Significance
Bacteria can communicate with each other by small diffusible molecules, a process termed quorum sensing. Many bacteria use acylated homoserine lactones (AHLs) as signals, which are sensed by so-called LuxR-type receptors. With the photopyrones from the insect pathogenic bacterium Photorhabdus luminescens, we recently identified the first quorum sensing molecules different from AHLs that are sensed by a LuxR-type receptor. Here we describe the second novel quorum sensing molecule sensed by a LuxR-type receptor of Photorhabdus species, PauR of the human pathogen Photorhabdus asymbiotica. We demonstrate that P. asymbiotica communicates via dialkylresorcinols (DARs) and cyclohexanediones (CHDs). As the synthesis pathway is widespread, and often present in human pathogens, we discuss DARs and CHDs as novel and widespread signaling molecules.
Keywords: quorum sensing, cell–cell communication, LuxR solos, Photorhabdus, pathogenic bacteria
Abstract
It is well recognized that bacteria communicate via small diffusible molecules, a process termed quorum sensing. The best understood quorum sensing systems are those that use acylated homoserine lactones (AHLs) for communication. The prototype of those systems consists of a LuxI-like AHL synthase and a cognate LuxR receptor that detects the signal. However, many proteobacteria possess LuxR receptors, yet lack any LuxI-type synthase, and thus these receptors are referred to as LuxR orphans or solos. In addition to the well-known AHLs, little is known about the signaling molecules that are sensed by LuxR solos. Here, we describe a novel cell–cell communication system in the insect and human pathogen Photorhabdus asymbiotica. We identified the LuxR homolog PauR to sense dialkylresorcinols (DARs) and cyclohexanediones (CHDs) instead of AHLs as signals. The DarABC synthesis pathway produces the molecules, and the entire system emerged as important for virulence. Moreover, we have analyzed more than 90 different Photorhabdus strains by HPLC/MS and showed that these DARs and CHDs are specific to the human pathogen P. asymbiotica. On the basis of genomic evidence, 116 other bacterial species are putative DAR producers, among them many human pathogens. Therefore, we discuss the possibility of DARs as novel and widespread bacterial signaling molecules and show that bacterial cell–cell communication goes far beyond AHL signaling in nature.
Bacterial communication via small diffusible molecules to mediate group-coordinated behavior, referred to as quorum sensing, is well recognized. The prototypical quorum sensing system of Gram-negative bacteria consists of a LuxI-like autoinducer synthase that produces acyl-homoserine lactones (AHLs) as signals and a LuxR-type receptor that detects the AHLs to control expression of specific genes (1). Usually, luxI/luxR pairs are genetically clustered; however, there are examples in which the luxI/luxR functional pairs are distantly located on the bacterial chromosome or on plasmids (2). The AHLs are synthesized by LuxI and are sensed by the cognate LuxR-type receptor when exceeding a threshold concentration. On AHL binding, LuxR binds to the promoter/operator regions of the target genes/operons, resulting in changes in gene expression in response to the number of cells, and has been shown to play an important role in virulence of animal and human pathogens (1). As luxI is usually included among the target genes, setting up a positive feedback loop between signal input and output, AHLs are also designated as autoinducers.
In addition to this prototypical arrangement, many proteobacteria have LuxR homologs with no cognate LuxI autoinducer synthase or possess additional LuxR homologs in addition to a functional LuxI/LuxR quorum sensing system (3). Those LuxR homologs are designated as LuxR orphans (4) or LuxR solos (2). LuxR solos have been found in AHL-producing as well as in non-AHL-producing bacteria. They might allow bacteria to respond to endogenous as well as exogenous signals produced by their neighbors, exemplified by SdiA of Escherichia coli and Salmonella enterica, detecting AHLs produced by other bacteria (5). LuxR-type proteins commonly have a modular domain organization consisting of a conserved C-terminal DNA-binding domain and an N-terminal signal-binding domain, which typically is an AHL domain in AHL sensors and is important for ligand binding (6). Recently, we described that the LuxR solo PluR of the insect pathogen Photorhabdus luminescens detects α-pyrones named photopyrones (PPYs) instead of AHLs as signals. The PPYs are produced by a ketosynthase-like enzyme named photopyrone synthase PpyS (7). Therefore, PluR was the first example of a LuxR solo detecting a non-AHL endogenous signal. It regulates expression of the pcfABCDEF operon, resulting in the production of Photorhabdus clumping factor (PCF) that contributes to the pathogenicity of the bacteria (7). Furthermore, the three known Photorhabdus species harbor an exceptionally high number of LuxR-type receptors potentially sensing as-yet-unknown signaling molecules and enable the study of their role in cell–cell communication (8,9). However, much of the existing data on quorum sensing in Gram-negative bacteria rely on AHL signaling, whereas our study expands the diversity of signaling molecules particularly used for cell–cell communication in animal and human pathogenic bacteria.
Results
The DarABC/PauR Pair Represents a Novel Quorum Sensing Circuit.
