Abstract
Five common tetracycline resistance genes tet(A), tet(B), tet(M), tet(O) and tet(S) were studied by polymerase chain reaction in 100 bacteria isolated from Iranian fish farms. In the antibiogram test most of the bacteria were either intermediately or completely resistant to tetracycline. Nine isolates out of 46 Aeromonas spp. contained either tet(A/M/S) resistant genes as follows: tet(A) in A. veronii/sobria (n = 1), A. media (n = 2), A. aquariorum (n = 1), and A. veronii (n = 3); tet(M) in one isolate of A. sobria and tet(S) in 1 isolate of A. jandaei. In other bacteria, tet(A) gene was detected in Citrobacter freundi (n = 1), Pseudomonas putida (n = 1); tet(S) was also identified in Yersinia ruckeri (n = 1), Arthrobacter arilaitensis (n = 1) and P. putida (n = 1). In total, 31 isolates (31.00%) contained the tetracycline resistance genes in which 21 bacteria (21.00%) showed the tet(S), nine bacteria (9.00%) contained the tet(A) and 1 bacteria (1.00%) was positive for tet(M). All of the L. garvieae isolates contained tet(S) in this study. The most widely distributed resistance gene was gene tet(A) and the least known resistance genes was tet(M) among the studied bacteria of the genus Aeromonas in this study.
Key Words: Bacteria, Fish farm, Polymerase chain reaction, Resistance gene, Tetracycline
Introduction
Aquaculture has been a growing activity for the last 30 years worldwide and this impressive development has been attended by some practices potentially damaging human and animal health.1 Synthetic antibiotics have been widely used for human and animals, and misuse of antibiotics in many countries potentially contributes to the emerging and spread of antibiotic resistant bacteria and antibiotics resistance genes in the environment.2
Tetracycline is a broad-spectrum antibiotic used in human and animal medicine as well as aquaculture industry and at least 40 different tetracycline resistance genes (tet) have been characterized to date.3 Resistance to tetracycline is governed by tet genes, which are involved in either active efflux of the drug, ribosomal protection or enzymatic drug modification.4 Among the various tet genes, the tet(A), tet(B), tet(D), tet(E) and tet(G) are reported in gram-negative bacteria.5 Whereas, the tet(K), tet(L), tet(M), tet(O), and tet(S) are significantly found in the gram-positive bacteria.6
Tetracyclines have been greatly used in aquaculture particularly to control furunculosis in salmonids and oxytetracycline is frequently used in most fish farming industries as a prophylactic agent.7 They are broad-spectrum agents including tetracycline, chlortetracycline, doxycycline and minocycline which exhibit activity against a wide range of gram-positive and -negative bacteria.8
In a study conducted in Japan and Korea, 34 isolates including Vibrio spp., Lactococcus garvieae, Photo-bacterium damsela subsp. piscicida, and unidentified gram-positive bacteria out of 151 isolates from fish and aquaculture sites contained tet(M) gene. The majority of these bacterial isolates displayed high-level resistance with a minimum inhibitory concentrations (MICs) equal to or greater than 250 µg mL-1 of oxytetracycline and only four isolates had MICs less than 31.3 µg mL-1. The tet(S) gene was also detected in L. garvieae from yellowtail in Japan and in Vibrio sp. from seawater in Korea.9
In Korea, 54 isolates of tetracycline-resistant S. parauberis contained the tet(M/O/S) genes, out of which 39 and 12 isolates contained the tet(M) and tet(S) genes, respectively, whereas three isolates contained both the tet(M) and tet(S) genes.10 These studies suggest that the tet(S), tet(M), tet(O) genes of fish and aquaculture from Asia are involved in the acquisition of high-level resistance to tetracycline. In a recent study, 63.3% of L. garvieae isolated from diseased rainbow trout in Iran were resistant to oxytetracycline.11 However, no molecular genetic studies were performed to investigate tetracycline resistance genes in the bacteria from fish farms and fish pathogenic bacteria in Iran. Therefore, the aim of this study was to detect the tetracycline resistant genes in bacterial isolates with aquaculture origin found to be resistant to tetracycline.
