Abstract
Methods for comparing associative relationships across ontologies often rely solely on lexical similarity between the names of the relationships, which may lead to missed matches and inaccurate matches. In this paper, we propose a novel method based on the analysis of paths between equivalent concepts across ontologies. Patterns of relationships are identified for each associative relationship. The most frequent patterns indicate a correspondence between an associative relationship in one ontology and one relationship (or combination thereof) in the other. We applied this method to two ontologies of anatomy. Our method was able to identify the correspondence between relationships even in the absence of lexical similarity between relationship names. The various types of matches identified are discussed as well as the application of this method to detecting inconsistencies across the ontologies.
Keywords: Ontology, associative relationship, hierarchical relationship, ontology matching, anatomy, GALEN, Foundational Model of Anatomy
Introduction
Knowledge representation systems generally consist of concepts modeled by hierarchical relationships. As importantly, concepts in ontologies are connected by associative relationships. While hierarchical relationships have been the object of careful inventories and standardization [1, 2], associative relationships tend to differ from system to system in names, semantics, and the constraints associated with their use. This is also because, unlike taxonomy and mereology which are required in virtually all ontologies, the theories expressed through associative relationships are generally specific to a subdomain. But even in a given subdomain, large differences may be observed in the use of associative relationships across systems, often corresponding to modeling choices.
Whether for merging, translating, or aligning, ontology matching techniques should consider not only the similarity among concepts, but also that among relationships, both hierarchical and associative. Due to the relatively limited and well-defined semantics of hierarchical relationships, their matching across ontologies is generally an easy task which can be carried out manually with limited domain knowledge. Associative relationships, in contrast, come in many different flavors dictated by the domain. Therefore, a different approach is required for matching them across ontologies. The objective of this paper is to identify equivalent expressions for associative relationships across two ontologies of anatomy. A secondary objective is to assess the consistency of associative relationships across ontologies.
We assume that one associative relationship in one ontology can be expressed in another ontology by either another associative relationship or a combination of associative and hierarchical relationships. Ideally, for a given associative relationship in one system, we expect to find a one-to-one correspondence in the other ontology. More realistically, correspondence to many or no relationships must also be considered. Finally, we assume the frequency with which a correspondence between two associative relationships is found to be a surrogate for the validity of the correspondence.
Our domain of interest for this study is anatomy. We selected two comprehensive ontologies representing anatomical knowledge: the Foundational Model of Anatomy (FMA) and the GALEN common reference model. This study logically follows previous work in which we identified equivalent concepts between FMA and GALEN using lexical resemblance between concept names and shared hierarchical relations [3, 4]. In this study, we focus on associative relationships with the objective of comparing their expression across systems. The expected benefit of this study is to provide additional clues for identifying equivalent concepts across systems.
Background
The general framework of this study is that of ontology matching. However, few of the tools and algorithms developed for ontology matching deal with the issue of comparing associative relationships. For example, those only considering taxonomical relationships include the Chimaera environment for merging and testing ontologies [5], the bottom-up FCA-MERGE method for structurally merging ontologies [6], and the machine learning based GLUE system for identifying similar concepts with higher probabilistic measure values [7]. PROMPT, as one the few tools to consider associative relationships, suggests merging relationships across ontologies when they have linguistically similar names [8]. Anchor-PROMPT searches for paths through associative and hierarchical relationships for finding semantically similar concepts, but ignores what relationships are on the paths or if they match across ontologies [9].
Methods for comparing associative relationships based solely on similarity among relationship names are notoriously weak. These methods fail to identify similar relationships having different names and may wrongly associate different relationships having resembling names. But more importantly here, the correspondence between one relationship and a combination of relationships could not be discovered by such methods. The major contribution of this paper is to propose a novel method for identifying semi-automatically the correspondence between associative relationships across ontologies.
Materials
The Foundational Model of Anatomy1 (FMA) [March 4, 2003 version] is an evolving ontology that has been under development at the University of Washington since 1994 [10, 11]. Its objective is to conceptualize the physical objects and spaces that constitute the human body. The underlying data model for FMA is a frame-based structure implemented with Protégé-2000. The 66,879 concepts in FMA cover the entire range of macroscopic, microscopic, and subcellular canonical anatomy.
The Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine2 (GALEN) [v. 6] has been developed as a European Union AIM project led by the University of Manchester since 1991 [12, 13]. The GALEN common reference model is a clinical terminology represented using GRAIL, a formal language based on description logics. GALEN contains 52,006 concepts and intends to represent the biomedical domain, of which canonical anatomy is only one part.
Both FMA and GALEN are modeled by ISA and PART_OF relationships and allow multiple inheritance. Relationships in GALEN are generally finer-grained than in FMA. For the purpose of this study, we considered as only one PART_OF relationship the various kinds of partitive relationships present in FMA (e.g., part of, general part of) and in GALEN (e.g., isStructuralComponentOf, isDivisionOf). ISA and PART_OF have inverse relationships, INVERSE_ISA and HAS_PART. Additionally, there are 59 kinds of associative relationships between concepts in FMA. While most of them have inverses (e.g., branch of and branch), a few do not (e.g., input from). GALEN has 562 associative relationships and all of them have inverses (e.g., isBranchOf and hasBranch, isServedBy and serves).
Methods
Methods for comparing associative relationships are based on a group of equivalent concept pairs across two ontologies. Using the lexical and structural alignment method described in [3], 2,604 equivalent pairs of concepts were identified between FMA and GALEN, accounting for about 4% of FMA concepts and 5% of GALEN concepts. For example, Pancreas in FMA and Pancreas in GALEN match as they have the same name and share some hierarchical relationships to other equivalent concepts (e.g., HAS_PART Exocrine pancreas and Head of pancreas). These concepts are also called anchors because they are going to be used for matching the associative relationships.
Under our hypothesis, a correspondence between relationships across ontologies is indicated by the frequent association between one inter-concept relationship in one ontology and either another relationship or a combination of relationships between the equivalent concepts in the other ontology. Thus, our method consists of identifying for each associative relationship in FMA the corresponding relationship (or combination thereof) in GALEN. The same process is applied starting with GALEN relationships.
Acquiring associative relations
Inter-concept associative relationships are generally represented by semantic relations <concept1, relationship, concept2>, where concept1 links to concept2 through relationship. Acquiring associative relations consists of extracting the relations explicitly represented and complementing the missing inverse relations. In canonical anatomy, the inverse relations are essentially always valid, although this may not necessarily be the case in the real world [14]. For example, <NasalCavity, isServedBy, ArteryOfNasalPassage> was complemented in GALEN from an explicit relation <ArteryOfNasalPassage, serves, NasalCavity>.
Instead of explicitly representing the relation of concept X to concept Y through an associative relationship such as <X, branch_of, Y>, ontologies sometimes reify this associative relationship in a hierarchical relation between X and a concept called Branch of Y, i.e., <X, ISA, Branch of Y>. These two relations are semantically equivalent. In order to facilitate comparisons across ontologies, in each ontology, we made explicit the relations implicitly embedded in the concept names (reified). We applied this augmentation technique to the reified branch of and tributary of relationships in FMA and isBranchOf in GALEN. For example, <Lateral cutaneous nerve of forearm, branch of, Musculocutaneous nerve> was added to FMA from an explicit hierarchical relation <Lateral cutaneous nerve of forearm, ISA, Branch of musculocutaneous nerve>.
Identifying relationship patterns
As anchors represent the correspondence between equivalent concepts across ontologies, relationship patterns represent the correspondence between relationships (or combination thereof) across ontologies. Such patterns are identified by investigating the relationships among anchors in the two systems. More precisely, for each associative relationship between two anchors in one ontology, we searched for all shortest paths between the same two anchors in the other ontology. We ignored paths involving more than six relationships because it would be both unlikely to find them with the high frequency sought and difficult to determine their semantics. Both hierarchical and associative relationships are allowed in the paths. However, we ignored the paths where an associative relationship and its inverse are present because such paths are usually not indicative of an associative relation of interest between the two anchors. For example, LiverÆ isServedBy Æ AutonomicNerveOfAbdomenÆ serves Æ SurfaceOfLiver was ignored for this reason. An associative relationship between two anchors in one ontology and a combination of relationships between the same two anchors in the other ontology compose a path pair.