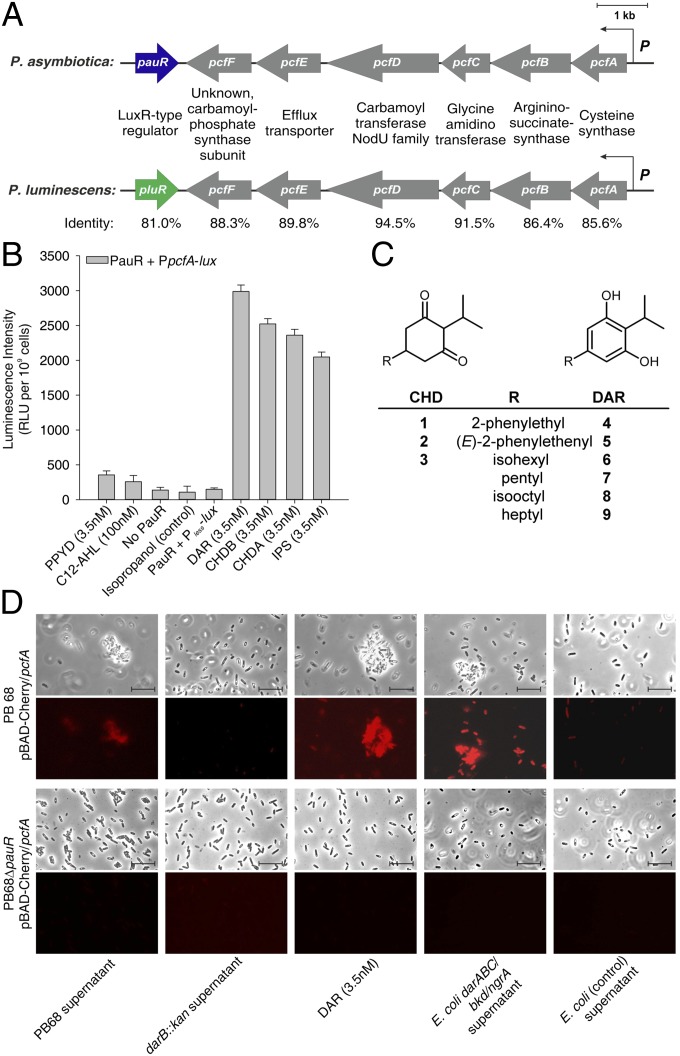
The insect and human pathogen P. asymbiotica harbors a pcf locus that is highly homologous to that of the closely related insect pathogen P. luminescens. It consists of the pcfABCDEF (pau_04068-pau_04063) operon and a neighboring gene encoding a LuxR solo we named PauR (Pau_04062). PauR shares 81% identity and 93% similarity with the LuxR solo PluR of P. luminescens (Fig. 1A). However, as a cognate signal synthase has been identified for PluR (PpyS) (7), it is disputable whether PluR can be still designated as a LuxR “solo.” Therefore, we designate PluR, as well as PauR, not as LuxR solos at this stage but, instead, simply as LuxR-type receptors. Interestingly, the P. asymbiotica genome does not encode a LuxI synthase and also lacks a ppyS gene. Hence, P. asymbiotica produces neither any AHLs nor PPYs. To test whether PauR can nevertheless sense exogenous PPYs or AHLs, we used an E. coli reporter strain similar to that which was already successfully used for PPY sensor PluR (7), which carried promoter fusion of PpcfA of P. asymbiotica with luxCDABE and PauR. Neither PPYs nor AHLs significantly induced PauR-mediated reporter gene expression (Fig. 1B and SI Appendix, Fig. S1). All Photorhabdus strains produce the dialkylresorcinol isopropylstilbene (compound 5) (Fig. 1C), which is important for nematode development (10) and acts as antibiotic (11). However, HPLC/MS analysis of several different Photorhabdus strains revealed a specific subset of dialkylresorcinols (DARs) (compounds 4 and 6–9; Fig. 1C) and their biochemical precursors cyclohexanediones (CHDs; compounds 1–3; Fig. 1C), which are exclusively produced in P. asymbiotica (SI Appendix, Fig. S2) and synthesized by a pathway encoded by the darABC operon (10, 12). Analysis of the previously isolated main derivatives 1, 3, 5, and 6 showed that all compounds were identified as specific inducers of PauR-mediated reporter gene activity (Fig. 1B). To elucidate whether DarABC/PauR is a cell–cell communication system, we used a P. asymbiotica PpcfA-mCherry reporter strain and investigated growth phase-dependent reporter gene activity. PpcfA activity was detectable in the early exponential growth phase and showed a significantly lower level in the late stationary growth phase (SI Appendix, Fig. S3). Furthermore, although P. asymbiotica is described as nonsusceptible to genetic manipulation in the literature (13), we were able to generate a ΔpauR deletion mutant in P. asymbiotica to test whether PpcfA activation is PauR-dependent, which was indeed the case (Fig. 1D and SI Appendix, Fig. S3). Neither fluorescence nor cell clumping was observed when conditioned medium of a P. asymbiotica darB::kan strain was used, proving that the darABC operon is essential for PauR-dependent pcfABCDEF activation (Fig. 1D and SI Appendix, Fig. S4). In addition, heterologous overproduction of the darABC operon in an E. coli strain harboring the bkd operon and ngrA resulted in production of DAR and CHD, which were released into the supernatant. This E. coli culture fluid then induced fluorescence and cell clumping in the P. asymbiotica PpcfA-mCherry reporter strain (Fig. 1D), revealing that DarABC/PauR constitutes a cell–cell communication circuit. This was not the case when using the ΔpauR PpcfA-mCherry strain as reporter (Fig. 1D), demonstrating that PauR is essential as a DAR/CHD signal receptor. Detailed analysis of selected DARs and CHDs to induce PpcfA activity revealed compound 6 (DAR) to be the most specific signal for PauR activation with a concentration as low as 3.5 nM (Fig. 1D and SI Appendix, Fig. S5).
Fig. 1.
The LuxR solo PauR specifically activates expression of the pcfABCDEF operon on induction with dialkylresorcinols. (A) Schematic presentation of the pcf/pauR and pcf/pluR locus of P. asymbiotica and P. luminescens, respectively. Identities of the respective protein sequences were generated by the ExPASy LALIGN Tool (embnet.vital-it.ch/software/LALIGN_form.html). (B) PauR specifically senses 2,5-dialkylresorcinol (DAR) and not photopyrones (PPYD) or acyl-homoserinelactones (C12-AHL). E. coli LMG194 strain harboring a PpcfA-luxCDABE (PpcfA-lux) fusion, as well as pBAD-pauR, were cultivated and exposed to different signaling molecules, PPYD 3.5 nM, 100 nM C12-AHL, isopropanol, 3.5 nM DAR (6), 3.5 nM CHDB (3), 3.5 nM CHDA (1), and 3.5 nM IPS (5), respectively. As controls, cells with no PauR or cells harboring a luxCDABE operon without a promoter (Pless) were used. Error bars represent SD of at least three independently performed experiments. RLU, relative light units. (C) Structures of known CHDs and DARs identified in Photorhabdus strains. (D) P. asymbiotica strain PB68.1 carrying plasmid pBAD-Cherry/pcfA from late stationary growth phase (PpcfA promoter activity is almost off) was exposed to different extracts (PB68.1 supernatant, PB68.1 darB::kan supernatant, E. coli LMG194 expressing darABC/bkd/ngrA, and E. coli LMG194 harboring empty plasmids) or pure DAR (6) and then analyzed for fluorescence as well as cell clumping in the microscope. The figure represents one characteristic of at least three independently performed experiments.