Materials and Methods
Bacteria and culture conditions. A total number of 100 bacterial isolates from diseased fish with apparent symptoms of fish diseases and from water samples where the fish were collected from Iranian fish farms were studied in this research. The bacteria were previously identified (unpublished data) based on their phenotypic, bio-chemical,12 and 16S rDNA sequencing characteristics. Some 16S rDNA sequencing data were already registered in the GenBank database. Aeromonas spp. as a main gram-negative bacteria were accounted for approximately 46.00%, Lactococcus garvieae as the main gram-positive bacteria for 17.00% and the other bacteria for 37.00% (Table 1). The isolates were stored at –70 ˚C in tryptic soy broth (TSB; Scharlau Chemie, Barcelona, Spain) containing 10% glycerol (Caldic Deutschland Chemie, Düsseldorf, Germany).
Table 1.
Characteristics of the bacterial isolates
| No. | Bacterial isolation codes (No. of isolates) | Bacteria | Source/Tissue | Accession No. |
| 1 | 1- 21 (21) | A. veronei bv. sobria | Fish/Kidney*, Aquarium/Water | JF313389-98 JF313414-15 JF313399 |
| 2 | 141-142 (2) | A. veronei | Fish/Kidney | - |
| 3 | 23-31 (8) | A. hydrophila | Fish/Kidney | JF313400-03 |
| 4 | 143-150 (7) | A. media | Fish/Kidney | JF313404-07 |
| 5 | 43-44 (2) | A. sobria | Fish/Kidney | - |
| 6 | 58 (1) | A. caviae | Aquarium/Water | - |
| 7 | 59 (1) | A. caviae/ A. media | Fish/Kidney | - |
| 8 | 78 (1) | A. aquariorum | Aquarium/Water | JF313412 |
| 9 | 112-113 (2) | A. bestarium/ A. piscicola | Fish/Kidney | - |
| 10 | 73 (1) | A. jandaei | Fish/Kidney | JF313413 |
| 11 | 82 (1) | Plesiomonas shigelloides | Fish/Kidney | - |
| 12 | 28-35 (7) | C. freundi | Fish/Kidney | - |
| 13 | 36 (1) | C. brakii | Fish/Kidney | - |
| 14 | 117-122 (6) | Y. ruckeri | Fish/Kidney | FJ870985 |
| 15 | 89-90 (2) | P. putida | Fish/Skin** | - |
| 16 | 91-94 (6) | Pseudomonas spp. | Fish/Skin | - |
| 17 | 301-306 (5) | Microbacterium spp. | Fish/Skin | - |
| 18 | 331 (1) | Vibrio anguillarum | Fish/Kidney | - |
| 19 | 311 (1) | Chryseobacterium aquaticum | Fish/Skin | FJ514480 |
| 20 | 312 (1) | Paenibacillus sp. | Aquarium/Water | FJ666319 |
| 21 | 512 (1) | Acinetobacter sp. | Fish/Skin | - |
| 22 | 513-514 (2) | Delftio acidovorans | Fish/Skin | - |
| 23 | 210-241 (17) | L. garvieae | Fish/Kidney | EU727199 |
| 24 | 245 (1) | Enterococcus faecium | Fish/Kidney | FJ870986 |
| 25 | 422 (1) | A. arilaitensis | Fish/Kidney | - |
| 26 | 77 (1) | Arthrobacter sp. | Fish/Kidney | - |
Bacteria were isolated from fish kidney of the diseased fish (with hemorrhagic septicemia).
Bacteria were isolated from fish skin of the diseased fish (with skin erosion and fin rot).
In preparation for antibiotic susceptibility tests, isolates were cultured for 48 hr on brain heart infusion agar (BHI; Quelab Labratories, Montreal, Quebec, Canada). Individual colonies were re-cultured on BHI for ensuring purity of the isolates. Colonies were then suspended in 3 mL TSB and adjusted to 0.5 McFarland (Becton Dickinson, Franklin Lakes, USA) using a colorimeter for use in the antimicrobial susceptibility test.
