Significance
When starved for nutrients, diatoms redirect carbon toward biosynthesis of storage lipids, triacylglycerols (TAGs). We examined how this modification is achieved in the diatom Phaeodactylum tricornutum. Under nitrogen stress, the cells cannibalized their photosynthetic apparatus while recycling intracellular nitrogen and redirecting it to synthesize nitrogen assimilation enzymes. Simultaneously, they allocated newly fixed carbon toward lipids. In contrast, a nitrate reductase knocked-down strain shunted ∼40% more carbon toward TAGs than the wild type without losing photosynthetic capacity. Our results show that diatoms can remodel their intermediate metabolism on environmental cues and reveal that a key signal in this remodeling is associated with nitrogen assimilation. This insight informs a strategy of developing a much more efficient pathway to produce algal-based biofuels.
Keywords: lipid, metabolism, stress, NR, RNAi
Abstract
Diatoms are unicellular algae that accumulate significant amounts of triacylglycerols as storage lipids when their growth is limited by nutrients. Using biochemical, physiological, bioinformatics, and reverse genetic approaches, we analyzed how the flux of carbon into lipids is influenced by nitrogen stress in a model diatom, Phaeodactylum tricornutum. Our results reveal that the accumulation of lipids is a consequence of remodeling of intermediate metabolism, especially reactions in the tricarboxylic acid and the urea cycles. Specifically, approximately one-half of the cellular proteins are cannibalized; whereas the nitrogen is scavenged by the urea and glutamine synthetase/glutamine 2-oxoglutarate aminotransferase pathways and redirected to the de novo synthesis of nitrogen assimilation machinery, simultaneously, the photobiological flux of carbon and reductants is used to synthesize lipids. To further examine how nitrogen stress triggers the remodeling process, we knocked down the gene encoding for nitrate reductase, a key enzyme required for the assimilation of nitrate. The strain exhibits 40–50% of the mRNA copy numbers, protein content, and enzymatic activity of the wild type, concomitant with a 43% increase in cellular lipid content. We suggest a negative feedback sensor that couples photosynthetic carbon fixation to lipid biosynthesis and is regulated by the nitrogen assimilation pathway. This metabolic feedback enables diatoms to rapidly respond to fluctuations in environmental nitrogen availability.
In plants, carbon and nitrogen are directed to specific tissues or structures in accordance with developmental programs. In contrast, unicellular algae flexibly direct carbon and nitrogen to various macromolecules associated with specific intracellular compartments to optimize growth under varying environmental conditions. The signals responsible for this optimization strategy are poorly understood. They clearly are not driven by a developmental program but rather, responses to environmental cues. For example, under optimal growth conditions, ∼40% of the photosynthetically fixed carbon in typical eukaryotic microalga is directed toward the synthesis of amino acids that ultimately are incorporated into proteins (1–3). Over 50 y ago, however, it was recognized that, when nitrogen limits growth, intermediate metabolism is altered, and many microalgae can accumulate storage lipids, mainly in the form of triacylglycerols (TAGs) (4–6). This phenomenon is especially pronounced in diatoms.
Diatoms, a highly successful class of eukaryotic algae that rose to ecological prominence during the past 30 My (7), often form massive blooms under turbulent conditions when nutrient supplies are highly variable (8). The ability of these organisms to optimize their growth under such conditions requires coordination of intermediate metabolism of carbon and nitrogen (9, 10). To optimize their growth, the first priority of the cells is to assimilate nitrogen into proteins, which also requires reducing equivalents and carbon skeletons that are primarily supplied by the tricarboxylic acid (TCA) cycle. However, when nitrogen availability decreases, the sink for TCA cycle metabolites declines, and acetyl-CoA, the source of carbon for the cycle, can be shunted toward fatty acid (FA) biosynthesis. Therefore, under nitrogen stress, cellular protein content decreases, whereas storage lipids increase (11, 12). This phenomenon has led to the hypothesis that overexpression of genes involved in lipid biosynthesis may increase the flux of carbon toward lipids (13, 14). Although this phenomenon is well-known, the signals that trigger the process remain unresolved. Genetic manipulations of lipid production in the model diatom, Phaeodactylum tricornutum, are ambiguous. Although there is one report showing that an overexpression of a type II diacylglycerol acyltransferase (DGAT; ProtID 49462) involved in TAG biosynthesis increases the accumulation of natural lipids in P. tricornutum (15), there are several reports indicating that manipulating FA biosynthesis does not significantly affect rates of lipid production (13, 14, 16).
Using biochemical, physiological, bioinformatic, and reverse genetic approaches, we examine here how a diatom remodels intermediate metabolism to rapidly respond to nitrogen stress and its resupply. Our results reveal how carbon is redirected toward lipid biosynthesis under nitrogen stress in P. tricornutum.
Results
Physiological Characteristics.
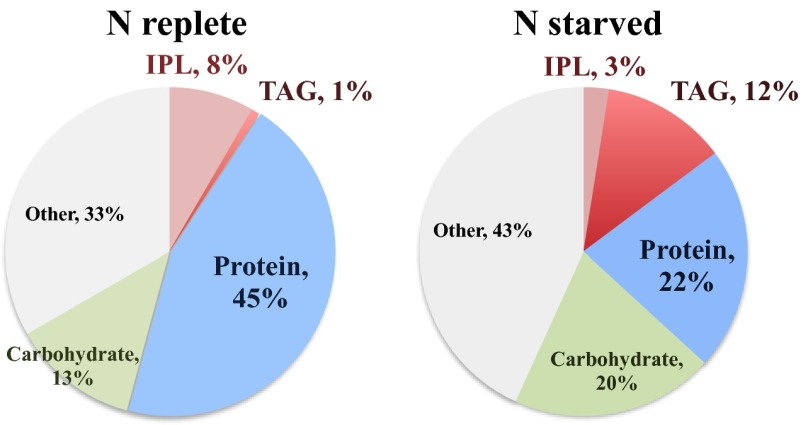
Under optimal growth conditions, when nitrate (as the sole nitrogen source) and other nutrients are not limiting and light is saturating for growth, ∼45% of the photosynthetically fixed carbon was incorporated into protein (Fig. 1). Under these conditions, only ∼9% of the cellular carbon was allocated into lipids, of which ∼1% was allocated for storage as TAGs. However, when being nitrogen-stressed, the fraction of the carbon that was shunted toward cellular protein decreased by 50%, whereas the carbon allocated to TAGs increased by an order or magnitude.