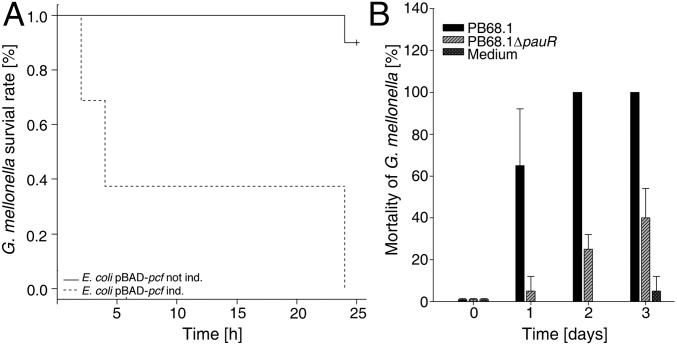
Analysis of the cell density-dependent promoter activities of pauR, pcfA, and darA in the wild-type and the ΔpauR strain (SI Appendix, Fig. S6) revealed an increase of PpauR activity in wild-type background in the exponential growth phase to reach a maximum in the stationary growth phase. Furthermore, a positive feedback regulation via PauR could be observed, indicating that DarABC/PauR constitutes a quorum sensing system, as PpauR activity was dramatically decreased in the ΔpauR background. PpcfA activity was maximal during the exponential growth phase and had a somewhat lower level in the stationary growth phase, which is comparable to the time-course of PpauR activity. In contrast, PdarA activity remained basal in both wild-type and ΔpauR cells. Reconstitution of the pcf operon, PauR, and the DAR biosynthesis in E. coli confirmed the role of all parts of this signaling circuit (SI Appendix, Fig. S7). Furthermore, the toxicity of E. coli overproducing PCF against the greater waxmoth Galleria mellonella revealed that PCF is also a virulence factor in P. asymbiotica, comparable with the P. luminescens PCF homolog (7) (Fig. 2A). Moreover, we observed that P. asymbiotica ΔpauR was less virulent towards G. mellonella (Fig. 2B). As we did not observe reduced virulence of the P. luminescens ΔpluR strain compared with the wild-type strain (7), we conclude that PauR-mediated cell clumping might have a greater effect on insect pathogenicity in P. asymbiotica than PCF in P. luminescens.
Fig. 2.
The DarABC/PauR quorum sensing circuit is important for pathogenicity. (A) Pathogenicity of E. coli harboring plasmid pBAD-pcfABCDEF, induced with 0.2% (wt/vol) arabinose or not induced (control), against G. mellonella. Ten larvae were infected, and the number of dead animals was recorded at the points after infection, as indicated. The portion of surviving animals was plotted versus the time according to the logrank method (31); P = 8.5 × 10−6. (B) Pathogenicity of P. asymbiotica PB68.1 and PB68.1 ΔpauR against G. mellonella. Cells were diluted in CASO medium, and ∼100 cells were injected in a volume of 10 μL into a G. mellonella last instar larva. Ten larvae were infected, and the number of dead animals was recorded at the points after infection, as indicated. The experiment was performed three times, and error bars represent the SDs.
Conserved Amino Acids in the Signal-Binding Domain of PauR Are Crucial for DAR Sensing.
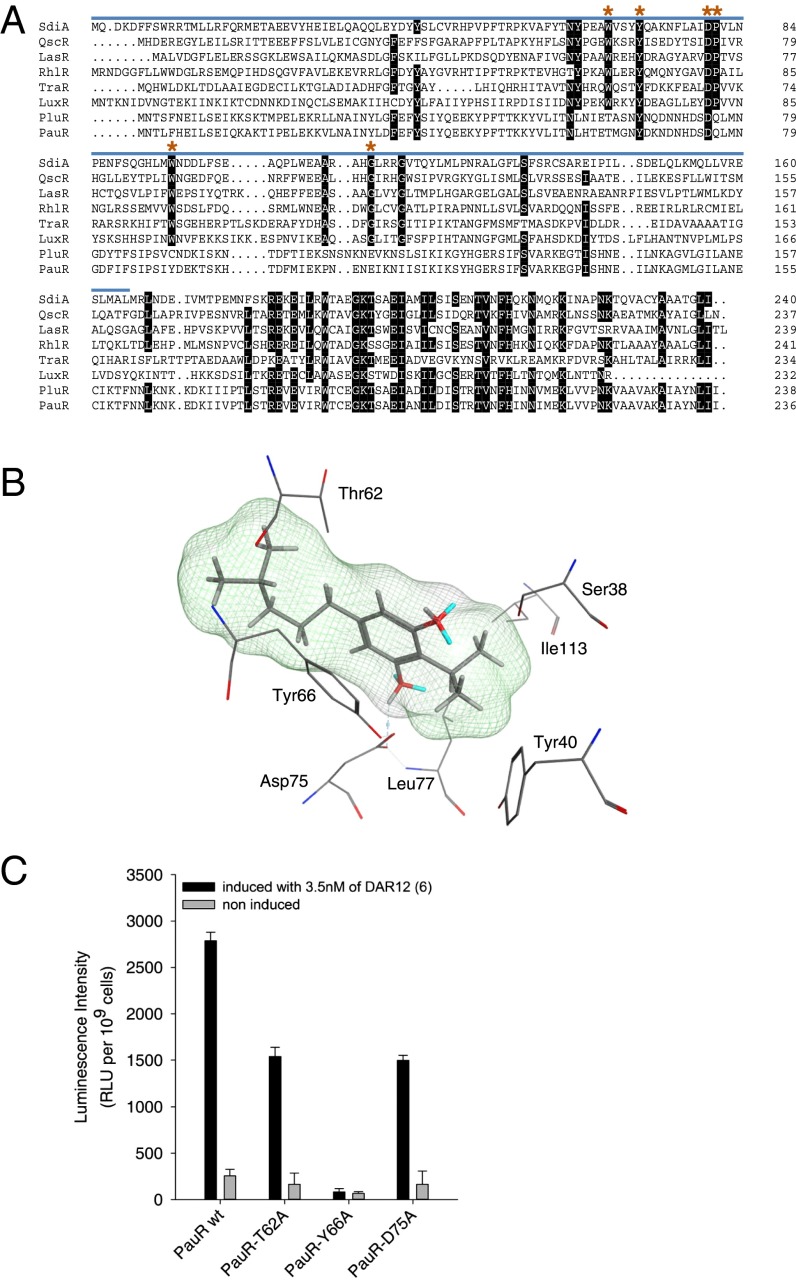
In general, the sequence identity of LuxR family members is quite low (18–25%), and as a consequence, the signal-binding domains of LuxR-type receptors (PFAM03472) are not highly conserved (Fig. 3A). Only six amino acids are classified as highly conserved in the AHL-binding domain of AHL sensors, which are Trp57, Tyr61, Asp70, Pro71, Trp85, and Gly113 (positions with respect to the AHL sensor TraR) (4, 14). The amino acids Trp57, Tyr61, and Asp70 (positions with respect to TraR) interact with the oxygen of the amide group and the carbonyl-oxygen of the lactone, as well as the amide group-nitrogen, respectively (15, 16). The observation that many