Antimicrobial susceptibility test. Antimicrobial susceptibility tests were performed according to the Muller Hinton agar methods described by the Clinical and Laboratory Standards Institute as previously reported.13 One hundred microliter of the 0.5 McFarland of each bacterial suspension were placed on Muller Hinton agar (Scharlau Chemie, Barcelona, Spain) and spread all over the plate. Antibiotic discs, each containing 30 µg tetracycline per disc (Padtan Teb Co., Tehran, Iran), were placed aseptically onto the surface of the seeded plates. Plates were incubated at 25 ˚C until 48 hr and the diameter of the inhibition zone for each bacterium was recorded. The breakpoint of inhibition zone was interpreted as follows: susceptible (S) ≥ 20 mm, intermediate (I) ≥ 8 mm, and resistant (R) ≥ 0 mm.
Polymerase chain reaction (PCR) and sequence analysis. Antimicrobial resistant genes were investigated using a single PCR assay. The stored isolates were cultured in TSB at 28 ˚C for 24 hr and DNA was extracted using boiling method.14 The DNA extracted by this method was visualized by gel electrophoresis on a 0.9% agarose gel before being stored at –20 ˚C. The oligonucleotide primer sets were used to detect tetracycline resistance genes including tet(A), tet(B), tet(M), tet(O), tet(S), (Table 2).15,16
Table 2.
Oligonucleotide sequences used as primers for polymerase chain reaction
| Gene | Primer name | Primer sequence | Product Size (bp) | References |
|---|---|---|---|---|
| tet (M) |
Tet(M)-F Tet(M)-R |
5'-GTG GAC AAA GGT ACA ACG AG-3' 5'-CGG TAA AGT TCG TCA CAC AC-3' |
406 | 15 |
| tet (O) |
Tet(O)-F Tet(O)-R |
5'-AAC TTA GGC ATT CTG GCT CAC-3' 5'-TCC CAC TGT TCC ATA TCG TCA-3' |
515 | 15 |
| tet (S) |
Tet(S)-F Tet(S)-R |
5'-CAT AGA CAA GCC GTT GAC C-3' 5'-ATG TTT TTG GAA CGC CAG AG-3' |
667 | 15 |
| tet (A) |
Tet(A)-F Tet(A)-R |
5'- GTGAAACCCAACATACCCC-3' 5'-GAAGGCAAGCAGGATGTAG-3' |
888 | 16 |
| tet (B) |
Tet(B)-F Tet(B)-R |
5'-CCTTATCATGCCAGTCTTGC-3' 5'-ACTGCCGTTTTTTCGCC-3' |
774 | 16 |
The primers were commercially synthesized by the CinnaGen Company (Tehran, Iran) (Table 2). Following PCR conditions were applied to each assay: 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 200 μM dNTPs, 20 pmol of each primer, and 2 U Taq DNA polymerase (Fermentas, St. Leon-Rot, Germany) per 50 μL reaction using 4 μL of DNA were extracted as the template. A gradient thermocycler (MG 5331; Eppendorf, Hamburg, Germany) was used to determine an optimal annealing temperature for the specific binding of the primer set to the template DNA. The optimal thermal parameters were as follows: initial denaturation at 94 ˚C for 5 min followed by 30 cycles of denaturation at 94 ˚C for 45 sec, annealing at 55 ˚C for 1 min and extension at 72 ˚C for 1 min. A final extension at 72 ˚C for 5 min at the end of the amplification cycles was included. Each sample was tested at least in duplicate and sterile water was used as a negative control.
The PCR products obtained from different tet genes were resolved by 1.5% agarose gel electrophoresis and stained with ethidium bromide (1 μg mL-1). The amplification products were visualized under a UV trans-illuminator (Armin Teb Novin Co., Tehran, Iran) and photographed.