Fig. 1.
Allocation of cellular carbon to different biosynthetic compounds in nitrogen-replete and -stressed P. tricornutum 48 h after inoculation.
Biophysical analysis of the kinetics of variable chlorophyll fluorescence on the microsecond timescale (17) revealed that nitrogen-stressed cells had ∼50% lower photosynthetic energy conversion efficiency [determined as the ratio of variable fluorescence (FV) to maximal fluorescence (FM), FV/FM] in photosystem II (PSII), but the effective absorption cross-section of PSII reaction center (σPSII) was 17% ± 1%.
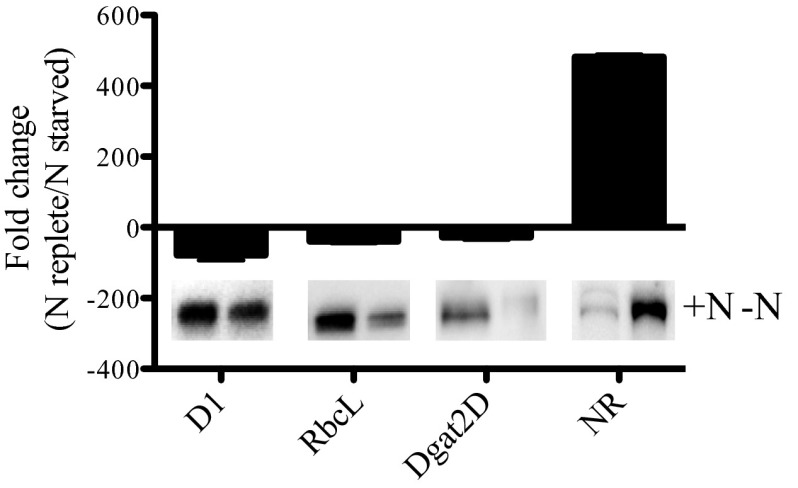
Although the abundance of total protein decreased by ∼50% under nitrogen stress, the losses were disproportionally distributed. Proteins involved in photosynthetic pathways decreased markedly. Western blot analysis revealed that the plastid-encoded proteins ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) and the D1 protein (PsbA), a core subunit of PSII, decreased by 60% and 20%, respectively. Despite the massive increase in carbon allocated to TAGs (Fig. 1 and Tables S1 and S2), DGAT type II (DGAT2D), a key rate-limiting enzyme associated with TAGs biosynthesis, decreased by 70% (Fig. 2). In contrast, nitrate reductase (NR), an enzyme essential for the assimilation of nitrate, increased by almost fourfold under nitrogen stress (Fig. 2).
Fig. 2.
Relative abundance of protein subunits related to photosynthesis, TAGs biosynthesis, and nitrogen assimilation in P. tricornutum under nitrogen-replete and -stressed conditions. The protein subunits are D1 (core protein of PSII), the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo, RbcL), type 2 diacylglycerol acyltransferase (DGAT2D), and nitrate reductase (NR). n = 3. Error bars represent ±1 SD.
Nitrogen stress also induced a change in the lipid profile; in conjunction with the marked increase in TAGs, nitrogen stress also led to threefold decrease in intact polar lipids (IPLs), which primarily are associated with membranes (Fig. S1 and Tables S1 and S2); consequently, the cellular IPL/TAG ratio was 25-fold higher in nitrogen-replete cells. In nitrogen-stressed cells, lipids associated with thylakoid membrane lipids, such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol (SQDG), accounted for >90% of the cellular IPLs and were dominated by DGDG, which can increase the structural stability of thylakoid membranes in diatoms (18) (Fig. S2 and Tables S1 and S2).
Transcriptome Analysis.
Analysis of the transcriptomes of axenic cultures in the two growth conditions yielded 10,234 sequences corresponding to 98% of the annotated genes (genome.jgi-psf.org/Phatr2), of which 5,620 genes were differentially expressed (DE) (Materials and Methods). Of the DE genes, 49% were up-regulated, whereas 51% were down-regulated in the nitrogen-stressed cells compared with the control. We further manually assigned genes related to metabolic pathways and functions to 1 of 16 categories and calculated the percentage of down-regulated, up-regulated, and non-DE (NDE) genes in each category (Fig. S2 and Dataset S1). This analysis revealed that genes related to nitrogen assimilation and the urea and TCA cycles were up-regulated, whereas genes related to lipid biosynthesis were mostly down-regulated (Figs. S2–S4).