LuxR homologs such as LuxR solos have substitutions of one or more of these six conserved amino acids originally prompted the idea that these proteins may sense signals other than AHLs, as recently demonstrated for PluR (7). In agreement with this observation, a sequence alignment of PauR and PluR against other receptors with homology to LuxR revealed that only two of six conserved amino acids (Tyr66, Asp75) are conserved in both proteins (Fig. 3A). As the third amino acid (Trp57 in TraR) that is involved in the AHL recognition is replaced with Thr in PluR, as well as PauR (position 62 in PluR and PauR, respectively), binding of a different signal molecule than AHL in both receptors seemed likely. To investigate a possible binding mode of DAR (6) in the active site of PauR, we generated a homology model of PauR based on the crystal structure of QscR from P. aeruginosa (17), which showed the highest sequence identity of available crystal structures to PauR (30.3%) (SI Appendix, Fig. S8). Docking experiments with this model using DAR (6) as ligand revealed a hydrogen bond formed between the DAR–hydroxy group and Asp75, as well as an arene–arene interaction between Thr62 and Tyr66 and the DAR aromatic ring (Fig. 3B). Replacement of Thr62 as well as Asp75 against Ala in PauR in fact reduced sensitivity of DAR (6) down to ∼50%, respectively. Moreover, replacement of Tyr66 against Ala caused complete loss of DAR (6) sensing (Fig. 3C), supporting the predictions of the in silico docking experiments. The loss in PauR binding to DAR (6) could also be demonstrated in silico, using the built-in residue scan function of Molecular Operating Environment (MOE) 2012.10 (SI Appendix, Table S4).
Fig. 3.
Dialkylresorcinol binding by PauR. (A) Alignment of SdiA of S. enterica var. Typhimurium; QscR, LasR, and RhlR of P. aeruginosa; TraR of Agrobacterium tumefaciens; LuxR of Vibrio fischeri; PluR of P. luminescens; and PauR of P. asymbiotica. Highly conserved amino acids (75–100%) are highlighted in black. The AHL-domain (“autoind_bind,” PFAM03472) is marked by the blue line. Amino acids involved in AHL-binding (Trp57, Tyr61, Asp70, Pro71, Trp85, and Gly113; positions numbered as in TraR) (4, 14, 19) are labeled with red asterisks. (B) A detailed view of the proposed PauR-binding pocket and the intermolecular interactions between the docked ligand 6 (DAR) and PauR. The cavity of the ligand is shown in a line representation, and possible hydrogen bonds are shown as dashed lines. (C) Influence of the amino acid replacements Thr62, Tyr66, and Asp75 against Ala in the signaling domain of PauR on sensing of DAR (6). E. coli LMG194 strains harboring a PpcfA-luxCDABE (PpcfA-lux) fusion and producing different PauR derivatives (PauR wild-type, PauR-T62A, PauR-Y66A, or PauR-D75A, respectively) were cultivated (noninduced) before 3.5 nM of 6 was added (induced). As controls, cells with no PauR or cells harboring a luxCDABE operon without a promoter (Pless) were used as shown in Fig. 1B. Error bars represent SD of at least three independently performed experiments. RLU, relative light units.
DAR-Dependent Quorum Sensing Systems in Other Pathogenic Bacteria.