The PCR products were directly sequenced using capillary DNA analyzer (ABI 3730; Applied Biosystems, Foster City, USA) after sequencing reactions with a cycle sequencing kit (BigDye Terminator V3.1; Applied Biosystems, Foster City, USA). Sequence generated from different tet genes were analyzed to assess the diversity of each gene compared to the GenBank data base using the BLAST program maintained by the National Center for Biotechnology Information (NCBI).17 Nucleotide sequences of each tet gene were aligned using the multiple sequence alignment program CLUSTALW provided by MEGA 4.0 software.18
Results
Resistance to tetracycline. Antimicrobial susceptibility test of the bacterial isolates using tetracycline discs demonstrated strong resistance to tetracycline in several isolates, i.e., Aeromonas spp. (5/46), Citrobacter freundi (3/7), Yersinia ruckeri (3/6), Pseudomonas putida (1/2), Penibacillus spp. (1/1), Acinetobacter sp. (1/1) and Lactococcus garvieae (9/17), (Table 3).
Table 3.
Resistance pattern of the bacterial isolates by antimicrobial susceptibility test and antibiotic resistance genes
| No. | Bacteria | Total No. of strains |
Tetracycline
|
tet genes | |||
|---|---|---|---|---|---|---|---|
| Sensitive (≥ 20 mm) | Intermediate (≥ 8 mm) | Resistant (0 mm < a < 8mm) | |||||
| 1 | A. veronei bv. sobria | 21 | 5 | 13 | 3 | 4 tet(A) | |
| 2 | A. veronei | 2 | 1 | - | 1 | - | |
| 3 | A. hydrophila | 8 | 5 | 3 | - | - | |
| 4 | A. media | 7 | 1 | 6 | - | 2 tet(A) | |
| 5 | A. sobria | 2 | - | 1 | 1 | 1 tet(M) | |
| 6 | A. caviae | 1 | 1 | - | - | ||
| 7 | A. caviae/ A. media | 1 | - | 1 | - | - | |
| 8 | A. aquariorum | 1 | - | 1 | - | 1 tet(A) | |
| 9 | A. bestarium/ A. piscicola | 2 | 1 | 1 | - | - | |
| 10 | A. jandaei | 1 | - | 1 | - | 1 tet(S) | |
| 11 | Plesiomonas shigelloides | 1 | - | 1 | - | - | |
| 12 | C. freundi | 7 | - | 4 | 3 | 1 tet(A) | |
| 13 | C. brakii | 1 | - | 1 | - | - | |
| 14 | Y. ruckeri | 6 | 3 | 3 | 1 tet(S) | ||
| 15 | P. putida | 2 | - | 1 | 1 | 1 tet(S) 1 tet(A) |
|
| 16 | Pseudomonas spp. | 6 | 2 | 4 | - | - | |
| 17 | Microbacterium spp. | 5 | 1 | 4 | - | - | |
| 18 | Vibrio anguillarum | 1 | 1 | - | - | ||
| 19 | Chryseobacterium aquaticum | 1 | - | 1 | - | - | |
| 20 | Paenibacillus sp. | 1 | - | - | 1 | - | |
| 21 | Acinetobacter sp. | 1 | - | - | 1 | - | |
| 22 | Delftio acidovorans | 2 | - | 2 | - | - | |
| 23 | L. garvieae | 17 | 1 | 7 | 9 | 17 tet(S) | |
| 24 | Enterococcus faecium | 1 | 1 | - | - | - | |
| 25 | A. arilaitensis | 1 | 1 | - | - | 1 tet(S) | |
| 26 | Arthrobacter sp. | 1 | 1 | - | - | - | |
Resistant genes. Antibiotic resistant genes detection on 100 bacterial isolates in this study showed 31 isolates (31.00%) contained the tetracycline resistant genes in which 21(21.00%) bacteria showed the tet(S), 9 (9.00%) bacteria contained the tet(A) and 1 (1.00%) bacteria was positive for tet(M). Nine isolates out of 46 Aeromonas spp. contained either tet(A/M/S) resistant genes as follows: tet(A) was detected in four isolate of A. veronii bv. sobria (n = 4), A. media (n = 2), A. aquariorum (n = 1); tet(M) in A. sobria (n = 1) and tet(S) in A. jandaei (n = 1). In other bacteria, tet(A) gene was identified in C. freundi (n = 1), P. putida (n = 1); tet(S) in Y. ruckeri (n = 1), P. putida (n = 1), Arthrobacter arilaitensis (n = 1) and L. garvieae (n = 17). The tet(B) and tet(O) genes were not detected in any of the examined isolates in our study (Table 3 and Fig. 1).