Concepts are removed from the paths to create relationship patterns. Additionally, these patterns are simplified by representing several successive relationships of the same kind by only one relationship. However, multiple occurrences of a relationship are left intact if separated by other relationships. These transformations generate pattern pairs from path pairs. For example, from the path pair:
FMA: Pancreas → arterial supply → Dorsal pancreatic artery
GALEN: Pancreas → isServedBy → CaudalPancreaticArtery → isBranchOf → InferiorPancreaticArtery → isBranchOf → Dorsal-PancreaticArtery
the following pattern pair is obtained:
FMA: arterial supply
GALEN: isServedBy - isBranchOf
This pattern pair is indirect as it involves more than one relationship. It would be direct otherwise. In order to assess the consistency of associative relationships across ontologies, we scrutinized the associations of a relationship in one ontology to both a given pattern and its inverse in the other ontology. Toward this endeavor, inverse patterns were systematically generated by reversing the order of the relationships in the pattern and replacing each relationship by its inverse. Examples of inverse patterns include isServedBy - isBranchOf and hasBranch - serves. Finally, the frequency of each pattern pair (i.e., the number of paths pairs this pattern pair comes from) was recorded in order to select only the most frequent pairs (as they are also expected to be the most significant ones), thus ignoring “accidental” pattern pairs.
Results
Associative relations acquired
The number of associative relations acquired in FMA and GALEN is listed in Table 1. Among the total number of associative relations, complemented relations have a much larger proportion in GALEN than in FMA. Conversely, augmentation techniques generated much more relations in FMA than in GALEN. The last row in Table 1 shows the number of associative relations among the 2,604 anchor concepts in FMA and GALEN.
Table 1.
Number of associative relations in FMA and GALEN
| Associative relations | FMA | GALEN |
|---|---|---|
| Explicit | 18,688 | 288,732 |
| Complemented | 1,057 | 249,938 |
| Augmented | 1,838 | 108 |
| Total | 21,583 | 538,778 |
| Among anchors | 847 | 6,922 |
Path pairs and pattern pairs identified
4,070 inter-anchor path pairs between FMA and GALEN were obtained. 350 pattern pairs were identified from these path pairs, and 47 of them are direct pattern pairs.
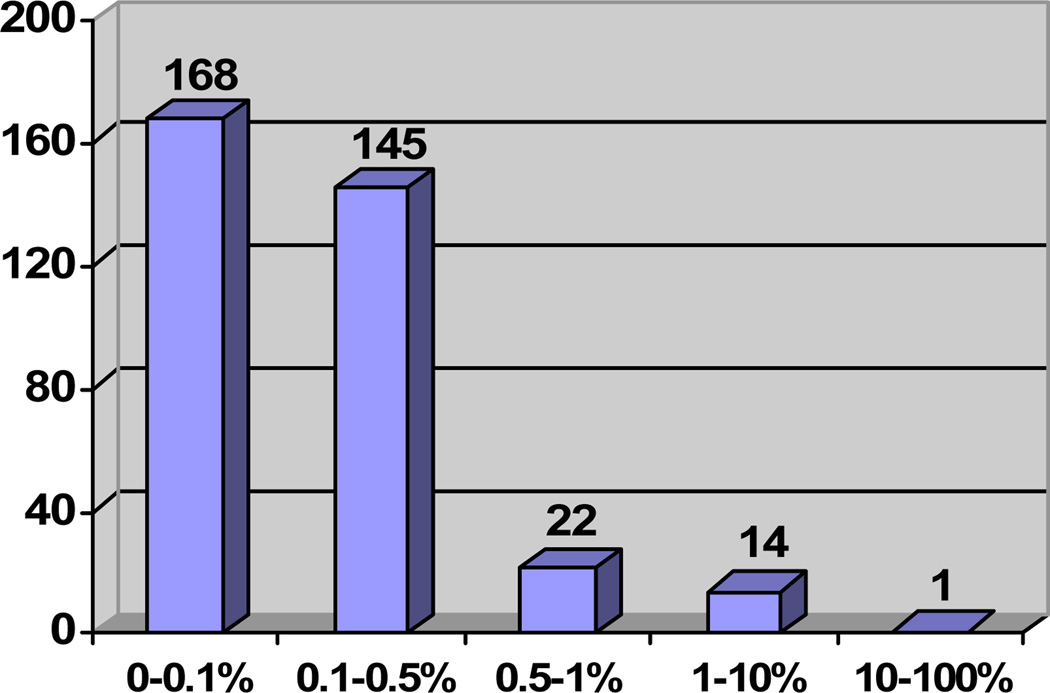