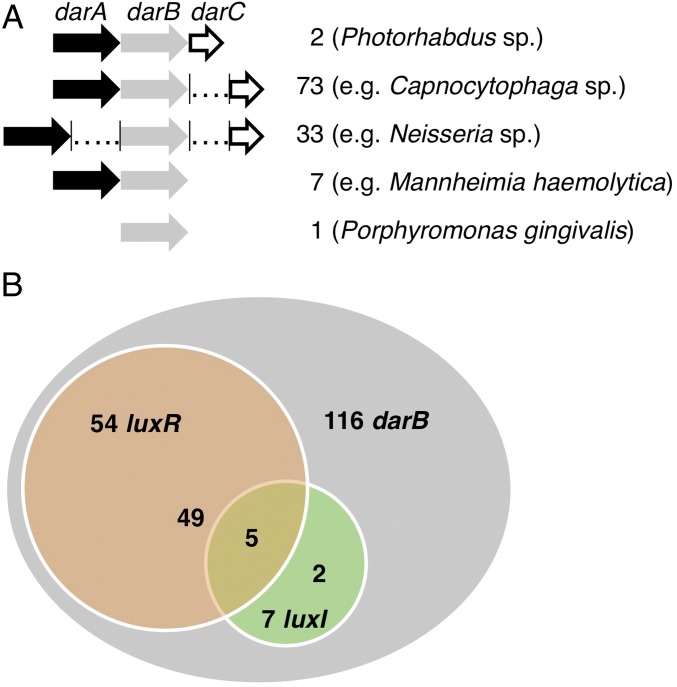
In this context, it is worth mentioning that the darABC operon responsible for the biosynthesis of DARs and CHDs has been detected in 116 bacteria (SI Appendix, Fig. S9 and SI Appendix, Table S5), several of which are pathogenic to humans, animals, or plants (12). Whereas darABC forms an operon in Photorhabdus strains, in most bacteria, darC is separate from darAB, and in approximately one-third of the analyzed bacteria, darA and darB are also separated from each other (Fig. 4A). Porphyromonas gingivalis ATCC 33277 is the only organism encoding DarB but not DarA, pointing to the exclusive presence of CHDs, but not DARs, in this strain. A detailed search of all darB-containing genomes for luxI and luxR homologs revealed that 46% of the bacteria encoding darABC also encode luxR homologs; however, LuxI homologs are rare in these bacteria and are mainly found in pseudomonads. In addition, 48% of bacterial genomes analyzed encode neither any LuxI nor a LuxR homolog, although possessing darB, suggesting a role for CHDs and DARs in these organisms other than acting as a quorum sensing signal in combination with LuxR-type receptors (Fig. 4B and SI Appendix, Fig. S10 and Table S7). Because LuxR-type receptors are not highly conserved (4), it is impossible to define whether a PauR solo, that is a LuxR homolog sensing DARs/CHDs in bacteria that do not harbor DarABC, exists.
Fig. 4.
Structure and assembly of darB containing operons (A) and correlation between darB, luxR, and luxI in darB encoding bacterial genomes (B).
Discussion
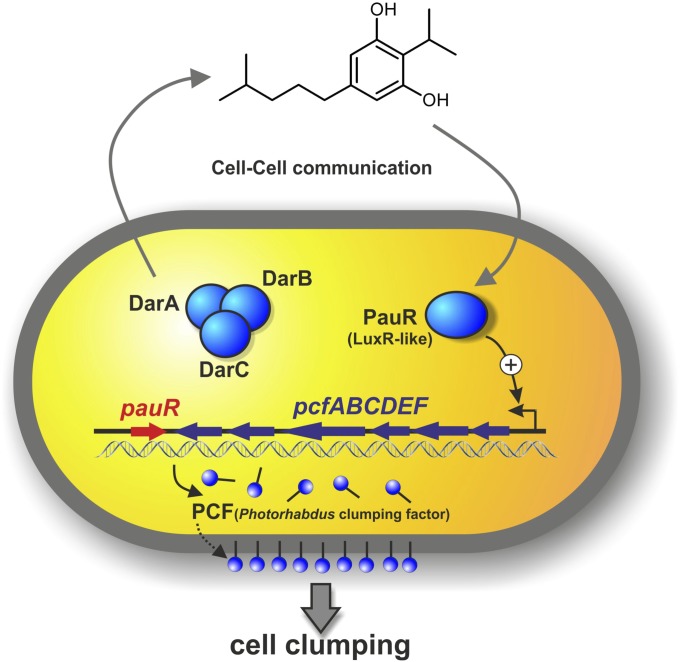
Here we identified a novel quorum sensing circuit that uses DARs/CHDs as signaling molecules. Thus, our findings reveal novel bacterial signaling molecules that are sensed by a LuxR homologous receptor. The LuxR-homologous receptor PauR does not sense AHLs or PPYs but, instead, DARs and CHDs, which are endogenously produced in a specific subset by the insect and human pathogenic bacterium P. asymbiotica. Furthermore, PauR most specifically detects DAR, and on ligand binding, the expression of the adjacent pcf operon is activated, leading to cell clumping. PCF-derived cell clumping then contributes to the high pathogenicity of P. asymbiotica toward insect larvae (Fig. 5). Indeed, quorum sensing-regulated processes are often linked with pathogenicity; for example, in Streptococcus pneumoniae or Vibrio cholerae, via regulating the expression of virulence factors at high cell density (1).
Fig. 5.
Model of the DarABC/PauR mediated quorum sensing system in P. asymbiotica, using dialkylresorcinol 6 as a signaling molecule.
PauR shows the typical domain structure of LuxR family proteins, consisting of a C-terminal DNA-binding domain and an N-terminal predicted AHL-binding domain. Recently, we have identified α-pyrones named PPYs as bacterial signaling molecules in P. luminescens used for quorum sensing, which are sensed by the LuxR homolog PluR (7). These two regulators, PauR and PluR, are highly homologous to each other; however, sensing different signaling molecules allows a species-specific cell–cell communication. P. asymbiotica is the only Photorhabdus species that is not only pathogenic toward insects but also toward humans. The use of DARs instead of PPYs as quorum sensing molecules might therefore be an important evolutionary step from invertebrate to human pathogenicity. Using quorum sensing molecules different from AHLs could be deeply rooted in the N-terminal signal-binding domain of PauR and PluR. In general, six conserved amino acids in signal-binding domain of AHL-sensing LuxR-type receptors are important for AHL binding. However, PauR and PluR only share two of them (Tyr66 and Asp75), which emerged as important for ligand sensing in all LuxR-type receptors (7, 9). Other than these subtle differences, PauR and PluR are very closely related to AHL-sensing LuxRs (SI Appendix, Fig. S11). In addition, the PauR-specific amino acid Thr62 in the signal-binding domain also emerged as important for DAR sensing. In addition to these, several other amino acids have been identified that shape the ligand pocket and are therefore crucial for signal-binding in AHL sensors (18). Moreover, a subfamily of LuxR-type proteins from plant-associated bacteria harbor substitutions within the six conserved amino acid motif in the signal-binding domain, suggesting the detection of plant-derived signal molecules, rather than AHLs (4, 19). Recently, these have been assumed to be part of a novel interkingdom signaling circuit and to be important for communication between plants and both pathogens and beneficial bacteria and might have undergone coevolution with the related host plant (20). This lends support to the idea that LuxR-homologous receptors have evolved to their distinct signaling molecules that are sensed for the adaptation to specific needs in different environments.