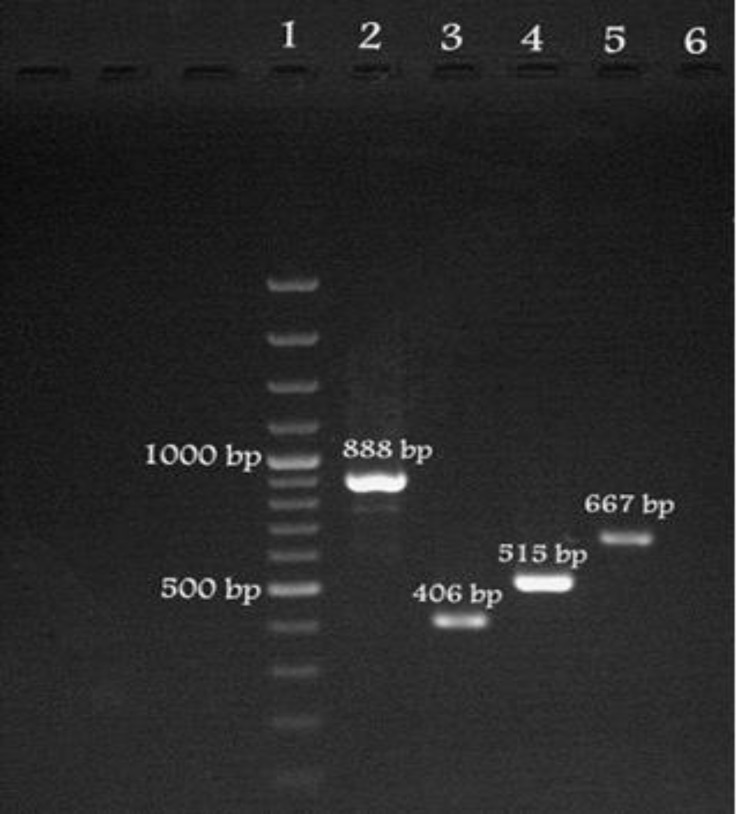
Fig. 1.
Electrophoretic analysis of PCR amplified products for tetracycline resistant genes (A, M, O and S). Lane 1, DNA ladder molecular markers; lane 2, tet(A) PCR product-888 bp produced from Pseudomonas putida strain Fars-110 ( JN937120); lane 3, tet(M) PCR product-406 bp obtained from Aeromonas sobria strain CW4 (JN806155); lane 4, tet(O), PCR product-515 bp obtained from Campylobacter jejuni strain Shiraz2 (JX853722); lane 5, tet(S) PCR product-667 bp produced from Lactococcus garvieae strain Iran.1S (JN998084).
Our results showed that the bacterial isolates contained tet(A) were completely resistant to tetracycline while those contained tet(S) were mostly resistant to tetracycline (Table 4).
Table 4.