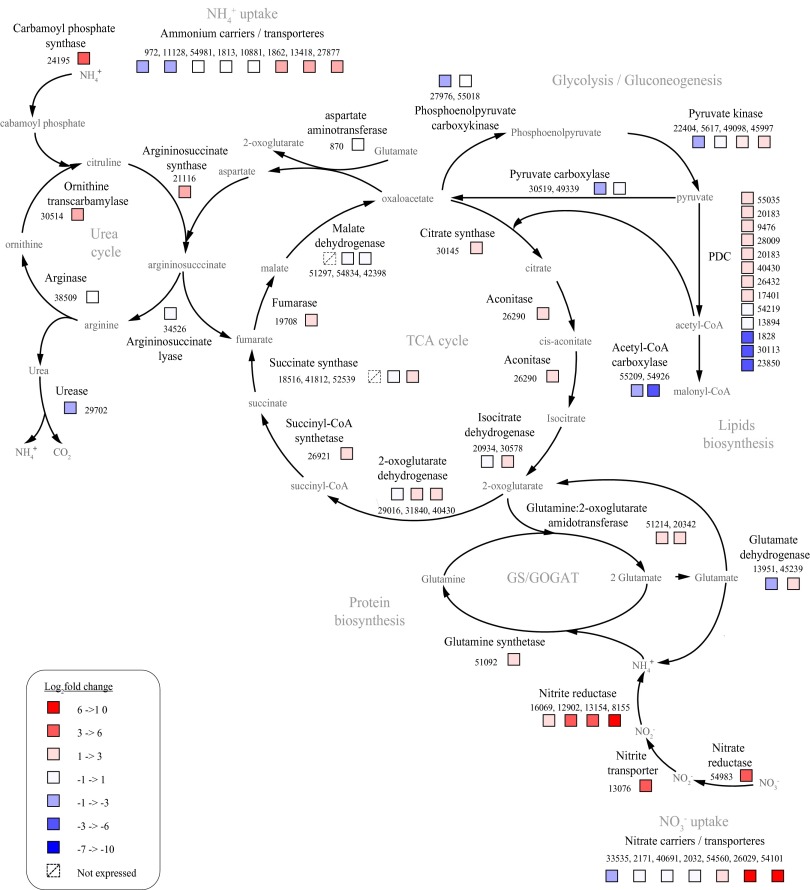
The most up-regulated genes in the transcriptome were associated with nitrate and urea uptake and assimilation [including the glutamine synthetase/glutamine 2-oxoglutarate aminotransferase (GOGAT) pathway], the pyruvate dehydrogenase complex, and the TCA cycle (Fig. 3). Genes related to chlorophyll biosynthesis and fucoxanthin chlorophyll a/c binding proteins were the most down-regulated; fucoxanthin chlorophyll a/c binding protein transcripts decreased by >300-fold. The expression of genes associated with photosynthetic carbon assimilation decreased markedly and was accompanied by down-regulation of the genes involved in both glycolytic and gluconeogenesis pathways. In addition, genes related to the pentose phosphate pathway and aerobic respiration exhibited a very high fraction of the NDE genes (Figs. S2 and S3 and Dataset S1).
Fig. 3.
Changes in transcript abundance of genes involved in carbon and nitrogen metabolism pathways between nitrogen-stressed and -replete conditions. A full description of the genes, exact fold-change values, and false detection rate can be found in Dataset S1. GOGAT, glutamine oxoglutarate aminotransferase; GS, glutamine synthetase; NH4+, ammonium; NO3−, nitrate; PDC, pyruvate dehydrogenase complex.
About one-third of all of the known genes related to lipids biosynthesis were down-regulated, including acetyl-CoA carboxylase, which represents a major branch point in intermediate metabolism that commits carbon to lipid biosynthesis (Fig. S4A and Dataset S1). Of the rest, 62% were NDE, and only 8% were up-regulated (Fig. S2 and Dataset S1). The genes responsible for TAGs metabolism in P. tricornutum are not well-annotated; however, based on the gene assignments, the expressions of genes related to TAGs biosynthesis were mostly NDE. Of five known DGAT type II, only dgat2D (43469) was up-regulated (Fig. S4B and Dataset S1). Fifty-seven percent of the genes related to biosynthesis of IPLs were NDE as were all genes related to biosynthesis of phospholipid, DGDG, and MGDG biosynthesis. The two genes related to SQDG biosynthesis were either NDE or down-regulated (Dataset S1).
Prediction of Metabolic Fluxes.
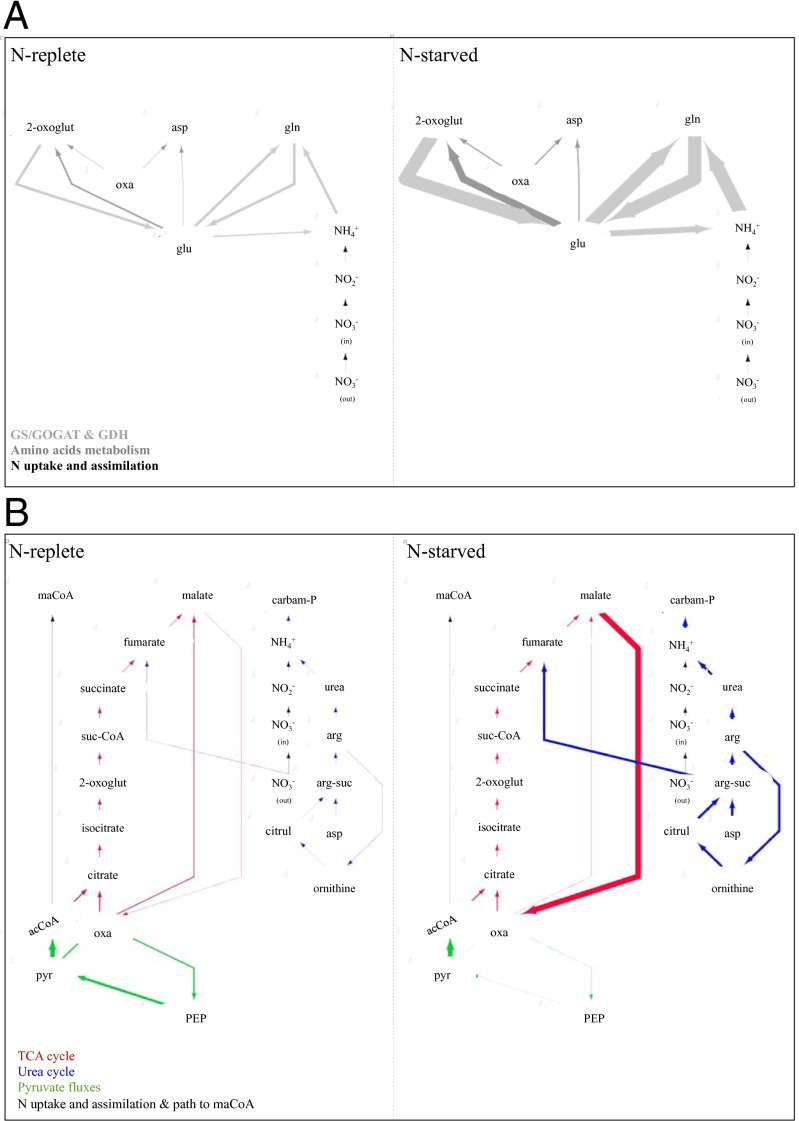
Based on an assumption of steady-state growth under the two conditions and the population of mRNAs of each of the genes in the transcriptomes, we calculated predicted fluxes of the intermediate metabolites of the cells. The analysis predicted that the fluxes of 92% of the reactions involving intermediate metabolites were lower than in nitrogen-replete conditions. The model further predicts that fluxes related to the production of glutamate, glutamine, and 2-oxoglutarate as well as those associated with the urea and TCA cycles increased significantly (Fig. 4).