Unfortunately, the subtle differences of LuxR protein sequences, resulting in high specificities in ligand binding (Fig. 3A and SI Appendix, Fig. S11), hampers the correlation between DAR or CHD ligand specificity of PauR homologs identified in P. asymbiotica and other strains just based on protein sequences (SI Appendix, Fig. S11). Nevertheless, the high abundance of LuxR but low abundance of LuxI homologs in these bacteria (Fig. 4B) might indicate that the DarABC-derived compounds are indeed also used as signals in these bacteria. As among these bacteria are several important human, animal, and plant pathogens such as Neisseria, Capnocytophaga, or Flavobacterium, the role of these compounds for pathogenicity has to be explored in the future. CHD derivatives are known as natural products for 10 y (21) and have originally been described as pollinator attractant of Chiloglottis orchids as well as sex pheromones of its pollinator, the thynnine wasp Neozeleboria cryptoides (22). DAR derivatives have been described as antibiotics (10, 23), cytotoxins (24), free radical scavengers (25), and growth-stimulating factors (26) and can also be part of flexirubins and related pigments (12, 27, 28), which might be involved in protection against lipid peroxidation and photooxidative damage (29). Thus, especially in bacteria without LuxR homologs, DARs and CHDs might have additional functions other than acting as signals. Recently, a LuxI solo named SscI has been identified in the marine sponge symbiont Ruegeria sp. (30), revealing that similar to DARs, AHLs might fulfill functions other than acting as quorum sensing signal in those bacteria.
In summary, our finding adds a new chapter to bacterial cell–cell communication and shows that signaling via LuxR family regulators extends beyond AHL-derived quorum sensing. The fact that at least the biosynthesis gene clusters for DAR and CHD biosynthesis is widespread in different bacterial taxa (SI Appendix, Fig. S10), including several pathogenic species, might point to a wide distribution of these compounds, or even the underlying signaling circuits.
Materials and Methods
Strains and Plasmids.
All strains used in this study are listed in SI Appendix, Table S1, the plasmids in SI Appendix, Table S2, and oligonucleotides in SI Appendix, Table S3. For details of strain and plasmid construction see SI Appendix.
Materials.
All AHLs used in this study were purchased in reagent grade from Fluka (Deisenhofen). Kanamycin, ampicillin, gentamycin sulfate, and l-arabinose were obtained in reagent grade from Roth (Karlsruhe), and chloramphenicol was from Sigma-Aldrich (Deisenhofen). Isopropyl-β-D-1-thiogalactopyranoside (IPTG) was purchased from AppliChem (Darmstadt). All other materials were reagent-grade and were obtained from commercial sources.
Promoter Activity Analyses.
Fluorescence of P. asymbiotica mCherry reporter strains was investigated with a fluorescence microscope (Leica), using an excitation wavelength of 546 nm and a 605-nm suppression filter with 75-nm bandwidth, and quantified in an Infinite 500 Plate reader (Tecan) with an excitation wavelength of 560 nm (20-nm bandwidth) and an emission wavelength of 610 nm (20-nm bandwidth). The integration time was set to 20 μs, and the number of measurements was 10 for measurement of fluorescence and for optical density. Raw fluorescence data were normalized with the optical density (OD600) of the respective culture. A colony of the respective P. asymbiotica strain (pcfA-promoter activity is turned off) carrying the pBAD-Cherry-pcfA reporter plasmid was suspended in 100 µL P. asymbiotica culture supernatant or Casein-Soya-Peptone (CASO) medium, respectively, with equal optical densities. HPLC-derived samples of culture supernatants were added to cells resuspended in CASO medium in a ratio of 1:100. Purified DARs/CHDs were added in concentrations of 3.5–350 nM. Cells were then incubated aerobically at 37 °C for 60 min before fluorescence was analyzed. Time courses of pcfA, pauR, or darA promoter activities were performed by cultivating P. asymbiotica PB68.1 or P. asymbiotica PB68.1 ΔpauR carrying plasmid pBAD-Cherry/pcfA, pBAD-Cherry/pauR, or pBAD-Cherry/darA, respectively, in 24-well micotiter plates at 37 °C and 130 × g in an Infinite 500 Reader (Tecan, Austria) for 50 h. Every 30 min, fluorescence and OD600 was determined and normalized as described earlier.
Specificity of PauR Sensing Toward Different Signaling Molecules.
To quantify the specificity of the sensing of PauR toward distinct signaling molecules, E. coli LMG194 was transformed with plasmid pBAD24 containing pauR and a reporter plasmid carrying a pcfA-luxCDABE promoter fusion. The ability of PauR to activate pcfA promoter activity in the presence of different signaling molecules was measured via luxCDABE expression, and therefore luminescence as readout. Briefly, E. coli LMG194 carrying plasmids pBAD24-pauR and pBBR1-MCS5-TT-RBS-PpcfA-lux was cultivated overnight in M9 minimal medium at 37 °C. As controls, E. coli LMG194 carrying plasmids pBAD24-pauR and pBBR1-MCS5-TT-RBS-lux (promoter-less) and pBAD24 (empty plasmid) and pBBR1-MCS5-TT-RBS-PpcfA-lux, respectively, was used and cultivated overnight in M9 minimal medium at 37 °C. The overnight cultures were adjusted to an OD620 of 0.05 and then aerobically cultivated in 96-well plates at 37 °C. At an OD620 of 0.1, 3.5 nM of 6 (DAR) and 3.5 nM of photopyrone D (PPYD) or different AHLs in concentrations of 1, 10, and 100 nM (N-butyryl-DL-homoserinelactone, N-butyryl-DL-homocysteinthiolactone, N -3-oxo-hexanoyl-DL-homoserinelactone, N-octanoyl-DL-homoserinelactone, N-decanoyl-DL-homoserinelactone, N-dodecanoyl-DL-homoserinelactone, and N-tetradecanoyl-DL-homoserinelactone) was added. Isopropanol, used as solvent for PPYD, and ethylacetate, used as solvent for AHLs, were used as negative controls and added to the E. coli LMG194 cells harboring a PpcfA-luxCDABE (PpcfA-lux) fusion, as well as pBAD-pauR. Every hour, the OD620, as well as the luminescence, was monitored in a Sunrise plate reader (Tecan) and a Centro luminometer (Berthold Technologies), respectively.