Figure 1 presents the number of pattern pairs with different frequency intervals, and Table 2 lists some examples of pattern pairs. Figure 1 shows that a small number of patterns occur with a high frequency, while the majority of patterns occur much less frequently (often only once or twice). The pattern pair with the highest frequency is {FMA: PART_OF, GALEN: isBranchOf}, and 518 path pairs have this pattern resulting in a frequency of nearly 13% of the total 4,070 path pairs. The pattern pair with the second highest frequency (8%) is {FMA: branch of, GALEN: isBranchOf}, shared by 310 path pairs. For the leftmost bar in Figure 1, each of the 168 patterns is supported by three path pairs or less. One of these low frequency pattern pairs is {FMA: contained in, GALEN: boundsSpace - INVERSE_ISA}, shared by two path pairs.
Figure 1.
Frequency distribution of pattern pairs
Table 2.
Example of pattern pairs
| FMA | GALEN | Frequency | |
|---|---|---|---|
| part_of | is Branch Of | 518 | 13 % |
| branch of | is Branch Of | 310 | 8 % |
| has_part | is To | 166 | 4 % |
| tributary of | is Branch Of | 104 | 3 % |
| member of | is a | 42 | 1 % |
| nerve supply | part_of - is Served By | 16 | 0.4 % |
| part_of - cont ained in | is Non Partitively Containe d In | 10 | 0.25 % |
| contained in | Bounds Space - inverse_is a | 2 | 0.05 % |
Relationship matches
Simple matches (1:1)
There are cases where one associative relationship in one ontology matches only one relationship pattern in another ontology. For example, as shown in Table 2, for the associative relationship member of in FMA, 42 path pairs were found in GALEN and all of them share one pattern, {FMA: member of, GALEN: ISA}.
Multiple matches (1:n)
On the other hand, many associative relationships correspond to multiple relationship patterns. For example, for the associative relationship arterial supply in FMA, 74 path pairs were found from which multiple patterns were extracted. High-frequency patterns are listed in Table 3. Among the inter-anchor arterial supply relations in FMA (74 in all), 24% are represented in GALEN by isServedBy, 46% by isServedBy combined with another relationship, and 30% by other combinations of relationships.
Table 3.
Multiple matches for arterial supply
| FMA | GALEN | Frequency | |
|---|---|---|---|
| arterial supply | is Served By | 18 | 24 % |
| Is Served By - is Branch Of | 16 | 22 % | |
| Is Served By - PART_OF | 12 | 16 % | |
| Is Served By - ISA | 4 | 5 % | |
| Is Served By -INVERSE_ISA | 2 | 3 % | |
| Other combinations | 22 | 30 % | |
Discussion
Lexical vs. semantic correspondence
Although we did not use lexical methods to match associative relationships, we can compare the results of our method to that of lexical techniques. The following three circumstances may occur between two associative relationships across ontologies.
Similar relationship names and semantics: three occurrences (e.g., {FMA: branch of, GALEN: isBranchOf}, and one of its path pairs is Perineal nerve → branch of → Pudendal nerve in FMA and Perineal-Nerve → isBranchOf → PudendalNerve in GALEN). This represents the ideal case.