. Comparison of active efflux pump pattern tet(A) with ribosomal protective protein pattern tet(S) in 30 bacterial isolates containing tet(A) and tet(S) genes
| tet (A, S) | Sensitive | Intermediate | Resistant | Total | |
|---|---|---|---|---|---|
| Total | - | 2 | 1 | 27 | 30 |
| 1 | tet(A) | 0 | 0 | 9 | 9 |
| 2 | tet(S) | 2 | 1 | 18 | 21 |
Sequence analysis. BLAST search of the Iranian tet(A) and tet(S) sequences in GenBank showed sequence identities ranging from 99.00% to 100% for these two genes. Whereas, sequence analysis of the obtained tet(M) gene from the A. sobria (NCBI accession no. JN806155) revealed sequence similarity ranging from 98.00% to 99.00%. Partial sequence obtained for tet(M) in this study (349 bp) had a high identity (99.00%) to the known tet(M) genes detected in Streptococcus pneumoniae (NCBI accession no. FM201786). Our data showed a new genotype for tet(M) in Iran that has not been previously described elsewhere in the world.
The sequences of tet(A), tet(S) and tet(M) genes obtained in this study have been deposited in GenBank under accession numbers JN937120, JN998084 and JN806155, respectively.
Discussion
Many mechanisms are involved in tetracycline resistance and three different specific mechanisms have been identified so far: Antibiotic efflux pumps, including tet(A), tet(B), tet(C), tet(E) and tet(L) target modification with ribosomal protection protein (RPP) including tet(M), tet(O), tet(Q), tet(S) and tet(W), and antibiotic inactivation tet(X).19 In this study, we examined 100 bacterial isolates from diseased fish and their aquatic environment in Iran for the presence of five tetracycline resistance genes (i.e., tet(A, B) as efflux pumps and tet(M, O, S) as RPPs class). One type of the efflux pump genes, tet(A) and two types of the RPP genes tet(M) and tet(S) were detected among the isolates. We found no genetic evidence for the presence of tet(B) and tet(O) in our experiments.
In the present study, the most frequent tetracycline-resistant gene was tet(S) (21.00%), followed by tet(A) (9.00%) and tet(M) (1.00%), (Table 3). In addition, the most widespread determinant in gram-positive and gram-negative bacteria were tet(S) and tet(A), respectively. Similarly, the tet(A) has a broad host range and is often carried by various environmental genera.20 Some authors previously believed that efflux pump genes (e.g., tet(A), tet(D), tet(E)) are often carried by Aeromonas spp. in fish farm ponds,21 and Vibrio spp. in marine environment,22 and suggested that tet genes might have host specificity in different environments. However, recent studies have also demonstrated that tet genes are often located on the plasmids and can be horizontally transferred among bacterial strains.23
Only one of the isolates, A. sobria contained tet(M) gene in this study. The limited occurrence of tet(M) in the isolates was surprising because this class of tetracycline resistant determinant has broadly been described in aquaculture,9,13 river and channel sediments.24 Nevertheless, this study reports the occurrence of tet(M) in the A. sobria for the first time. The occurrence of tet(M) in Enterococcus spp. isolated from integrated and traditional freshwater fish farm pond in Thailand is also reported.25 According to current insights, tet(M) is the most widely distributed tet gene, being detected in at least eight gram-negative and 18 gram-positive genera, including the lactic acid bacterial genera Enterococcus, Streptococcus, and Bifidobacterium.8 Some studies suggested that the origin of tet(M) is most probably the tetracycline-producing species of Streptomyces and that its integration into mobile genetic elements (plasmids and transposons) has led to its widespread distribution.8 The presence of this gene has also been described in a range of bacteria including Vibrio spp., L. garvieae, P. damselae subsp. piscicida and some gram-positive bacteria isolated from healthy and diseased fish as well as from seawater in Japan and Korea.9 These results support the fact that increasing numbers of gram-negative bacteria carry what has been previously considered as gram-positive specific tet genes, such as tet(M). Therefore, further epidemiological and molecular investigations are needed to evaluate the presence of genetically mobile antibiotic resistant genes in human and animal food chain.