Fig. 4.
Predicted metabolic fluxes of intermediate metabolites related to intermediate carbon and nitrogen metabolic pathways under nitrogen-replete and -stressed conditions. The flux line width represents the relative volume of the flux as calculated by the model. (A) Fluxes that represent recycling of internal nitrogen recycling. Dark gray, glutamate dehydrogenase- and aspartate aminotransferase-related fluxes; light gray, GS/GOGAT pathway; black, nitrogen uptake and assimilation related fluxes. (B) Fluxes that are mainly related to the TCA and the urea cycles. Fluxes are color-coded according to their pathway: black, nitrogen uptake and assimilation and shunting carbon toward FA metabolism; blue, urea cycle; green, pyruvate-related; red, TCA cycle. acCoA, acetyl-CoA; arg, arginine; arg-suc, argininosuccinate; asp, aspartate; carbam-P, carbamoyl-phosphate; citrul, citrulline; GDH, glutamate dehydrogenase; gln, glutamine; glu, glutamate; GS, glutamine synthetase; maCoA, malonyl CoA; NH4+, ammonium; NO2−, nitrite; NO3−, nitrate; oxa, oxaloacetate; 2-oxoglut, 2-oxoglutarate; PEP, phosphoenolpyruvate; pyr, pyruvate; suc-CoA, succinyl CoA.
Overall, this computational analysis suggests that nitrogen stress leads to a remodeling of intermediate metabolism centered around glutamate. This metabolic hub involves transfer of an amino group to form glutamine and subsequent reactions involving both glutamic dehydrogenase and GOGAT (Fig. 4A) and seems to critically conserve intracellular nitrogen by recycling protein degradation products through the urea cycle (Fig. 4B). Our metabolic flux analysis further predicts that this hub is coupled to the intermediate metabolism of carbon in the TCA cycle. Specifically, the analysis predicts an increase in the oxidation of malate to oxaloacetate, resulting in an increased flux of catabolically produced reductant (Fig. 4A). In contrast, the central hub involving pyruvate, phosphoenolpyruvate, and oxaloacetate is down-regulated in nitrogen-stressed cells, and this intermediate metabolic pathway is unlikely to be a significant source of carbon for lipid biosynthesis.
Characterization of an NR Knock-Down Strain.
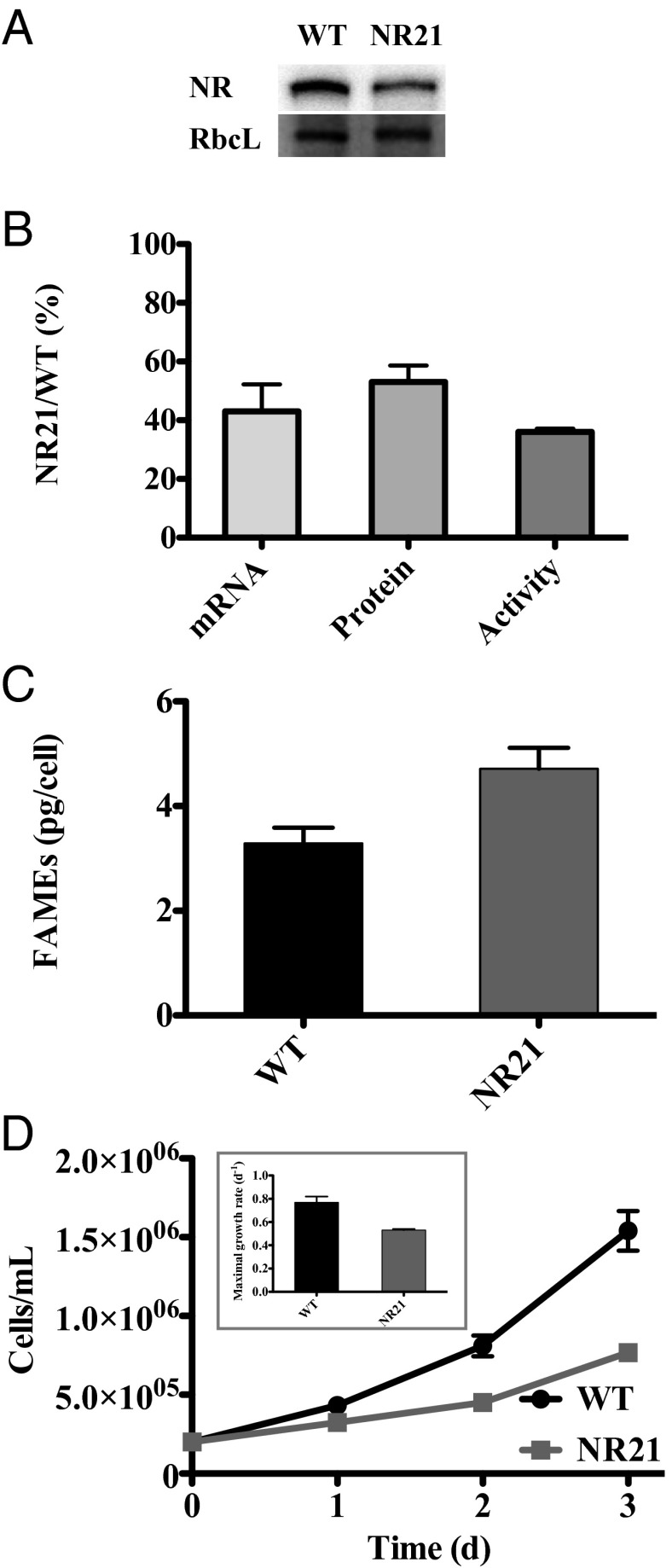
To further examine the cellular responses of diatoms to nitrogen stress, we generated an NR knock-down (KD) transformant strain, NR21. This transformant had ∼40% of the NR mRNA copy numbers, 50% of the NR protein amount, and 50% of the NR activity rates compared with the wild type (WT) under nutrient-replete conditions (Fig. 5 A and B). The change in NR is specific; the transformant had the same amount of total protein as the WT and the amounts of RuBisCo for both cell lines that were unchanged between the strains (Fig. 5A). Moreover, the strain’s FV/FM was identical to that of the WT. However, the strain grew slower with maximal growth rates that were 70% of the WT under nutrient-replete conditions (Fig. 5C). In effect, strain NR21 is a nitrogen-deficient phenotype, which is reflected by a 43% increase in the amount of lipids produced under nutrient-replete conditions (Fig. 5D).
