. doi: 10.1097/00005537-200302000-00019. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Matsuwaki Y, Shin SH, Ponikau JU, Kita H. Nonpathogenic, environmental fungi induce activation and degranulation of human eosinophils. J Immunol. 2005;175:5439–5447. doi: 10.4049/jimmunol.175.8.5439. [DOI] [PubMed] [Google Scholar]
- Douglas R, Bruhn M, Tan LW, Ooi E, Psaltis A, Wormald PJ. Response of peripheral blood lymphocytes to fungal extracts and staphylococcal superantigen B in chronic rhinosinusitis. Laryngoscope. 2007;117:411–414. doi: 10.1097/MLG.0b013e31802c0707. [DOI] [PubMed] [Google Scholar]
- Orlandi RR, Marple BF, Georgelas A, Durtschi D, Barr L. Immunologic response to fungus is not universally associated with rhinosinusitis. Otolaryngol Head Neck Surg. 2009;141:750–756. doi: 10.1016/j.otohns.2009.09.016. e751–752. [DOI] [PubMed] [Google Scholar]
- Ebbens FA, Scadding GK, Badia L, et al. Amphotericin B nasal lavages: not a solution for patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2006;118:1149–1156. doi: 10.1016/j.jaci.2006.07.058. [DOI] [PubMed] [Google Scholar]
- Isaacs S, Fakhri S, Luong A, Citardi MJ. A meta-analysis of topical amphotericin B for the treatment of chronic rhinosinusitis. Int Forum Allergy Rhinol. 2011;1:250–254. doi: 10.1002/alr.20056. [DOI] [PubMed] [Google Scholar]
- Fokkens WJ, Lund VJ, Mullol J, et al. European position paper on rhinosinusitis and nasal polyps 2012. Rhinol Suppl. 2012;(23):1–298. 3 p preceding table of contents. [PubMed] [Google Scholar]
- Saitoh T, Kusunoki T, Yao T, et al. Role of interleukin-17A in the eosinophil accumulation and mucosal remodeling in chronic rhinosinusitis with nasal polyps associated with asthma. Int Arch Allergy Immunol. 2010;151:8–16. doi: 10.1159/000232566. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Shiozawa A, Ono N, et al. Subclassification of chronic rhinosinusitis with nasal polyp based on eosinophilic and neutrophilic infiltration. Laryngoscope. 2013;123:E1–E9. doi: 10.1002/lary.24154. [DOI] [PubMed] [Google Scholar]
- Okano M, Fujiwara T, Haruna T, et al. Prostaglandin E(2) suppresses staphylococcal enterotoxin-induced eosinophilia-associated cellular responses dominantly through an E-prostanoid 2-mediated pathway in nasal polyps. J Allergy Clin Immunol. 2009;123:868–874. doi: 10.1016/j.jaci.2009.01.047. [DOI] [PubMed] [Google Scholar]
- Ebbens FA, Fokkens WJ. The mold conundrum in chronic rhinosinusitis: where do we stand today? Curr Allergy Asthma Rep. 2008;8:93–101. doi: 10.1007/s11882-008-0018-6. [DOI] [PubMed] [Google Scholar]
- Scheuller MC, Murr AH, Goldberg AN, Mhatre AN, Lalwani AK. Quantitative analysis of fungal DNA in chronic rhinosinusitis. Laryngoscope. 2004;114:467–471. doi: 10.1097/00005537-200403000-00015. [DOI] [PubMed] [Google Scholar]
- Kim ST, Choi JH, Jeon HG, Cha HE, Hwang YJ, Chung YS. Comparison between polymerase chain reaction and fungal culture for the detection of fungi in patients with chronic sinusitis and normal controls. Acta Otolaryngol. 2005;125:72–75. doi: 10.1080/00016480410018133. [DOI] [PubMed] [Google Scholar]
- Murr AH, Goldberg AN, Vesper S. Fungal speciation using quantitative polymerase chain reaction (QPCR) in patients with and without chronic rhinosinusitis. Laryngoscope. 2006;116:1342–1348. doi: 10.1097/01.mlg.0000225896.91392.6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens WJ, Ebbens F, van Drunen CM. Fungus: a role in pathophysiology of chronic rhinosinusitis, disease modifier, a treatment target, or no role at all? Immunol Allergy Clin North Am. 2009;29:677–688. doi: 10.1016/j.iac.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Gosepath J, Brieger J, Vlachtsis K, Mann WJ. Fungal DNA is present in tissue specimens of patients with chronic rhinosinusitis. Am J Rhinol. 2004;18:9–13. [PubMed] [Google Scholar]