Recent Iranian genotype of tet(M) was placed in genetically distant group, suggesting different origins for dissemination of resistance in this region. According to the published data on bacterial species in marine sediments, the majority of tet(M) possessing isolates were belonged to Bacillales (121 strains) Actinomycetales (three strains), Flavobacteriales (one strain) and Pseudomonadales (one strain).26 This indicates that tet(M) is present in various bacterial species in marine sediments, which is the natural reservoir of the tet(M) gene. Moreover, our results supported that tet(M) might be derived from different origins in environment. Recently, Wu et al. carried out a phylogenetic analysis of tet(M) in soil and found that all the five tet(M) types obtained matched known genes in GenBank, with sequence identities ranging from 98.00% to 100%.26
In our study, the tet(A) gene was found in 7 isolates of Aeromonas spp. one isolate of C. freundi and one isolate of P. putida in which all of the strains were resistant to tetracycline. Conversely, the tet(S) was also detected in tetracycline-sensitive isolates (n = 2) as well as tetracycline-intermediate isolates (n = 1). These results suggested that there was no direct correlation between the presence of tet(S) gene and simultaneous antibiotic resistance. Hence, it was likely that further genetic elements played an important role in antibiotic resistance.
The isolates of L. garvieae used in this study were either susceptible (6.00%), intermediate resistance (41.00%) or resistant (52.00%) to tetracycline. In a recent study conducted on L. garvieae isolated from diseased rainbow trout culture in 10 fish farms in Chaharmahal and Bakhtyari province of Iran, which is a major trout producing area, it was exhibited that 65.30% and 76.90% of the isolates were resistant to oxytetracycline and doxycycline, respectively.11 Also in a previous study, it was demonstrated a high level of resistance to tetracycline among L. garvieae isolated from rainbow trout farms in the south and southwest of Iran during summer 2002 to winter 2008.27 Our molecular investigations indicated that the continuous rise in the rate of tetracycline resistance in L. garvieae in Iran might contribute to the high frequency of tet(S) in 100% of our L. garvieae isolates in this investigation.
Tian et al. showed that Streptococci use two major mechanisms of antibiotic resistance, including efflux by proton antiporters, encoded by the tet(L) gene, and ribosomal protection; mediated by the tet(M), tet(O) and tet(S) genes.28 In the present study, tet(S) incidence was only described in L. garvieae isolates among ribosome protection types of resistance genes.28 Historically, the tet(S) gene has predominantly been found in gram-positive bacteria such as Listeria monocytogenes and Enterococcus faecalis isolated from humans and L. lactis isolated from cheese.9 In gram-negative bacteria, only Veillonella spp., an oral bacterium, is known to possess tet(S).29 Now this range is extended and includes a marine Vibrio species as well.9 In the present study we also found tet(S) in additional gram-negative bacteria, i.e., Y. ruckeri, P. putida and A. jandaei.
In conclusion, our findings showed the importance of reducing antibiotic use and the need for appropriate vaccines to prevent the widespread emergence of resistance L. garvieae. Fortunately the development of fish vaccines and the use of selected disease resistant stocks nowadays have greatly limited the utilization of antibiotics. This study further emphasized the potential for aquaculture sources to act as a reservoir of antibiotic resistance genes, which could contaminate the environment and water sources as well as aquaculture-produced food products.
Acknowledgements
We would like to thank the authorities of the Veterinary School, Shiraz University for their financial support. The authors are also grateful to Mr. Mohammad Saied Fereidouni for his technical assistance.