Fig. 5.
Physiological and NR-related characteristics of exponentially grown WT P. tricornutum and the NR21 strain under nitrogen-replete conditions with nitrate as a sole nitrogen source. (A) NR and RbcL protein abundance. (B) NR characteristics: mRNA copies, protein abundance, and enzyme activity. (C) Fatty acid methyl esters (FAMEs). (D) Cell growth. (Inset) Maximal growth rates. n = 3. Error bars represent ±1 SD.
Discussion
The remodeling of intermediate metabolism resulting from nitrogen stress in the marine diatom, P. tricornutum, is analogous to physiological changes associated with the developmentally programmed process of grain filling in terrestrial grasses. During grain filling in wheat, for example, nitrogen stored in the photosynthetic proteins in the flag leaf is catabolized and redirected to the seeds as the plant matures (19, 20). Similar to the loss of nitrogen and impaired photosynthetic abilities of the wheat flag leaf (21), P. tricornutum becomes chlorotic under N stress and cannibalizes and remobilizes plastid proteins and polar lipids toward energy storage, primarily in the form of TAGs. In effect, diatom plastids function like an intracellular flag leaf under nitrogen-stressed conditions. Diatoms are able to accumulate lipids as a storage product, albeit within a single cell rather than a fruiting body (Fig. 1 and Tables S1 and S2). However, the remodeling of intermediate metabolism in nitrogen-stressed diatoms is not developmentally programmed. How is it executed in these single-celled algae?
Nitrogen stress sets a global constraint on protein translation by limiting amino acid biosynthesis (22). Thus, on nitrogen stress, P. tricornutum cells redirect their photosynthetically fixed carbon toward lipids, while concurrently, shunting their internal pools of nitrogen toward the nitrogen assimilation machinery (Fig. 2, Fig. S1, and Tables S1 and S2). The net effect of this intracellular reorganization is that cells can quickly respond to changes in external nitrogen availability, thereby affording a great competitive advantage in many aquatic environments where nitrogen can be introduced into the environment rapidly through turbulence (23). Thus, instead of filling grains with proteins and lipids, diatoms redirect intracellular nitrogen to enhance their nitrogen assimilation capacity at the expense of photosynthetic carbon assimilation (Fig. 2). Choreographing this intermediate metabolic dance requires one or more signal transduction pathways that lead to changes in both nuclear and plastid gene expression. Indeed, our transcriptomic data and computational flux model reveal that several central metabolic pathways, such as lipid biosynthesis, are down-regulated, whereas others, such as the TCA and nitrogen assimilatory cycles, are markedly up-regulated. Because internal cellular nitrogen is not shunted into synthesis of more lipid biosynthetic machinery, we propose a mechanism that leads to increased lipid accumulation in nitrogen-stressed cells.
Like in all other eukaryotic microalgae, in diatoms, the intermediate metabolism of carbon and nitrogen metabolism is closely coupled with a hub centered around glutamate and 2-oxoglutarate (Fig. 4). Our results suggest that this hub operates to redirect intracellular nitrogen derived from the catabolism of enzymes that are temporarily incapable of supporting growth toward a set of enzymes that confers selective advantage under nitrogen stress. The latter is primarily associated with nitrogen uptake and assimilation (including NR and GS/GOGAT) degradation of proteins, including the urea cycle and glutamate dehydrogenase, and energy-generating/anaplarotic reactions, such as the TCA cycle. This observation is consistent with other studies with WT P. tricornutum reporting the up-regulation of TCA cycle genes (24, 25) and the nitrogen uptake and assimilation genes under nitrogen stress (25). We hypothesize that the selection for the up-regulated pathways is so fundamental to the cell’s survival that suppressing a diatom’s ability to incorporate nitrogen must influence the cell’s carbon allocation. This hypothesis is supported by the phenotype of the NR21 KD transformant, where the restriction of nitrogen assimilation leads to massive accumulation of lipids without causing chlorosis or a significant change in photosynthetically capacity.
The most noteworthy change in the lipid profile of nitrogen-stressed P. tricornutum was the increase in TAGs accumulation and diversity paralleled by a decrease in IPLs (Fig. S1 and Tables S1 and S2). Regardless of whether TAGs biosynthesis is a result of the catabolism of IPLs or de novo synthesis of FAs, the final step is catalyzed by DGATs, which convert diacylglycerols to TAGs (26). Previously, it was reported that a DGAT gene could be up-regulated under nitrogen stress (24). In our study, only one of five DGAT paralogs, dgat2D (ProtID 43469), had a significant increase in transcript abundance (Fig. S4B), but its relative protein amount was 70% lower under N stress (Fig. 2). This apparent paradox can be simply explained by the decreased overall metabolic flux of carbon in the cell caused by a higher quantum requirement for growth and lipid biosynthesis (11). Thus, based on our results and previously published studies, we suggest that the subsequent accumulation of TAGs under nitrogen stress seems to be a consequence of allocation of carbon and reductant, rather than an up-regulation of lipid biosynthesis genes (Figs. 1 and 4 and Figs. S1–S4) (24, 25, 27).