- Rao AK, Mathers PH, Ramadan HH. Detection of fungi in the sinus mucosa using polymerase chain reaction. Otolaryngol Head Neck Surg. 2006;134:581–585. doi: 10.1016/j.otohns.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T, Nishikawa A, Ikeda R, Shinoda T. Identification of medically relevant Trichosporon species based on sequences of international transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol. 1999;37:1985–1993. doi: 10.1128/jcm.37.6.1985-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth F, Goldani LZ. Epidemiology of Rhodotorula: an emerging pathogen. Interdiscip Perspect Infect Dis. 2012;2012:465717. doi: 10.1155/2012/465717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CW, Scheynius A, Heitman J. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 2012;8:e1002701. doi: 10.1371/journal.ppat.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy DY, Leid JG, Sanderson AR, Hunsaker DH. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2008;138:641–647. doi: 10.1016/j.otohns.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Kawano K, Kusunoki T, Ono N, et al. Heme oxygenase-1 expression in chronic rhinosinusitis with eosinophilic infiltration. Auris Nasus Larynx. 2012;39:387–392. doi: 10.1016/j.anl.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Shin SH, Lee YH, Jeon CH. Protease-dependent activation of nasal polyp epithelial cells by airborne fungi leads to migration of eosinophils and neutrophils. Acta Otolaryngol. 2006;126:1286–1294. doi: 10.1080/00016480500395179. [DOI] [PubMed] [Google Scholar]
- Kauffman HF, Tomee JF, van de Riet MA, Timmerman AJ, Borger P. Protease-dependent activation of epithelial cells by fungal allergens leads to morphologic changes and cytokine production. J Allergy Clin Immunol. 2000;105:1185–1193. doi: 10.1067/mai.2000.106210. [DOI] [PubMed] [Google Scholar]
- Rudack C, Steinhoff M, Mooren F, et al. PAR-2 activation regulates IL-8 and GRO-alpha synthesis by NF-kappaB, but not RANTES, IL-6, eotaxin or TARC expression in nasal epithelium. Clin Exp Allergy. 2007;37:1009–1022. doi: 10.1111/j.1365-2222.2007.02686.x. [DOI] [PubMed] [Google Scholar]
- Zelante T, De Luca A, D'Angelo C, Moretti S, Romani L. IL-17/Th17 in anti-fungal immunity: what's new? Eur J Immunol. 2009;39:645–648. doi: 10.1002/eji.200839102. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Dong Z, Yang Z, Wang C. Expression of TLR2 and TLR4 messenger RNA in the epithelial cells of the nasal airway. Am J Rhinol. 2005;19:236–239. [PubMed] [Google Scholar]
- Claeys S, Van Hoecke H, Holtappels G, et al. Nasal polyps in patients with and without cystic fibrosis: a differentiation by innate markers and inflammatory mediators. Clin Exp Allergy. 2005;35:467–472. doi: 10.1111/j.1365-2222.2005.02215.x. [DOI] [PubMed] [Google Scholar]
- Ramanathan M, Jr, Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. Sinonasal epithelial cell expression of toll-like receptor 9 is decreased in chronic rhinosinusitis with polyps. Am J Rhinol. 2007;21:110–116. doi: 10.2500/ajr.2007.21.2997. [DOI] [PubMed] [Google Scholar]
- Lane AP, Truong-Tran QA, Schleimer RP. Altered expression of genes associated with innate immunity and inflammation in recalcitrant rhinosinusitis with polyps. Am J Rhinol. 2006;20:138–144. [PMC free article] [PubMed] [Google Scholar]
- Kato A, Schleimer RP. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr Opin Immunol. 2007;19:711–720. doi: 10.1016/j.coi.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson A, Bogefors J, Cervin A, Uddman R, Cardell LO. NOD-like receptors in the human upper airways: a potential role in nasal polyposis. Allergy. 2011;66:621–628. doi: 10.1111/j.1398-9995.2010.02527.x. [DOI] [PubMed] [Google Scholar]