Similar relationship names and differing semantics: four occurrences (e.g., no paths were found to support the pattern {FMA: bounded by, GALEN: isSpaceBoundedBy} that could have been suggested lexically). This illustrates why we did not want to rely on lexical similarity for matching relationships.
Different relationship names and similar semantics: eleven occurrences (e.g., {FMA: nerve supply, GALEN: isServedBy}, and one of its path pairs is Pronator tere → nerve supply → Median nerve in FMA and PronatorTeres → isServedBy → MedianNerve in GALEN). This mapping would have been missed by methods relying solely on lexical similarity.
Analysis of relationship matches
In 87% of the cases, one associative relationship corresponds to a combination of hierarchical and associative relationships in another ontology (e.g., {FMA: arterial supply, GALEN: isServedBy - ISA}, and one of its path pairs is Liver → arterial supply → Hepatic artery in FMA and Liver → isServedBy → Intermediate- HepaticArtery → ISA → HepaticArtery in GALEN). This is indicative of different levels of granularity or modeling choices in the two ontologies. Here, for example, the concept IntermediateHepaticArtery is not represented in FMA.
In 9% of the cases, pattern pairs consist of one associative relationship in one ontology and a hierarchical relationship in the other. For example, {FMA: bounded by, GALEN: HAS_PART} was extracted from path pairs such as Liver → bounded by → Surface of liver in FMA and Liver → HAS_PART → SurfaceOfLiver in GALEN. Actually all inter-anchor bounded by relations in FMA were represented as HAS_PART in GALEN. The two relationships in the pair must not be interpreted as being semantically equivalent. More likely, this indicates that no associative relationships in GALEN match bounded by in FMA. Here, bounded by and HAS_PART simply happen to co-occur between the same anchors.
Most matches are multiple matches, in which one associative relationship in one ontology matches several patterns in the other. In the case of arterial supply illustrated in Table 3, it is clear that the relationship arterial supply in FMA corresponds to isServedBy in GALEN, alone or combined with other relationships, in a large majority of cases. Not shown in Table 3, the relationship isServedBy in GALEN, corresponds to three different relationships in FMA: arterial supply, venous drainage and nerve supply. Here, the three relationships in FMA are finer-grained than the unique relationship in GALEN.
Not surprisingly, no match was found for some associative relationships. About 56% of the associative relationships in FMA and 84% in GALEN do not appear in the patterns (e.g., fascicular architecture in FMA and isPositionedDistalTo in GALEN). Logically, there is no correspondence in FMA for isPositionedDistalTo in GALEN since no associative relationships in FMA describe topological relationships among concepts.
Inverse patterns matching the same relationship
An associative relationship is not expected to match one pattern and its inverse in the other ontology. Actually, we found a small number of such occurrences, which we examined, suspecting that they may reveal inconsistencies.
One such case, illustrated in Table 4, is represented by the two pattern pairs: {GALEN: isSpaceDefinedBy, FMA: PART_OF} (with 102 path pairs) and {GALEN: isSpaceDefinedBy, FMA: HAS_PART} (with 6 path pairs). Paths corresponding to these patterns include OrbitalCavity → isSpaceDefinedBy → Orbit in GALEN and Orbital cavity → PART_OF → Orbit in FMA for the former and ConjunctivalSac → isSpaceDefinedBy → Conjunctiva in GALEN and Conjunctival sac → HAS_PART → Conjunctiva in FMA for the latter. What happens is that, unlike in GALEN, Conjunctival sac is not considered a cavity in FMA. Although supported by lexical and structural similarity based on the hierarchical relationships, Conjunctival sac does not seem to be an anchor, as indicated by its associative relations to other anchors.
Table 4.