Our results strongly suggest that the remodeling of intermediate metabolism due to nitrogen stress is a result of at least two different processes. The first process is related to shuffling of preformed protein nitrogen as reflected by the loss of photosynthetic machinery. The second process is related to redirecting photosynthetically fixed carbon from amino acids to other sinks, especially lipids, similar to the findings reported by Ge et al. (27). Although nitrogen-stressed cells recycle the nitrogen from preexisting photosynthetically fixed carbon toward nitrogen-deficient storage molecules, they continuously synthesize carbon skeletons, primarily in the forms of 2-oxoglutarate, fumarate, and malate (Figs. 3 and 4). These relatively oxidized intermediate metabolites become potential sinks for photosynthetically produced reductant, leading to the formation of lipids.
There is a clear difference in the allocation of reductant between the nitrogen-stressed WT and NR21. Although NR21 shifts only 4% of its cellular reductant from nitrate toward carbon reduction, we calculate a 70% increase in the fraction of NADPH that is used to reduce carbon to FA (Tables S3–S5) (28). In contrast, nitrogen-stressed cells shift 16% of their cellular reductants from nitrate to carbon reduction and increase the amount of reductant that is used for FA biosynthesis by 160%. This significant increase strongly suggests a massive accumulation of cellular reductant; because reductants are primarily synthesized in the plastid, it is highly likely that a retrograde signal alters the pattern of nuclear gene expression associated with nitrogen stress (29). Indeed, the phenotype of the NR21 KD strain clearly indicates that lipid biosynthesis can be uncoupled from protein shuffling (Fig. 5). Analysis of the transcriptome in the NR21 strain grown under nitrogen-replete conditions provides additional evidence that the increased lipid accumulation is not accompanied by up-regulation of the glutamate and 2-oxoglutarate hub gene. We further hypothesize that NR itself is a signal that is involved in the allocation of reductant toward lipids, whereas the plastid controls the shuffling of intercellular nitrogen. In addition, P. tricornutum operates a redox-dependent posttranslational mechanism that can support rapid responses to variable environmental conditions (30). Specifically, it was suggested that the redox regulation of nitrogen assimilation enzymes, including NR, could act as a feedback mechanism; the chloroplast redox state is monitored by those enzymes, and posttranslational modification of the enzyme plays a key role in regulating intermediate metabolism. The combined retrograde and NR feedback signals potentially allow the cell to rapidly respond to changes in nutrient availability in the environment (23). The physiological phenomenon has been described several times in shift-up metabolic experiments (31), where diatoms rapidly assimilate and horde inorganic N under a boom and bust strategy. This strategy has allowed diatoms to become extremely successful in highly turbulent environments (32, 33), where survivor cells become the seeds of new populations. However, the underlying mechanisms responsible for the responses have never been explained. Finally, the results presented here suggest that redirecting carbon toward lipid biosynthesis can be achieved genetically by altering the expression of a relatively small number of genes not directly involved with lipid biosynthesis.
Materials and Methods
Cultivation and Experimental Planning.
Cultivation of the P. tricornutum strain (accession Pt1 8.6, the Provasoli–Guillard National Center for Culture of Marine Phytoplankton) (34), the NR21 transformant, and nitrogen stress experiments were done following our previously described experiments (11) and as described in SI Materials and Methods. For characterizing the NR21 strain vs. the WT, optically thin cultures were sampled during exponential growth under nitrogen-replete conditions with nitrate as their sole nitrogen source.
Analytical Methods.
Exponentially growing cultures were analyzed for their IPLs TAGs and FAMEs using previously described methods (11, 12, 35–39). The percentage of carbon allocated to each biosynthetic compound was calculated given the following information: 16.5 and 9.4 pg C per cell, 13.6 and 3.6 pg protein per cell, and 7.7 and 5.5 pg carbohydrate per cell for the nitrogen-replete and -stressed cultures, respectively (11, 12); TAG and IPL percentages are calculated based on the data provided in Tables S1 and S2. Total protein determination was performed as previously described (11). The abundance of four selected proteins (PsbA, RbcL, NR, and DGAT2D) was determined by Western blots. PSII biophysical characteristics were measured on a custom-built fluorescence induction and relaxation instrument (17, 40). The kinetics of the single-turnover saturating flash were analyzed to obtain the maximum quantum efficiency of photochemistry (FV/FM) and the functional absorption cross-section of PSII (σPSII). Samples for quantitative PCR were collected, extracted, processed, and analyzed as previously described (11). The NR activity was measured following the method described by Eppley et al. (41) with modifications (3). Additional details are available in SI Materials and Methods.
RNA-Seq.
Samples for RNA-Seq were harvested and extracted from both the nitrogen-replete and -stressed cultures as described for the quantitative PCR samples. TruSeq RNA (Illumina) was used to prepare mRNA libraries for each of six samples according to the manufacturer’s instructions. The 50-bp single-ended libraries were multiplexed and sequenced on an Illumina MiSeq platform. The raw reads were trimmed for adaptor and low-quality sequences and then aligned to P. tricornutum’s version 2.0 set of 10,402 filtered gene models (genome.jgi.doe.gov/Phatr2/Phatr2.info.html) using CLC Genomics Workbench (v6.02) (42). Functional metabolic assignment for the different gene models were done according to KEGG database (43), Diatomcyc database (www.diatomcyc.org), and published literature (44, 45). Additional details on the data analysis can be found in SI Materials and Methods.
Computational Metabolic Flux Prediction.
Computational prediction of metabolic fluxes was done using an extension of flux balance analysis that infers a metabolic flux distribution from transcriptomic data (46–48). In our analysis, the minimum and maximum reaction rates of the flux balance analysis were set based on the expression level of the genes associated with each reaction. In addition, based on a given limited translational efficiency and a limited accumulation of enzyme over the time, we set the objective function to maximize the correlation between the flux vector and a vector of corresponding gene expression data. Additional details are available in SI Materials and Methods.