Cell Clumping Assay.
To visualize cell clumping, the bacteria were analyzed by phase contrast microscopy. Briefly, E. coli LMG194 carrying plasmid pBAD24, pBAD-pcfABCDEF, or pBAD-pcfABCDEF/pauR, respectively, was cultivated overnight in M9 minimal medium, adjusted to an OD620 of 0.05, and then further cultivated. Expression of the pcfABCDEF operon was then induced by addition of 0.2% (wt/vol) arabinose or via the native promoter with 3.5 nM of 6 (DAR) at an OD620 of 0.5. After 1 h of further incubation at 37 °C under shaking with 200 × g in a Thermomixer (Eppendorf), 20 µL of each sample was analyzed for cell clumping in a DCI microscope (Zeiss), using phase contrast.
Effects of Amino Acid Replacements in PauR on DAR Sensing.
To quantify the influence of amino acid replacements in the signaling domain of PauR, E. coli LMG194 was cultivated with plasmid pBAD24 containing pauR or pauR derivatives and a reporter plasmid carrying a pcfA-luxCDABE promoter fusion. The ability of PauR and its derivatives to activate pcfA promoter activity in the presence of 6 (DAR) was measured via luxCDABE expression, and therefore luminescence as readout. Briefly, E. coli LMG194 carrying plasmids pBAD24-pauR and pBBR1-MCS5-TT-RBS-PpcfA-lux, pBAD-pauR-T62A and pBBR1-MCS5-TT-RBS-PpcfA-lux, pBAD-pauR-Y66A and pBBR1-MCS5-TT-RBS-PpcfA-lux, or pBAD-pauR-D75A and pBBR1-MCS5-TT-RBS-PpcfA-lux, respectively, was cultivated overnight in M9 minimal medium. As controls, E. coli LMG194 carrying plasmids pBAD24-pauR and pBBR1-MCS5-TT-RBS-lux (promoter-less) and pBAD24 (empty plasmid) and pBBR1-MCS5-TT-RBS-PpcfA-lux, respectively, were used and cultivated over night in M9 minimal medium. The overnight cultures were adjusted to an OD620 of 0.05 and were then aerobically cultivated in 96-well plates at 37 °C. At an OD620 of 0.1, 3.5 nM of 6 (DAR) was added and the OD620 as well as the luminescence were monitored in a Sunrise plate reader (Tecan) and a Centro luminometer (Berthold Technologies), respectively.
Pathogenicity Bioassays.
G. mellonella larvae (Terraristika Express) were surface sterilized in a 70% (vol/vol) ethanol bath followed by washing with sterile water and then incubated on ice for 10 min to reduce movements. Larvae were infected with cell suspensions by injection of 10 µL cell suspensions containing ∼100 P. asymbiotica cells or 4,000 E. coli cells, respectively, s.c., using a sterilized microsyringe (Hamilton 1702 RN, 25 µL), and incubated at room temperature. Mortality rate was determined by counting dead and alive animals after points indicated. Survival rate of E. coli-infected larvae was evaluated according to the log rank test (31).
Molecular Modeling of PauR and Docking.
The protein sequence of PauR was loaded into the MOE 2012.10. Then a BLAST search was performed to find an appropriate template crystal structure (32). For homology modeling, the crystal structure coordinates of QscR cocrystalized with N-3-oxo-dodecanoyl-l-homoserine lactone from Pseudomonas aeruginosa (PDB ID code 3SZT) was used. The sequence identity of PauR with its reference structure QscR was 30.3%. To avoid deletions or insertions in conserved regions, the alignment was inspected and corrected manually if necessary. A series of 10 models was constructed with MOE, using a Boltzmann-weighted randomized procedure combined with specialized logic for the handling of sequence insertions and deletions (33, 34). The model with the best packing quality function was selected for full-energy minimization. MOE packing score for PauR was 2.2882798, using Merck Molecular Force Field 99 × (MMFF99X). The stereo-chemical qualities of the model were assessed using Ramachandran plot: 4.2% outliner, 7.2% allowed, and 88.6% core.
Protein-ligand docking calculations were carried out using the program GOLD (version 5.1) (35), using the empirical scoring function for advanced protein-ligand docking CHEMPLP (36). The binding site of PauR was centered at D75, and the default docking parameters were used.
The virtual mutagenesis of PauR was carried out with the built-in residue scan function of MOE 2013.0802. The effect of the introduced mutations is then predicted by MOE, using the GBVI/WSA dG (37) scoring function to estimate the loss of affinity of the protein–ligand complex. The protein stability is predicted by MOE, using an energy equation to predict the change of stability of the wild-type and the mutant. To evaluate the residue scan function, the procedure was performed with the available crystal structure of QscR bound to N-3-oxo-dodecanoyl-homoserine lactone and the experimentally measured loss of affinity (17). The scores for ligand affinity and protein stability for the QscR derivatives are shown in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
Work in the R.H. laboratory was financially supported by the Deutsche Forschungsgemeinschaft (HE 5247/4-1 and SPP 1617). Work in the H.B.B. laboratory was supported by the Deutsche Forschungsgemeinschaft (SPP 1617) and a European Research Council starting grant under grant agreement 311477.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All DNA and protein sequences from P. asymbiotica PB68.1 and P. asymbiotica ATCC 43949 have been deposited in the GenBank database, www.ncbi.nlm.nih.gov/genbank/ [accession nos. KP258225 (ATCC 43949, PauR), KP258226 (ATCC 43949, PCF operon), KP258227 (ATCC 43949, DarABC), KP258228 (PB68.1, PauR), KP258229 (PB68.1, PCF operon), KP258230 (PB68.1, DarABC)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417685112/-/DCSupplemental.