Associative relationship matching inverse patterns
| FMA | GALEN | Frequency | |
|---|---|---|---|
| PART_OF | is Space Defined By | 102 | 2.5 % |
| HAS_PART | 6 | 0.15 % | |
| branch of | ISA - has Branch | 2 | 0.05 % |
| Is Branch Of -INVERSE_ISA | 2 | 0.05 % | |
Nevertheless, as illustrated in Table 4, not all cases of inverse patterns are indicative of inconsistencies. Paths corresponding to these patterns include ExternalNasalNerve → ISA → PeripheralNerve → hasBranch → AnteriorEthmoidalNerve and ExternalNasalNerve → isBranchOf → PeripheralNerve → INVERSE_ISA → AnteriorEthmoidalNerve in GALEN match External nasal nerve → branch of →Anterior ethmoidal nerve in FMA. These two paths in GALEN do not conflict.
Limitations and future work
In our method, the identification of equivalent associative relationships is based on the existence of anchors, i.e., equivalent concepts across ontologies. Therefore, the associative relationships that do not participate in any paths between anchors cannot be matched by this method (e.g., fascicular architecture in FMA). The equivalent concepts used for identifying equivalent relationships were extracted automatically and have not been validated by domain experts yet. The inaccurate identification of equivalent concepts may lead to the inaccurate identification of equivalent relationships (see the Conjunctival sac example presented earlier).
Beside the validation, we plan to take advantage of the equivalent relationships identified here for discovering more equivalent concepts. Finally, we will move to our broader objective, i.e., to investigate the reasoning capabilities of the two ontologies.
Acknowledgments
The research was supported in part by an appointment to the National Library of Medicine Research Participation Program administrated by the Oak Ridge Institute of Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine.
Footnotes
Contributor Information
Songmao Zhang, Email: szhang@nlm.nih.gov.
Olivier Bodenreider, Email: olivier@nlm.nih.gov.
References
- 1.Winston M, Chaffin R, Herrmann D. A Taxonomy of Part-Whole Relations. Cognitive Science. 1987;(11):417–444. [Google Scholar]
- 2.Guarino N. Some Ontological Principles for Designing Upper Level Lexical Resources. Proceedings of the First International Conference on Language resources and Evaluation. 1998:527–534. [Google Scholar]
- 3.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. Proc AMIA Symp. 2003:753–757. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, Bodenreider O. Knowledge Augmentation for Aligning Ontologies: An Evaluation in the Biomedical Domain. Proceedings of the Semantic Integration Workshop at the Second International Semantic Web Conference (ISWC 2003) 2003:109–114. [Google Scholar]
- 5.McGuinness DL, Fikes R, Rice J, Wilder S. The Chimaera ontology environment. Proc of AAAI. 2000:1123–1124. [Google Scholar]
- 6.Stumme G, Alexander M. FCA-MERGE: Bottom-up merging of ontologies. Proceedings of IJCAI 2001. 2001:225–230. [Google Scholar]
- 7.Doan A, Madhavan J, Dhamankar R, Domingos P, Halevy A. Learning to match ontologies on the semantic web. VLDB Journal (Special Issue on the Semantic Web) 2003;12(4):303–319. [Google Scholar]
- 8.Noy NF, Musen MA. PROMPT: algorithm and tool for automated ontology merging and alignment. Proc of AAAI. 2000:450–455. [Google Scholar]
- 9.Noy N, Musen M. Anchor-PROMPT: Using non-local context for semantic matching. Proceedings of the IJCAI 2001 Workshop on Ontologies and Information Sharing. 2001 http://www-smi.stanford.edu/pubs/SMI_Reports/SMI-2001-0889.pdf. [Google Scholar]
- 10.Rosse C, Mejino JL, Modayur BR, Jakobovits R, Hinshaw KP, Brinkley JF. Motivation and organizational principles for anatomical knowledge representation: the digital anatomist symbolic knowledge base. J Am Med Inform Assoc. 1998;5(1):17–40. doi: 10.1136/jamia.1998.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9(2):139–171. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 13.Rogers J, Rector A. GALEN's model of parts and wholes: experience and comparisons. Proc AMIA Symp. 2000:714–718. [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz S. Bidirectional mereological reasoning in anatomical knowledge bases. Proc AMIA Symp. 2001:607–611. [PMC free article] [PubMed] [Google Scholar]