Construction of an Inverted Repeat Vector for Silencing NR, Genetic Transformation, and Screening.
An inverted repeat construct for silencing of the NR gene was generated using standard molecular cloning methods for RNAi based on the pKS-Sh ble-FA plasmid (49). The inverted repeat sequence was designed to match the C terminus of the NADH binding domain (50). The pKS-Sh ble nrIR-FA vector was inserted into P. tricornutum using a PDS-1000/He Particle Delivery System (Bio-Rad) as previously described (51). The insertion was verified by PCR of the antibiotic-resistant marker. To select for the best clone, survival curves were obtained by growing the transformants on 17 mM chlorate (ClKO3) (52). The best strain, NR21, was chosen as the most suitable strain for additional studies. Additional details about the transformation and screening methods are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Angela Falciatore for the gift of the pKS-Sh ble-FA plasmid, Andrew Allen for providing a nitrate reductase antibody for our initial work, and Helen Fredricks (Woods Hole Oceanographic Institution) for assistance with lipid analysis. We also thank Benjamin Bailleul (University of Liege) for constructive discussions and Arye Harel (Rutgers University) for advice on visualizing metabolic fluxes. This research was supported by the US Department of Energy Consortium of Algal Biofuels Commercialization Program, a gift from James G. Gibson (to P.G.F.), the Bennett L. Smith Endowment, and the Rutgers Energy Institute. L.T.G. was supported by a doctoral fellowship from the Portuguese Foundation for Science and Technology (FCT-MCTES) (SFRH/BD/61387/2009).
Footnotes
The authors declare no conflict of interest.
Data deposition: The DESeqs output for all 10,402 genes reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE56346), and all reads reported in this paper have been deposited in the National Center for Biotechnology Information’s Short Read Archive (accession no. SRP040703.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419818112/-/DCSupplemental.
References
- 1.Parsons TR, Stephens K, Strickland JDH. On the chemical composition of eleven species of marine phytoplankters. Journal of the Fisheries Research Board of Canada. 1961;18(6):1001–1016. [Google Scholar]
- 2.Myers J. In: On the Algae: Thoughts About Physiology and Measurements of Efficiency. Primary Productivity in the Sea, Environmental Science Research. Vol 19. Falkowski PG, editor. Springer; New York: 1980. pp. 1–16. [Google Scholar]
- 3.Berges JA, Harrison PJ. Nitrate reductase-activity quantitatively predicts the rate of nitrate incorporation under steady-state light limitation - a revised assay and characterization of the enzyme in three species of marine-phytoplankton. Limnol Oceanogr. 1995;40(1):82–93. [Google Scholar]
- 4.Orcutt DM, Patterson GW. Sterol, fatty acid and elemental composition of diatoms grown in chemically defined media. Comp Biochem Physiol B. 1975;50(4):579–583. doi: 10.1016/0305-0491(75)90093-0. [DOI] [PubMed] [Google Scholar]
- 5.Opute FI. Studies on fat accumulation in Nitzschia palea Kütz. Ann Bot (Lond) 1974;38(4):889–902. [Google Scholar]
- 6.Badour SS, Gergis MS. Cell division and fat accumulation in Nitzschia sp. grown in continuously illuminated mass cultures. Arch Mikrobiol. 1965;51(1):94–102. doi: 10.1007/BF00406853. [DOI] [PubMed] [Google Scholar]
- 7.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305(5682):354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 8.Falkowski PG, Oliver MJ. Mix and match: How climate selects phytoplankton. Nat Rev Microbiol. 2007;5(10):813–819. doi: 10.1038/nrmicro1751. [DOI] [PubMed] [Google Scholar]
- 9.Allen AE, et al. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature. 2011;473(7346):203–207. doi: 10.1038/nature10074. [DOI] [PubMed] [Google Scholar]
- 10.Zehr JP, Falkowski PG. Pathway of ammonium assimilation in a marine diatom determined with the radiotracer N13. J Phycol. 1988;24(4):588–591. [Google Scholar]
- 11.Guerra LT, et al. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy. 2013;59:306–315. [Google Scholar]
- 12.Frada MJ, Burrows EH, Wyman KD, Falkowski PG. Quantum requirements for growth and fatty acid biosynthesis in the marine diatom Phaeodactylum tricornutum (Bacillariophyceae) in nitrogen replete and limited conditions. J Phycol. 2013;49(2):381–388. doi: 10.1111/jpy.12046. [DOI] [PubMed] [Google Scholar]
- 13.Dunahay TG, Jarvis EE, Dais SS, Roessler PG. Manipulation of microalgal lipid production using genetic engineering. Appl Biochem Biotechnol. 1996;(57-58):223–231. [Google Scholar]
- 14.Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng. 2011;13(1):89–95. doi: 10.1016/j.ymben.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Niu YF, et al. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar Drugs. 2013;11(11):4558–4569. doi: 10.3390/md11114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitan O, Dinamarca J, Hochman G, Falkowski PG. Diatoms: Fossil fuel of the future. Trends Biotechnol. 2014;32(3):117–124. doi: 10.1016/j.tibtech.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Kolber ZS, Prášil O, Falkowski PG. Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: Defining methodology and experimental protocols. Biochimica et Biophysica Acta Bioenergetics. 1998;1367(1-3):88–106. doi: 10.1016/s0005-2728(98)00135-2. [DOI] [PubMed] [Google Scholar]
- 18.Mock T, Kroon BM. Photosynthetic energy conversion under extreme conditions—I: Important role of lipids as structural modulators and energy sink under N-limited growth in Antarctic sea ice diatoms. Phytochemistry. 2002;61(1):41–51. doi: 10.1016/s0031-9422(02)00216-9. [DOI] [PubMed] [Google Scholar]
- 19.Barbottin A, Lecomte C, Bouchard C, Jeuffroy M-H. Nitrogen remobilization during grain filling in wheat. Crop Science. 2005;45(3):1141–1150. [Google Scholar]