References
- 1.Naylor R, Burke M. Aquaculture and ocean resources: Raising tigers of the sea. Annu Rev Environ Resour. 2005;30:185–218. [Google Scholar]
- 2.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 4.Giovanetti E, Brenciani A, Lupidi R, et al. Presence of the tet(O) gene in erythromycin- and tetracycline resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob Agents Chemother. 2003;47:2844–2849. doi: 10.1128/AAC.47.9.2844-2849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CH, Tuckman M, Murphy E, et al. Identification and sequence of a tet(M) tetracycline resistance determinant homologue in clinical isolates of Escherichia coli. J Bacteriol. 2006;188:7151–7164. doi: 10.1128/JB.00705-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts MC. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 7.Miranda CD, Kehrenberg C, Ulep C, et al. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob Agents Chemother. 2003;47:883–888. doi: 10.1128/AAC.47.3.883-888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Nonaka L, Suzuki S. Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett. 2004;237:147–156. doi: 10.1016/j.femsle.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Park YK, Nho SW, Shin GW, et al. Antibiotic susceptibility and resistance of Streptococcus iniae and Streptococcus parauberis isolated from olive flounder (Paralichthys oliveceus) Vet Microbiol. 2008;136:76–81. doi: 10.1016/j.vetmic.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Raissy M, Ansari M. Antibiotic susceptibility of Lactococcus garvieae Isolated from rainbow trout (Oncorhynchus mykiss) in Iran fish farms. African J Biotechnol. 2011;10:1473–1476. [Google Scholar]
- 12.Austin B, Austin DA. Bacterial fish pathogens, diseases of farmed and wild fish. 5th ed. London, UK: Springer; 2012. pp. 147–173. [Google Scholar]
- 13.Akinbowale OL, Peng H, Barton MD. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J Appl Microbiol. 2007;103:2016–2025. doi: 10.1111/j.1365-2672.2007.03445.x. [DOI] [PubMed] [Google Scholar]
- 14.Holmes DS, Quiqley M. A rapid boiling method for the preparation of bacterial plasmids. Ann Rev Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 15.Ng LK, Martin I, Alfa M, et al. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 16.Maynard C, Fairbrother JM, Bekal S, et al. Anti-microbial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47:3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.BLAST: Basic local alignment search tool. [Accessed Oct 5, 2012]. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 18.Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software. Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Schweizer HP. Bacterial resistance to antibiotics: Active efflux and reduced uptake. Adv Drug Deliv Rev. 2005;57:1486–1513. doi: 10.1016/j.addr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XX, Zhang T, Fang HHP. Antibiotic resistance genes in water environment. Appl Microbiol Biotechno. 2009;82:397–414. doi: 10.1007/s00253-008-1829-z. [DOI] [PubMed] [Google Scholar]
- 21.Dang HY, Zhang XX, Song L, et al. Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J Appl Microbiol. 2007;103:2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim YH, Jun LJ, Park SH, et al. Prevalence of tet(B) and tet(M) genes among tetracycline-resistant Vibrio spp. in the aquatic environments of Korea. Dis Aquat Org. 2007;75:209–216. doi: 10.3354/dao075209. [DOI] [PubMed] [Google Scholar]
- 23.Szczepanowski R, Krahn I, Linke B, et al. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multi-drug transport system. Microbiology. 2004;150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Suehiro F, Tuyen BC, et al. Distribution and diversity of tetracycline resistance genes encoding ribosomal protection proteins in Mekong river sediments in Vietnam. FEMS Microbiol Ecol. 2007;59:729–737. doi: 10.1111/j.1574-6941.2006.00244.x. [DOI] [PubMed] [Google Scholar]
- 25.Petersen A, Dalsgaard A. Species composition and antimicrobial resistance genes of Enterococcus spp., isolated from integrated and traditional fish farms in Thailand. Environ Microbiol. 2003;5:395–402. doi: 10.1046/j.1462-2920.2003.00430.x. [DOI] [PubMed] [Google Scholar]
- 26.Wu N, Qiao M, Zhang B, et al. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ Sci Technol. 2010;44:6933–6939. doi: 10.1021/es1007802. [DOI] [PubMed] [Google Scholar]
- 27.Sharifiyazdi H, Akhlaghi M, Tabatabaei M, et al. Isolation and characterization of Lactococcus garvieae from diseased rainbow trout (Oncorhynchus mykiss, Walbaum) cultured in Iran. Iranian J Vet Res. 2011;11:342–350. [Google Scholar]
- 28.Tian Y, Aarestrup FM, Lu CP. Characterization of Streptococcus suis serotype 7 isolates from diseased pigs in Denmark. Vet Microbiol. 2004;103:55–62. doi: 10.1016/j.vetmic.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Villedieu A, Diaz-Torres ML, Hunt N, et al. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. 2003;47:878–888. doi: 10.1128/AAC.47.3.878-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]