References
- 1.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Subramoni S, Venturi V. LuxR-family ‘solos’: Bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155(Pt 5):1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- 3.Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: Their frequency and function among the Proteobacteria. ISME J. 2008;2(4):345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- 4.Patankar AV, González JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33(4):739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- 5.Ahmer BMM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52(4):933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A, et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(Database issue):D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brachmann AO, et al. Pyrones as bacterial signaling molecules. Nat Chem Biol. 2013;9(9):573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 8.Heermann R, Fuchs TM. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: Uncovering candidate genes involved in insect pathogenicity. BMC Genomics. 2008;9:40. doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brameyer S, Kresovic D, Bode HB, Heermann R. LuxR solos in Photorhabdus species. Front Cell Infect Microbiol. 2014;4:166. doi: 10.3389/fcimb.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joyce SA, et al. Bacterial biosynthesis of a multipotent stilbene. Angew Chem Int Ed Engl. 2008;47(10):1942–1945. doi: 10.1002/anie.200705148. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Chen G, Wu H, Webster JM. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol. 1995;61(12):4329–4333. doi: 10.1128/aem.61.12.4329-4333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs SW, et al. Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occurring ketosynthase. Angew Chem Int Ed Engl. 2013;52(15):4108–4112. doi: 10.1002/anie.201210116. [DOI] [PubMed] [Google Scholar]
- 13.Jones RT, et al. Photorhabdus adhesion modification protein (Pam) binds extracellular polysaccharide and alters bacterial attachment. BMC Microbiol. 2010;10:141. doi: 10.1186/1471-2180-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chugani SA, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98(5):2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem Biol. 2009;16(9):961–970. doi: 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R-G, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417(6892):971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 17.Lintz MJ, Oinuma K, Wysoczynski CL, Greenberg EP, Churchill MEA. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc Natl Acad Sci USA. 2011;108(38):15763–15768. doi: 10.1073/pnas.1112398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahumedo M, Díaz A, Vivas-Reyes R. Theoretical and structural analysis of the active site of the transcriptional regulators LasR and TraR, using molecular docking methodology for identifying potential analogues of acyl homoserine lactones (AHLs) with anti-quorum sensing activity. Eur J Med Chem. 2010;45(2):608–615. doi: 10.1016/j.ejmech.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 19.González JF, Venturi V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 2013;18(3):167–174. doi: 10.1016/j.tplants.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Covaceuszach S, Degrassi G, Venturi V, Lamba D. Structural insights into a novel interkingdom signaling circuit by cartography of the ligand-binding sites of the homologous quorum sensing LuxR-family. Int J Mol Sci. 2013;14(10):20578–20596. doi: 10.3390/ijms141020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiestl FP, et al. The chemistry of sexual deception in an orchid-wasp pollination system. Science. 2003;302(5644):437–438. doi: 10.1126/science.1087835. [DOI] [PubMed] [Google Scholar]
- 22.Franke S, et al. The discovery of 2,5-dialkylcyclohexan-1,3-diones as a new class of natural products. Proc Natl Acad Sci USA. 2009;106(22):8877–8882. doi: 10.1073/pnas.0900646106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanda N, Ishizaki N, Inoue N, Oshima M, Handa A. DB-2073, a new alkylresorcinol antibiotic. I. Taxonomy, isolation and characterization. J Antibiot (Tokyo) 1975;28(12):935–942. doi: 10.7164/antibiotics.28.935. [DOI] [PubMed] [Google Scholar]
- 24.Kronenwerth M, Brachmann AO, Kaiser M, Bode HB. Bioactive derivatives of isopropylstilbene from mutasynthesis and chemical synthesis. ChemBioChem. 2014;15(18):2689–2691. doi: 10.1002/cbic.201402447. [DOI] [PubMed] [Google Scholar]
- 25.Kato S, Shindo K, Kawai H, Matsuoka M, Mochizuki J. Studies on free radical scavenging substances from microorganisms. III. Isolation and structural elucidation of a novel free radical scavenger, resorstatin. J Antibiot (Tokyo) 1993;46(6):1024–1026. doi: 10.7164/antibiotics.46.1024. [DOI] [PubMed] [Google Scholar]
- 26.Imai S, et al. Studies on cell growth stimulating substances of low molecular weight. Part 3. Resorcinin, a mammalian cell growth stimulating substance produced by Cytophaga johnsonae. J Antibiot (Tokyo) 1993;46(8):1319–1322. doi: 10.7164/antibiotics.46.1319. [DOI] [PubMed] [Google Scholar]
- 27.Schöner TA, Fuchs SW, Reinhold-Hurek B, Bode HB. Identification and biosynthesis of a novel xanthomonadin-dialkylresorcinol-hybrid from Azoarcus sp. BH72. PLoS ONE. 2014;9(3):e90922. doi: 10.1371/journal.pone.0090922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schöner TA, Fuchs SW, Schönau C, Bode HB. Initiation of the flexirubin biosynthesis in Chitinophaga pinensis. Microb Biotechnol. 2014;7(3):232–241. doi: 10.1111/1751-7915.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poplawsky AR, Urban SC, Chun W. Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microbiol. 2000;66(12):5123–5127. doi: 10.1128/aem.66.12.5123-5127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zan J, et al. (October 29, 2014) A Solo LuxI-Type Gene Directs Acylhomoserine Lactone Synthesis and Contributes to Motility Control in the Marine Sponge Symbiont Ruegeria sp. KLH11. Microbiology, 10.1099/mic.0.083956-0. [DOI] [PMC free article] [PubMed]
- 31.Bland JM, Altman DG. The logrank test. BMJ. 2004;328(7447):1073. doi: 10.1136/bmj.328.7447.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fechteler T, Dengler U, Schomburg D. Prediction of protein three-dimensional structures in insertion and deletion regions: A procedure for searching data bases of representative protein fragments using geometric scoring criteria. J Mol Biol. 1995;253(1):114–131. doi: 10.1006/jmbi.1995.0540. [DOI] [PubMed] [Google Scholar]
- 34.Levitt M, Sharon R. Accurate simulation of protein dynamics in solution. Proc Natl Acad Sci USA. 1988;85(20):7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 36.Korb O, Stützle T, Exner TE. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model. 2009;49(1):84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 37.Corbeil CR, Williams CI, Labute P. Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des. 2012;26(6):775–786. doi: 10.1007/s10822-012-9570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.