- 20.Simmons SR, Wheat S. Nitrogen and dry matter accumulation by kernels formed at specific florets in spikelets of spring wheat. Crop Science. 1978;18(1):139–143. [Google Scholar]
- 21.Bahrani A, Joo MH. Flag leaf role in N accumulation and remobilization as affected by nitrogen in a bread and durum wheat cultivars. Amer-Eurasian J Agric & Environ Sci. 2010;8(6):728–735. [Google Scholar]
- 22.Herzig R, Falkowski PG. Nitrogen limitation in Isochrysis galbana (haptophyceae). I. photosynthetic energy conversion and growth efficiencies. J Phycol. 1989;25(3):462–471. [Google Scholar]
- 23.Cermeño P, Lee J-B, Wyman K, Schofield O, Falkowski PG. Competitive dynamics in two species of marine phytoplankton under non-equilibrium conditions. Mar Ecol Prog Ser. 2011;429:19–28. [Google Scholar]
- 24.Yang Z-K, et al. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol Biofuels. 2013;6(1):67. doi: 10.1186/1754-6834-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valenzuela J, et al. Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol Biofuels. 2012;5(1):40. doi: 10.1186/1754-6834-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JE, Smith AG. A look at diacylglycerol acyltransferases (DGATs) in algae. J Biotechnol. 2012;162(1):28–39. doi: 10.1016/j.jbiotec.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Ge F, et al. Methylcrotonyl-CoA Carboxylase Regulates Triacylglycerol Accumulation in the Model Diatom Phaeodactylum tricornutum. Plant Cell. 2014;26(4):1681–1697. doi: 10.1105/tpc.114.124982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falkowski PG, Dubinsky Z, Wyman K. Growth-irradiance relationships in phytoplankton. Limnol Oceanogr. 1985;30(2):311–321. [Google Scholar]
- 29.Giordano M, Chen YB, Koblížek M, Falkowski PG. Regulation of nitrate reductase in Chlamydomonas reinhardtii by the redox state of the plastoquinone pool. Eur J Phycol. 2005;40(4):345–352. [Google Scholar]
- 30.Rosenwasser S, et al. Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc Natl Acad Sci USA. 2014;111(7):2740–2745. doi: 10.1073/pnas.1319773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dugdale R, Wilkerson E, Morel A. Realization of new production in coastal upwelling areas: A means to compare relative performance. Limnol Oceanogr. 1990;35(4):822–829. [Google Scholar]
- 32.Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanologica Acta. 1978;1(4):493–509. [Google Scholar]
- 33.Tozzi S, Schofield O, Falkowski P. Historical climate change and ocean turbulence as selective agents for two key phytoplankton functional groups. Mar Ecol Prog Ser. 2004;274:123–132. [Google Scholar]
- 34.Martino AD, Meichenin A, Shi J, Pan K, Bowler C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions1. J Phycol. 2007;43(5):992–1009. [Google Scholar]
- 35.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 36.Popendorf KJ, Fredricks HF, Van Mooy BA. Molecular ion-independent quantification of polar glycerolipid classes in marine plankton using triple quadrupole MS. Lipids. 2013;48(2):185–195. doi: 10.1007/s11745-012-3748-0. [DOI] [PubMed] [Google Scholar]
- 37.Holčapek M, Lísa M, Jandera P, Kabátová N. Quantitation of triacylglycerols in plant oils using HPLC with APCI-MS, evaporative light-scattering, and UV detection. J Sep Sci. 2005;28(12):1315–1333. doi: 10.1002/jssc.200500088. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Ruiz J, Belarbi E-H, Sánchez JLG, Alonso DL. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnology Techniques. 1998;12(9):689–691. [Google Scholar]
- 39.Holčapek M, Jandera P, Zderadička P, Hrubá L. Characterization of triacylglycerol and diacylglycerol composition of plant oils using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A. 2003;1010(2):195–215. doi: 10.1016/s0021-9673(03)01030-6. [DOI] [PubMed] [Google Scholar]
- 40.Falkowski PG, Koblížek M, Gorbunov M, Kolber Z. Development and application of variable chlorophyll fluorescence techniques in marine ecosystems. In: Papageorgiou GC, Govindjee, editors. Chlorophyll and Fluorescence. Springer; Berlin: 2004. pp. 757–778. [Google Scholar]
- 41.Eppley RW, Rogers JN, McCarthy JJ, Sournia A. Light/dark periodicity in nitrogen assimilation of the marine phytoplankters Skeletonema costatum and coccolithus huxleyi in N-limited chemostat culture. J Phycol. 1971;7(2):150–154. [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue D1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Martino A, et al. Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist. 2011;162(3):462–481. doi: 10.1016/j.protis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Rayko E, Maumus F, Maheswari U, Jabbari K, Bowler C. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 2010;188(1):52–66. doi: 10.1111/j.1469-8137.2010.03371.x. [DOI] [PubMed] [Google Scholar]
- 46.Brandes A, et al. Inferring carbon sources from gene expression profiles using metabolic flux models. PLoS ONE. 2012;7(5):e36947. doi: 10.1371/journal.pone.0036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colijn C, et al. Interpreting expression data with metabolic flux models: Predicting Mycobacterium tuberculosis mycolic acid production. PLOS Comput Biol. 2009;5(8):e1000489. doi: 10.1371/journal.pcbi.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Riso V, et al. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009;37(14):e96. doi: 10.1093/nar/gkp448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen AE, Ward BB, Song B. Characterization of diatom (bacillariophyceae) nitrate reductase genes and their detection in marine phytoplankton communities. J Phycol. 2005;41(1):95–104. [Google Scholar]
- 51.Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY) 1999;1(3):239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- 52.Cove DJ. Cholorate toxicity in Aspergillus nidulans: The selection and characterisation of chlorate resistant mutants. Heredity (Edinb) 1976;36(2):191–203. doi: 10.1038/hdy.1976.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.